Genome-Wide Analysis of Sugar Transporters Identifies the gtsA Gene for Glucose Transportation in Pseudomonas stutzeri A1501
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Computer Analyses and Screening Strategies
2.3. Monitoring Growth
2.4. Reverse Transcription PCR (RT-PCR) and Quantitative Real-Time PCR (qPCR)
2.5. Phylogenetic Analysis
2.6. Construction of the gtsA Deletion Mutant and Complementation
2.7. Measurement of Glucose Uptake
3. Results
3.1. Prediction of Sugar-Transport Systems
3.2. Growth in Different Sugars
3.3. Effect of Glucose on the Expression of Sugar Transporters
3.4. Functional Analysis of Glucose-Binding Protein GtsA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Daniels, C.; Godoy, P.; Duque, E.; Molina-Henares, M.A.; De La Torre, J.; Del Arco, J.M.; Herrera, C.; Segura, A.; Guazzaroni, M.E.; Ferrer, M.; et al. Global regulation of food supply by Pseudomonas putida DOT-T1E. J. Bacteriol. 2010, 192, 2169–2181. [Google Scholar] [CrossRef]
- Antonovsky, N.; Gleizer, S.; Noor, E.; Zohar, Y.; Herz, E.; Barenholz, U.; Zelcbuch, L.; Amram, S.; Wides, A.; Tepper, N.; et al. Sugar synthesis from CO2 in Escherichia coli. Cell 2016, 166, 115–125. [Google Scholar] [CrossRef]
- Thattai, M.; Shraiman, B.I. Metabolic switching in the sugar phosphotransferase system of Escherichia coli. Biophys. J. 2003, 85, 744–754. [Google Scholar] [CrossRef]
- Tchieu, J.H.; Norris, V.; Edwards, J.S.; Saier, J. The complete phosphotransferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2001, 3, 329–346. [Google Scholar] [PubMed]
- Saier, M.H. Families of transmembrane sugar transport proteins. Mol. Microbiol. 2000, 35, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Elferink, M.G.L.; Albers, S.V.; Konings, W.N.; Driessen, A.J.M. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol. 2001, 39, 1494–1503. [Google Scholar] [CrossRef] [PubMed]
- Verimeiren, H.; Willems, A.; Schoofs, G.; de Mot, R.; Keijers, V.; Hai, W.L.; Vanderleyden, J. The rice inoculant strain Alcaligenes faecalis A15 is a nitrogen-fixing Pseudomonas stutzeri. Syst. Appl. Microbiol. 1999, 22, 215–224. [Google Scholar] [CrossRef]
- Han, Y.; Wang, R.; Yang, Z.; Zhan, Y.; Ma, Y.; Ping, S.; Zhang, L.; Lin, M.; Yan, Y. 1-aminocyclopropane-1-carboxylate deaminase from Pseudomonas stutzeri A1501 facilitates the growth of rice in the presence of salt or heavy metals. J. Microbiol. Biotechnol. 2015, 25, 1119–1128. [Google Scholar] [CrossRef]
- Yan, Y.; Yang, J.; Dou, Y.; Chen, M.; Ping, S.; Peng, J.; Lu, W.; Zhang, W.; Yao, Z.; Li, H.; et al. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc. Natl. Acad. Sci. USA 2008, 105, 7564–7569. [Google Scholar] [CrossRef]
- Desnoues, N.; Lin, M.; Guo, X.; Ma, L.; Carreño-Lopez, R.; Elmerich, C. Nitrogen fixation genetics and regulation in a Pseudomonas stutzeri strain associated with rice. Microbiology 2003, 149, 2251–2262. [Google Scholar] [CrossRef]
- Staskawicz, B.; Dahlbeck, D.; Keen, N.; Napoli, C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 1987, 169, 5789–5794. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbachb, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Figurski, D.H.; Helinski, D.R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 1979, 76, 1648–1652. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Mortimer, M.; Devarajan, N.; Li, D. Multiwall carbon nanotubes induce more pronounced transcriptomic responses in Pseudomonas aeruginosa PG201 than graphene, exfoliated boron nitride, or carbon black. ACS Nano 2018, 12, 2728–2740. [Google Scholar] [CrossRef]
- Yaagoubi, A.E.I.; Kohiyama, M.; Richarme, G. Defect in export and synthesis of the periplasmic galactose receptor MglB in dnaK mutants of Escherichia coli, and decreased stability of the mglB mRN. Microbiology 1996, 142, 2595–2602. [Google Scholar] [CrossRef][Green Version]
- Boos, W.; Shuman, H. Maltose/maltodextrin system of Escherichia coli: Transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 1998, 62, 204–229. [Google Scholar] [CrossRef]
- Bell, A.W.; Buckel, S.D.; Groarkes, M.; Hope, J.N.; David, H.; Hermodson, M.A. The nucleotide sequences of the rbsD, rbsA, and rbsC genes of Escherichia coli K12. J. Biol. Chem. 1986, 261, 7652–7658. [Google Scholar]
- Small, D.A.; Chang, W.; Toghrol, F.; Bentley, W.E. Comparative global transcription analysis of sodium hypochlorite, peracetic acid, and hydrogen peroxide on Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2007, 76, 1093–1105. [Google Scholar] [CrossRef]
- Del Castillo, T.; Duque, E.; Ramos, J.L. A set of activators and repressors control peripheral glucose pathways in Pseudomonas putida to yield a common central intermediate. J. Bacteriol. 2008, 190, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Carolé, S.; Pichoff, S.; Bouché, J.-P. Escherichia coli gene ydeA encodes a major facilitator pump which exports l-arabinose and isopropyl-β-d-thiogalactopyranoside. J. Bacteriol. 1999, 181, 5123–5125. [Google Scholar] [CrossRef][Green Version]
- Sweet, G.; Gandor, C.; Voegele, R.; Wittekindt, N.; Beuerle, J.; Truniger, V.; Lin, E.C.C.; Boos, W. Glycerol facilitator of Escherichia coli: Cloning of glpF and identification of the glpF product. J. Bacteriol. 1990, 172, 424–430. [Google Scholar] [CrossRef]
- Schulein, K.; Benz, R. LamB (maltoporin) of Salmonella typhimurium: Isolation, purification and comparison of sugar binding with LamB of Escherichia Coli. Mol. Microbiol. 1990, 4, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Wylie, J.L.; Worobec, E.A. Cloning and nucleotide sequence of the Pseudomonas aeruginosa glucose-selective OprB porin gene and distribution of OprB within the family Pseudomonadaceae. Eur. J. Biochem. 1994, 220, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, H.L. Routes for fructose utilization by Escherichia coli. J. Mol. Microbiol. Biotechnol. 2001, 3, 355–359. [Google Scholar]
- Maleszka, R.; Wang, P.Y.; Schneider, H. A Col E1 hybrid plasmid containing Escherichia coli genes complementing d-xylose negative mutants of Escherichia coli and Salmonella typhimurium. Can. J. Biochem. 1982, 60, 144–151. [Google Scholar] [CrossRef]
- Helling, R.B.; Weinberg, R. Complementation studies of arabinose genes in Escherichia coli. Genetics 1963, 48, 1397–1410. [Google Scholar]
- Hope, J.N.; Bell, A.W.; Hermodson, M.A.; Groarke, J.M. Ribokinase from Escherichia coli K12. Nucleotide sequence and overexpression of the rbsK gene and purification of ribokinase. J. Biol. Chem. 1986, 261, 7663–7668. [Google Scholar]
- Herman, R.H. Mannose metabolism. I. Am. J. Clin. Nutr. 1971, 24, 488–498. [Google Scholar] [CrossRef]
- Cooper, R.A. The utilisation of d-galactonate and d-2-oxo-3-deoxygalactonate by Escherichia coli K-12. Arch. Microbiol. 1978, 118, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Modak, A.; Phale, P.S.; Bhaumik, P. High resolution structures of periplasmic glucose-binding protein of Pseudomonas putida CSV86 reveal structural basis of its substrate specificity. J. Biol. Chem. 2016, 291, 7844–7857. [Google Scholar] [CrossRef] [PubMed]
- Cuneo, M.J.; Changela, A.; Warren, J.J.; Beese, L.S.; Hellinga, H.W. The crystal structure of a thermophilic glucose binding protein reveals adaptations that interconvert mono and di-saccharide binding sites. J. Mol. Biol. 2006, 362, 259–270. [Google Scholar] [CrossRef]
- Tian, Y.; Cuneo, M.J.; Changela, A.; Höcker, B.; Beese, L.S.; Hellinga, H.W. Structure-based design of robust glucose biosensors using a Thermotoga maritima periplasmic glucose-binding protein. Protein Sci. 2007, 16, 2240–2250. [Google Scholar] [CrossRef] [PubMed]
- Rojo, F. Carbon catabolite repression in Pseudomonas: Optimizing metabolic versatility and interactions with the environment. Fems Microbiol. Rev. 2010, 34, 658–684. [Google Scholar] [CrossRef] [PubMed]
- Hylemon, P.B.; Phibbs, P.V., Jr. Independant regulation of hexose catabolizing enzymes and glucose transport activity in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 1972, 48, 1041–1048. [Google Scholar] [CrossRef]
- Collier, D.N.; Hager, P.W.; Phibbs, P.V. Catabolite repression control in the Pseudomonads. Res. Microbiol. 1996, 147, 551–561. [Google Scholar] [CrossRef]
- Tiwari, N.P.; Campbell, J.J.R. Enzymatic control of the metabolic activity of Pseudomonas aeruginosa grown in glucose or succinate media. BBA Gen. Subj. 1969, 192, 395–401. [Google Scholar] [CrossRef]
- Bhuiyan, S.H.; Ami, Y.I.T.; Izumori, K. Isolation of an L-rhamnose isomerase-constitutive mutant of Pseudomonas sp. Strain LL172: Purification and characterization of the enzyme. J. Ferment. Bioeng. 1997, 84, 319–323. [Google Scholar] [CrossRef]
- Leang, K.; Takada, G.; Fukai, Y.; Morimoto, K.; Granström, T.B.; Izumori, K. Novel reactions of L-rhamnose isomerase from Pseudomonas stutzeri and its relation with D-xylose isomerase via substrate specificity. Biochim. Biophys. Acta Gen. Subj. 2004, 1674, 68–77. [Google Scholar] [CrossRef]
- Liu, Y.; Rainey, P.B.; Zhang, X. Molecular mechanisms of xylose utilization by Pseudomonas fluorescens: Overlapping genetic responses to xylose, xylulose, ribose and mannitol. Mol. Microbiol. 2015, 98, 553–570. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; You, C.B. Root exudates of rice (Oryza Sativa L.) and its interaction with Alcaligenes faecalis. Sci. Agric. Sin. 1989, 6. [Google Scholar]
- Kamilova, F.; Kravchenko, L.V.; Shaposhnikov, A.I.; Azarova, T.; Makarova, N.; Lugtenberg, B. Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol. Plant Microbe Interact. 2006, 19, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Malfanova, N.; Kamilova, F.; Validov, S.; Chebotar, V.; Lugtenberg, B. Is L-arabinose important for the endophytic lifestyle of Pseudomonas spp.? Arch. Microbiol. 2013, 195, 9–17. [Google Scholar] [CrossRef]
- Albalat, R.; Cañestro, C. Evolution by gene loss. Nat. Rev. Genet. 2016, 17, 379–391. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Economy, speed and size matter: Evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. 2005, 95, 147–175. [Google Scholar] [CrossRef]
- Sly, L.M.; Worobec, E.A.; Perkins, R.E.; Phibbs, P.V., Jr. Reconstitution of glucose uptake and chemotaxis in Pseudomonas aeruginosa glucose transport defective mutants. Can. J. Microbiol. 1993, 39, 1079–1083. [Google Scholar] [CrossRef]
- Fuentes, L.G.; Lara, A.R.; Martínez, L.M.; Ramírez, O.T.; Martínez, A.; Bolívar, F.; Gosset, G. Modification of glucose import capacity in Escherichia coli: Physiologic consequences and utility for improving DNA vaccine production. Microb. Cell Fact. 2013, 12, 1–11. [Google Scholar] [CrossRef]
- Sensen, C.W.; Charlebois, R.L.; Chow, C.; Clausen, I.G.; Curtis, B.; Doolittle, W.F.; Duguet, M.; Erauso, G.; Gaasterland, T.; Garrett, R.A. Completing the sequence of the Sulfolobus solfataricus P2 genome. Extremophiles 1998, 2, 305–312. [Google Scholar] [CrossRef]
- Koning, S.M.; Albers, S.V.; Konings, W.N.; Driessen, A.J.M. Sugar transport in (hyper)thermophilic archaea. Res. Microbiol. 2002, 153, 61–67. [Google Scholar] [CrossRef]
- Nelson, K.E.; Clayton, R.A.; Gill, S.R.; Gwinn, M.L.; Dodson, R.J.; Haft, D.H.; Hickey, E.K.; Peterson, J.D.; Nelson, W.C.; Ketchum, K.A. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 1999, 399, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Gosset, G. Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Fact. 2005, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Badyaev, A.V. Stress-induced variation in evolution: From behavioural plasticity to genetic assimilation. Proc. R. Soc. B Biol. Sci. 2005, 272, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Konings, W.N.; Albers, S.-V.; Koning, S.; Driessen, A.J.M. The cell membrane plays a crucial role in survival of bacteria and archaea in extreme environments. Antonie Van Leeuwenhoek 2002, 81, 61–72. [Google Scholar] [CrossRef]
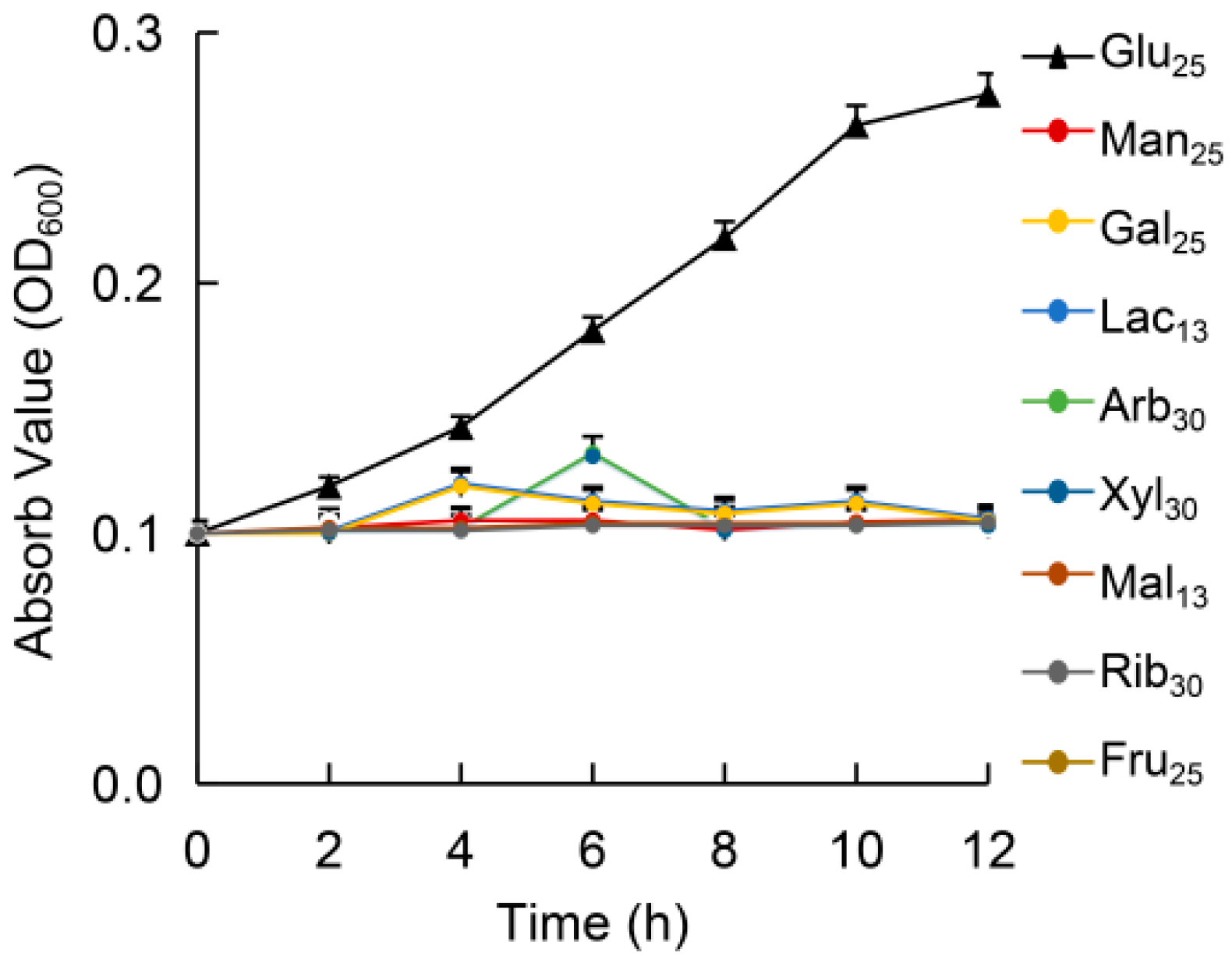

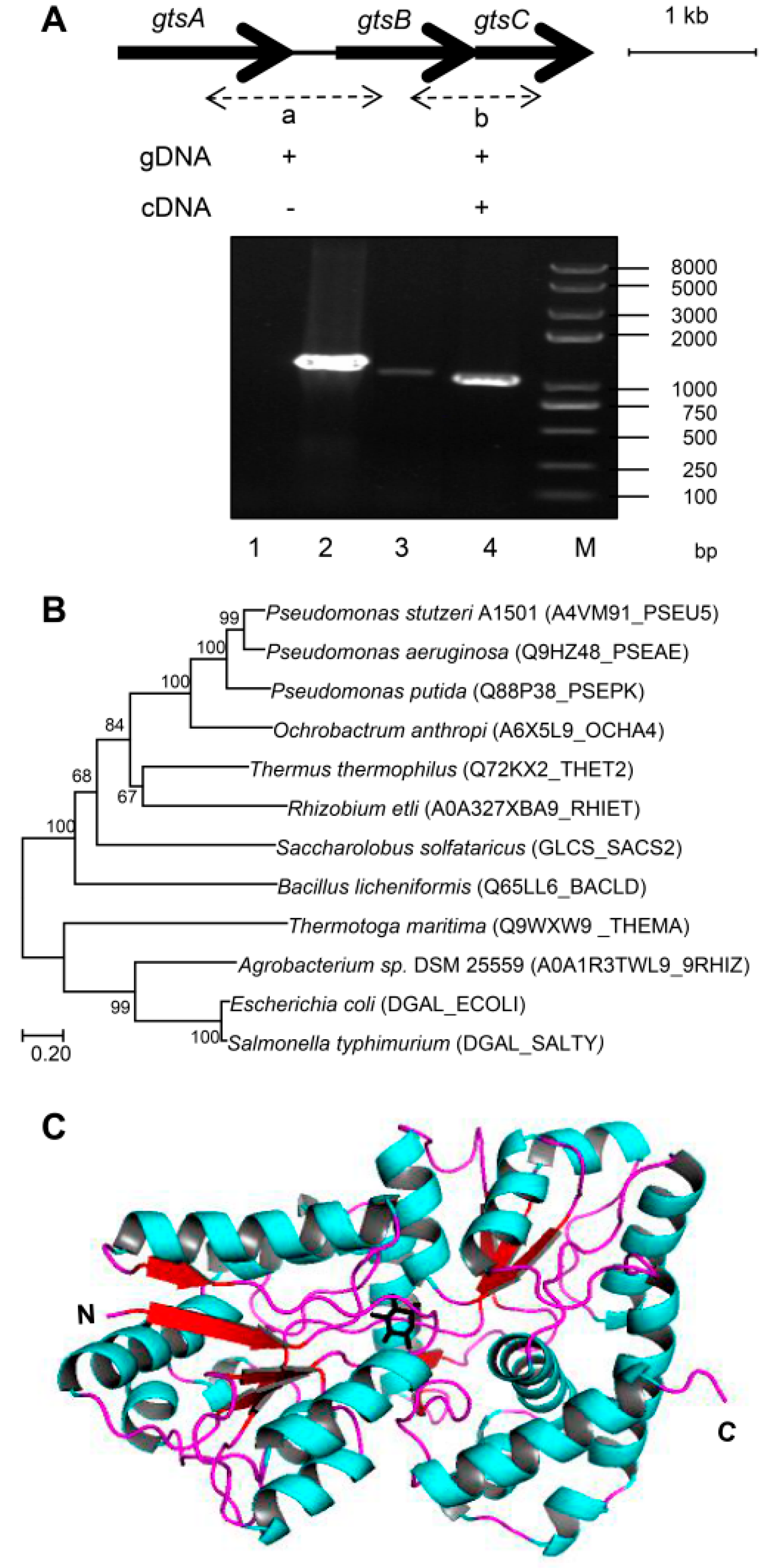
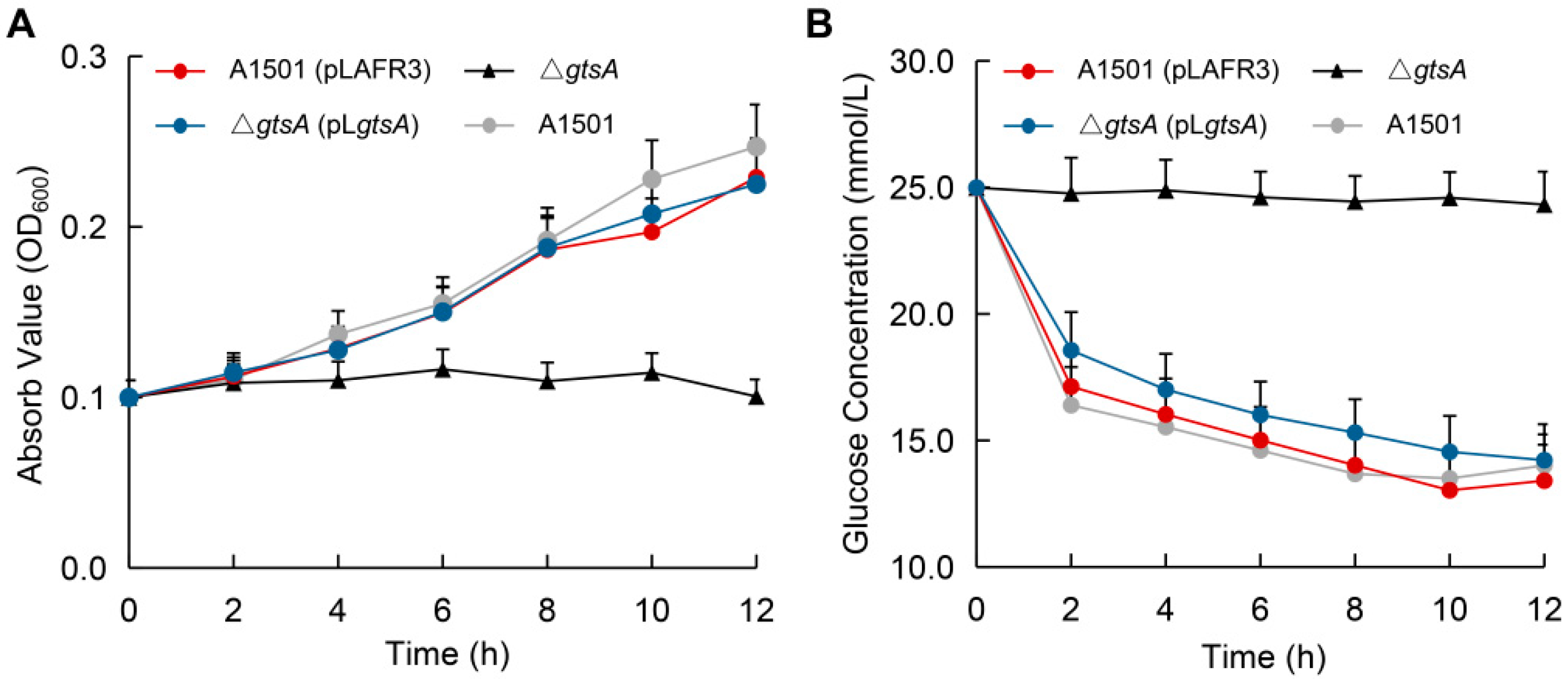
| Strains or Plasmids | Relevant Characteristics | Source or Reference |
|---|---|---|
| Strains | ||
| P. stutzeri A1501 | Wild type, Chinese Culture Collection: CGMCC (China General Microbiological Culture Collection Center) 0351 | [10] |
| ΔgtsA | gtsA deleted mutant strain, Cmr | This study |
| ΔgtsA (pLgtsA) | ΔgtsA complemented with pLgtsA, Kmr, Tcr | This study |
| A1501 (pLgtsA) | A1501 complemented with pLgtsA, Kmr | This study |
| Plasmids | ||
| pLAFR3 | Broad host range cloning vector, Tcr | [11] |
| pK18mobsacB | Suicide plasmid for gene knockout, Kmr | [12] |
| pRK2013 | Used as mobilizing plasmid in triparental crosses, Kmr | [13] |
| pK18gtsA | Deleted gtsA fragment cloned into pK18mobsacB, Kmr, Cmr | This study |
| pLgtsA | pLAFR3 derivative carried a fragment encoding the gtsA gene, used to complement, Tcr | This study |
| Family/Substrate | Locus Tag(s) | Fold Change a | Homologue/References * | |
|---|---|---|---|---|
| Glucose25 | Glucose3 | |||
| ATP-binding Cassette Family | ||||
| Maltose/Mannitol/Lactose | PST2190-2193 | NSS b | NSS b | mtlEFGK, P. aeruginosa [20] |
| Glucose/Mannose | PST2437-2440 | +727.96 | +24.21 | gtsABCD, P. putida [21] |
| +181.73 | +25.23 | |||
| +241.87 | +81.08 | |||
| +222.30 | +42.64 | |||
| Unknown | PST2907-2909 | NSS b | NSS b | |
| Maltose/Maltodextrin/Maltooligosaccharide | PST3478 | +2.33 | NSS b | malEFGK, E. coli [18] |
| PST3484-3486 | NSS b | |||
| Ribose/Glucose | PST3579-3583 | NSS b | NSS b | rbsABC, E. coli [19] tsgABCD13, H. volcanii DS2 |
| Phosphotransferase System | ||||
| Fructose | PST0987-0990 | NSS b | NSS b | fruAB, E. coli [4] |
| Major Facilitator Superfamily | ||||
| Sugar/Arabinose | PST1613 | NSS b | NSS b | ydeA, E. coli [22] |
| Melibiose/Galactose | PST1972 | +2.81 | NSS b | |
| Major Intrinsic Protein Family | ||||
| Glycerol | PST1604 | NSS b | NSS b | glpF, E. coli [23] |
| Sodium Solute Superfamily | ||||
| Glucose | PST1574 | NSS b | NSS b | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Shang, L.; Zhan, Y.; Lin, M.; Liu, Z.; Yan, Y. Genome-Wide Analysis of Sugar Transporters Identifies the gtsA Gene for Glucose Transportation in Pseudomonas stutzeri A1501. Microorganisms 2020, 8, 592. https://doi.org/10.3390/microorganisms8040592
Liu Y, Shang L, Zhan Y, Lin M, Liu Z, Yan Y. Genome-Wide Analysis of Sugar Transporters Identifies the gtsA Gene for Glucose Transportation in Pseudomonas stutzeri A1501. Microorganisms. 2020; 8(4):592. https://doi.org/10.3390/microorganisms8040592
Chicago/Turabian StyleLiu, Yaqun, Liguo Shang, Yuhua Zhan, Min Lin, Zhu Liu, and Yongliang Yan. 2020. "Genome-Wide Analysis of Sugar Transporters Identifies the gtsA Gene for Glucose Transportation in Pseudomonas stutzeri A1501" Microorganisms 8, no. 4: 592. https://doi.org/10.3390/microorganisms8040592
APA StyleLiu, Y., Shang, L., Zhan, Y., Lin, M., Liu, Z., & Yan, Y. (2020). Genome-Wide Analysis of Sugar Transporters Identifies the gtsA Gene for Glucose Transportation in Pseudomonas stutzeri A1501. Microorganisms, 8(4), 592. https://doi.org/10.3390/microorganisms8040592





