Characterization of Streptomyces piniterrae sp. nov. and Identification of the Putative Gene Cluster Encoding the Biosynthesis of Heliquinomycins
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Phenotypic Characterization
2.3. Chemotaxonomic Characterization
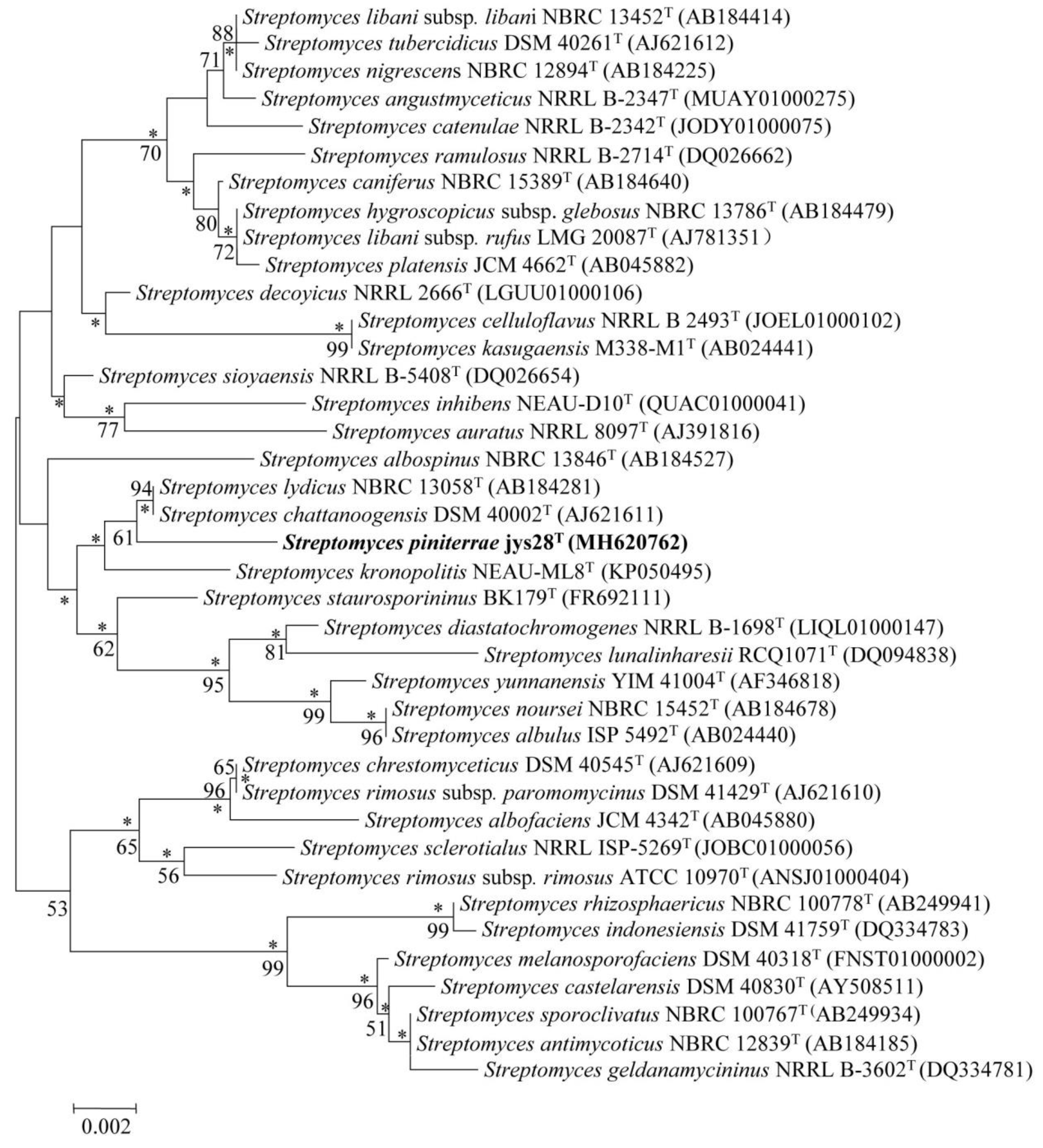
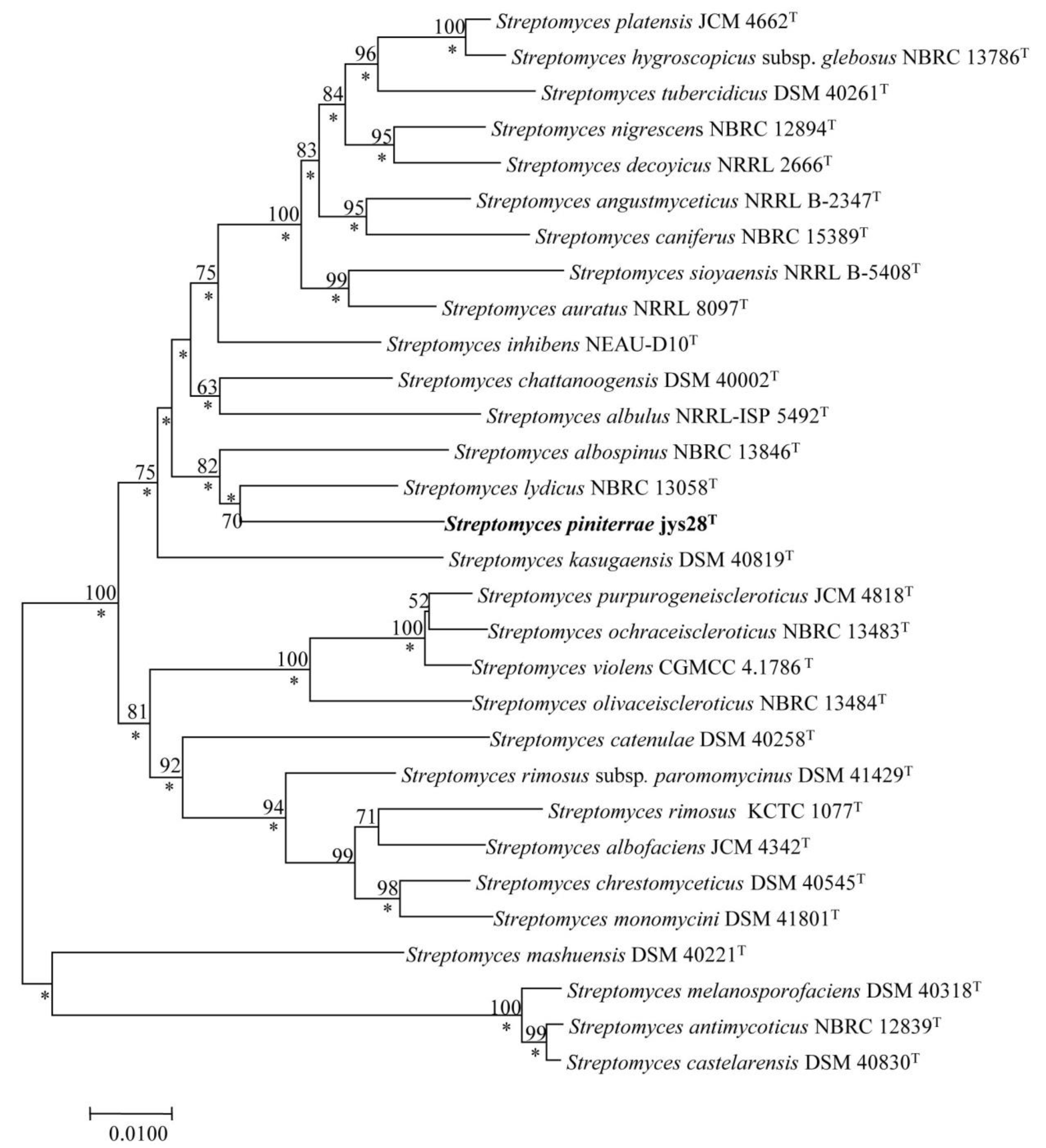
2.4. Phylogenetic Analysis
2.5. Genome Analysis
3. Results and Discussion
3.1. Polyphasic Taxonomic Characterization of Strain jys28T
3.2. Description of Streptomyces piniterrae sp. nov.
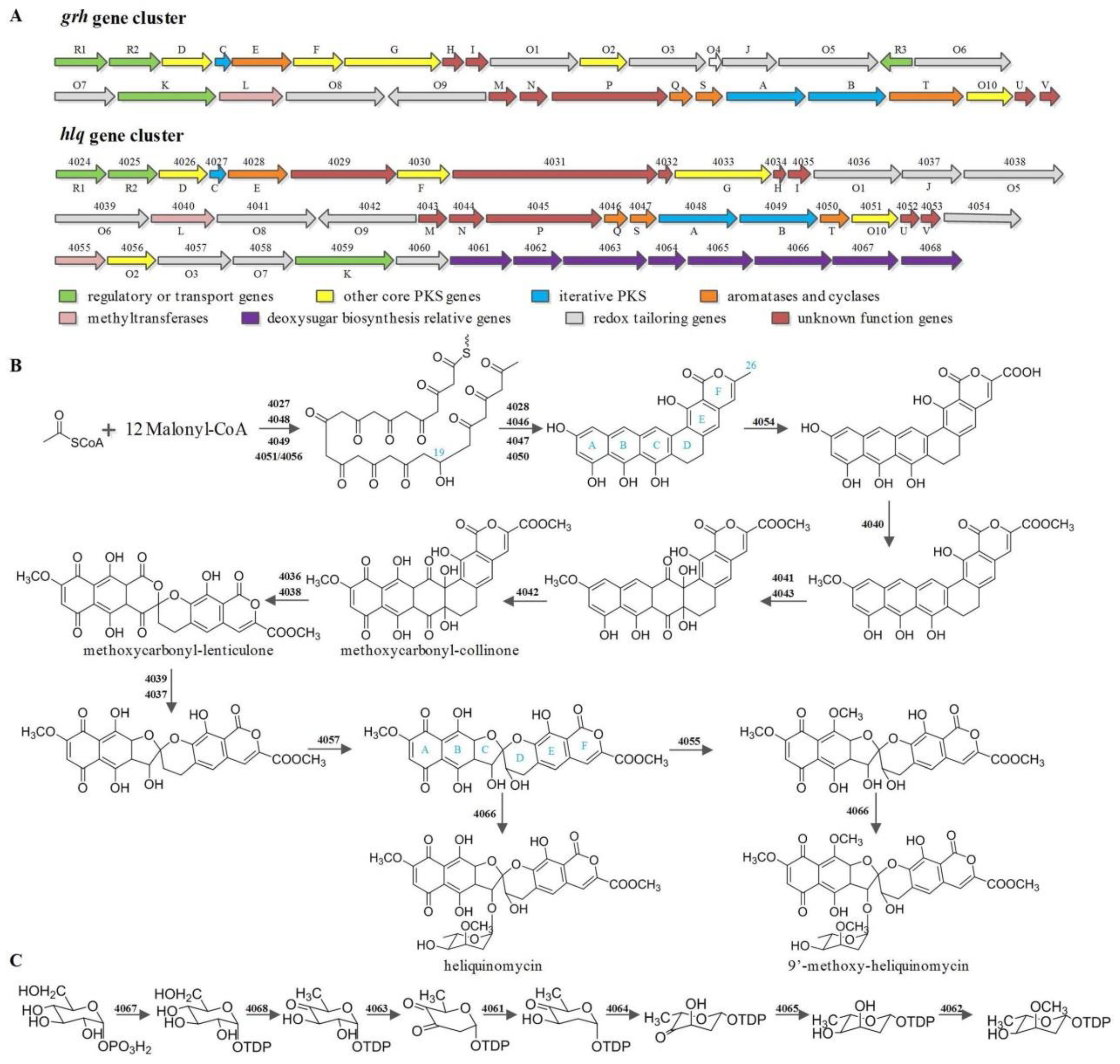
3.3. Identification of Secondary Metabolic Biosynthetic Gene Clusters, Including the Putative BGC for Heliquinomycins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Procópio, R.E.; Silva, I.R.; Martins, M.K.; Azevedo, J.L.; Araújo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Liu, C.X.; Wang, Y.J.; Xiang, W.S.; Zou, C.G.; Huang, S.X. A new heliquinomycin analogue with immunosuppressive activity from Streptomyces sp. jys28. Rec. Nat. Prod. 2019, 13, 456–461. [Google Scholar] [CrossRef]
- Brockmann, H.; Renneberg, K.H. Rubromycin, ein rotes antibiotikum aus actinomyceten. Sci. Nat.-Heidelberg. 1953, 40, 59–60. [Google Scholar] [CrossRef]
- Waters, S.P.; Fennie, M.W.; Kozlowski, M.C. Convergent route to the purpuromycin bisphenolic spiroketal: Hydrogen bonding control of spiroketalization stereochemistry. Tetrahedron. Lett. 2006, 47, 5409–5413. [Google Scholar] [CrossRef]
- Eckardt, K.; Tresselt, D.; Ihn, W. The structure of the antibiotic griseorhodin C. J. Antibiot. 1978, 31, 970–973. [Google Scholar] [CrossRef]
- Chino, M.; Nishikawa, K.; Umekita, M.; Hayashi, C.; Yamazaki, T.; Tsuchida, T.; Sawa, T.; Hamada, M.; Takeuchi, T. Heliquinomycin, a new inhibitor of DNA helicase, produced by Streptomyces sp. MJ929-SF2. I. taxonomy, production, isolation, physico-chemical properties and biological activities. J. Antibiot. 1996, 49, 752–757. [Google Scholar] [CrossRef]
- Yuen, T.Y.; Ng, Y.P.; Ip, F.C.F.; Chen, J.L.Y.; Atkinson, D.J.; Sperry, J.; Ip, N.Y.; Brimble, M.A. Telomerase inhibition studies of novel spiroketal containing rubromycin derivatives. Aust. J. Chem. 2013, 66, 530–533. [Google Scholar] [CrossRef]
- Takamasa, U.; Hirotada, T.; Masako, O.; Mizunuma, M.; Yokoyama, A.; Goto, Y.; Mizushina, Y.; Sakaguchi, K.; Hayashi, H. Inhibition of human telomerase by rubromycins:? Implication of spiroketal system of the compounds as an active moiety. Biochemistry 2000, 39, 5995–6002. [Google Scholar]
- Mizushinaa, Y.; Ueno, T.; Oda, M.; Yamaguchi, T.; Saneyoshi, M.; Sakaguchi, K. The biochemical mode of inhibition of DNA polymerase β by α-rubromycin. BBA-Gen. Subjects. 2000, 1523, 172–181. [Google Scholar] [CrossRef]
- Wu, K.L.; Mercado, E.V.; Pettus, T.R. A convergent total synthesis of (±)-γ-rubromycin. J. Am. Chem. Soc. 2011, 133, 6114–6117. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.P.; Xue, J.J.; Liu, H.B.; Wang, W.J.; Li, Y. Synthesis of (±)-γ-rubromycin via a new hypoiodite-catalytic oxidative cycloetherification. Org. Lett. 2012, 20, 5302–5305. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.L.; Schenk, A.; Hertweck, C. Molecular analysis of the benastatin biosynthetic pathway and genetic engineering of altered fatty acid-polyketide hybrids. J. Am. Chem. Soc. 2007, 129, 6022–6030. [Google Scholar] [CrossRef] [PubMed]
- Lackner, G.; Schenk, A.; Xu, Z.L.; Reinhardt, K.; Yunt, Z.S.; Piel, J.; Hertweck, C. Biosynthesis of pentangular polyphenols: Deductions from the benastatin and griseorhodin pathways. J. Am. Chem. Soc. 2007, 129, 9306–9312. [Google Scholar] [CrossRef]
- Yunt, Z.; Reinhardt, K.; Li, A.Y.; Engeser, M.; Dahse, H.M.; Gutschow, M.; Bruhn, T.; Bringmann, G.; Piel, J. Cleavage of four carbon-carbon bonds during biosynthesis of the griseorhodin A spiroketal pharmacophore. J. Am. Chem. Soc. 2009, 131, 2297–2305. [Google Scholar] [CrossRef]
- Liu, C.X.; Zhuang, X.X.; Yu, Z.Y.; Wang, Z.Y.; Wang, Y.J.; Guo, X.W.; Xiang, W.S.; Huang, S.X. Community structures and antifungal activity of root-associated endophytic actinobacteria of healthy and diseased soybean. Microorganisms 2019, 7, 243. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Waksman, S.A. The Actinomycetes. In A Summary of Current Knowledge; Ronald Press: New York, NY, USA, 1967. [Google Scholar]
- Jones, K.L. Fresh isolates of actinomycetes in which the presence of sporogenous aerial mycelia is a fluctuating characteristic. J. Bacteriol. 1949, 57, 141–145. [Google Scholar] [CrossRef]
- Zhao, J.W.; Han, L.Y.; Yu, M.Y.; Cao, P.; Li, D.M.; Guo, X.W.; Liu, Y.Q.; Wang, X.J.; Xiang, W.S. Characterization of Streptomyces sporangiiformans sp. nov., a novel soil actinomycete with antibacterial activity against Ralstonia solanacearum. Microorganisms 2019, 7, 360. [Google Scholar] [CrossRef]
- Gordon, R.E.; Barnett, D.A.; Handerhan, J.E.; Pang, C. Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int. J. Syst. Bacteriol. 1974, 24, 54–63. [Google Scholar] [CrossRef]
- Yu, Z.Y.; Han, C.Y.; Yu, B.; Zhao, J.W.; Yan, Y.J.; Huang, S.X.; Liu, C.X.; Xiang, W.S. Taxonomic characterization, and secondary metabolite analysis of Streptomyces triticiradicis sp. nov.: A novel actinomycete with antifungal activity. Microorganisms 2020, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- McKerrow, J.; Vagg, S.; McKinney, T.; Seviour, E.M.; Maszenan, A.M.; Brooks, P.; Sevious, R.J. A simple HPLC method for analyzing diaminopimelic acid diastereomers in cell walls of Gram-positive bacteria. Lett. Appl. Microbiol. 2000, 30, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Lechevalier, M.P.; Lechevalier, H.A. The Chemotaxonomy of Actinomycetes. In Actinomycete Taxonomy; Dietz, A., Thayer, D.W., Eds.; Special Publication for Society of Industrial Microbiology: Arlington, TX, USA, 1980; pp. 227–291. [Google Scholar]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods. 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Collins, M.D. Isoprenoid Quinone Analyses in Bacterial Classification and Identification. In Chemical Methods in Bacterial Systematics; Goodfellow, M., Minnikin, D.E., Eds.; Academic Press: Cambridge, MA, USA, 1985; pp. 267–284. [Google Scholar]
- Zhuang, X.X.; Peng, C.H.; Wang, Z.Y.; Zhao, J.W.; Shen, Y.; Liu, C.X.; Xiang, W.S. Actinomadura physcomitrii sp. nov., a novel actinomycete isolated from moss [Physcomitrium sphaericum (Ludw) Fuernr]. Antonie. Leeuwenhoek. 2020. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Yu, Z.Y.; Zhao, J.W.; Zhuang, X.X.; Cao, P.; Guo, X.W.; Liu, C.X.; Xiang, W.S. Community composition, antifungal activity and chemical analyses of ant-derived actinobacteria. Front. Microbiol. 2020, 11, 201. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Nikodinovic, J.; Barrow, K.D.; Chuck, J.A. High yield preparation of genomic DNA from Streptomyces. Biotechniques 2003, 35, 932–934. [Google Scholar] [CrossRef] [PubMed]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Duran, H.G.S.; Santos, E.L.C.D.L.; Kim, H.U.; Nave, M.; et al. Antismash 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic. Acids. Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Goker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC. Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large scale evaluation of algorithms to calculate average nucleotide identity. Antonie. Leeuwenhoek. 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Rong, X.; Huang, Y. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst. Appl. Microbiol. 2012, 35, 7–18. [Google Scholar] [CrossRef]
- Kämpfer, P.; Genus, I. Streptomyces Waksman and Henrici 1943, 339 AL. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: New York, NY, USA, 2012; pp. 1679–1680. [Google Scholar]
- Kim, B.Y.; Zucchi, T.D.; Fiedler, H.P.; Goodfellow, M. Streptomyces staurosporininus sp. nov., a staurosporine-producing actinomycete. Int. J. Syst. Evol. Microbiol. 2012, 62, 966–970. [Google Scholar] [CrossRef]
- Lechevalier, M.P.; Lechevalier, H.A. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Bacteriol. 1970, 20, 435–443. [Google Scholar] [CrossRef]
- Li, A.; Piel, J. A gene cluster from a marine Streptomyces encoding the biosynthesis of the aromatic spiroketal polyketide griseorhodin A. Chem. Biol. 2002, 9, 1017–1026. [Google Scholar] [CrossRef]
- Puder, C.; Loya, S.; Hizi, A.; Zeeck, A. Structural and biosynthetic investigations of the rubromycins. Eur. J. Org. Chem. 2000, 5, 729–735. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Chandler, P.M.; Poole, A.; Dennis, E.S.; Peacock, W.J. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 2065–2070. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Sterner, O.; Alvarez, M.A.; Clercq, E.D.; Bailey, J.E.; Minas, W.G. Collinone, a new recombinant angular polyketide an engineered Streptomyces strain. J. Antibiot. 2001, 54, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Nonomiya, T.; Usami, M.; Ohta, T.; Omura, S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA 1999, 96, 9509–9514. [Google Scholar] [CrossRef]
- Wohlert, S.E.; Lomovskaya, N.; Kulowski, K.; Fonstein, L.; Occi, J.L.; Gewain, K.M.; MacNeil, D.J.; Hutchinson, C.R. Insights about the biosynthesis of the avermectin deoxysugar L-oleandrose through heterologous expression of Streptomyces avermitilis deoxysugar genes in Streptomyces lividans. Chem. Biol. 2001, 8, 681–700. [Google Scholar] [CrossRef]

| Characteristic | 1 | 2 | 3 |
|---|---|---|---|
| Spore surface | Spiny | Spinya | Smootha |
| Hydrolysis of aesculin | – | + | + |
| Production of H2S | – | + | – |
| Carbon source utilization | |||
| L-arabinose | – | + | + |
| Lactose | + | – | + |
| L-rhamnose | + | – | + |
| D-ribose | + | – | + |
| D-xylose | – | – | + |
| Nitrogen source utilization | |||
| L-aspartic acid | + | – | + |
| L-threonine | – | + | + |
| L-tryptophan | – | – | + |
| pH range for growth | 4–8 | 5–9 | 5–9 |
| Growth at 40 °C | + | – | – |
| Tolerance of NaCl (%, w/v) | 11 | 2 | 2 |
| Fatty Acid | 1 | 2 | 3 |
|---|---|---|---|
| Saturated fatty acids | |||
| C14:0 | – | – | 14.3 |
| C15:0 | 15.8 | – | 18.2 |
| C16:0 | 5.8 | 30.0 | 28.4 |
| C17:0 | 6.9 | 1.7 | – |
| C18:0 | 1.0 | 41.4 | 1.8 |
| Unsaturated fatty acids | |||
| C16:1 ω7c | 11.9 | 16.2 | 2.8 |
| C17:1 ω7c | 7.0 | – | 1.3 |
| C18:1 ω7c | 2.6 | 7.9 | 1.9 |
| Branched fatty acids | |||
| anteiso-C15:0 | 11.7 | – | 5.6 |
| anteiso-C17:0 | 8.9 | – | 11.1 |
| iso-C14:0 | 3.1 | – | – |
| iso-C16:0 | 25.3 | 1.1 | 12.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, X.; Wang, Z.; Peng, C.; Su, C.; Gao, C.; Wang, Y.; Huang, S.; Liu, C. Characterization of Streptomyces piniterrae sp. nov. and Identification of the Putative Gene Cluster Encoding the Biosynthesis of Heliquinomycins. Microorganisms 2020, 8, 495. https://doi.org/10.3390/microorganisms8040495
Zhuang X, Wang Z, Peng C, Su C, Gao C, Wang Y, Huang S, Liu C. Characterization of Streptomyces piniterrae sp. nov. and Identification of the Putative Gene Cluster Encoding the Biosynthesis of Heliquinomycins. Microorganisms. 2020; 8(4):495. https://doi.org/10.3390/microorganisms8040495
Chicago/Turabian StyleZhuang, Xiaoxin, Zhiyan Wang, Chenghui Peng, Can Su, Congting Gao, Yongjiang Wang, Shengxiong Huang, and Chongxi Liu. 2020. "Characterization of Streptomyces piniterrae sp. nov. and Identification of the Putative Gene Cluster Encoding the Biosynthesis of Heliquinomycins" Microorganisms 8, no. 4: 495. https://doi.org/10.3390/microorganisms8040495
APA StyleZhuang, X., Wang, Z., Peng, C., Su, C., Gao, C., Wang, Y., Huang, S., & Liu, C. (2020). Characterization of Streptomyces piniterrae sp. nov. and Identification of the Putative Gene Cluster Encoding the Biosynthesis of Heliquinomycins. Microorganisms, 8(4), 495. https://doi.org/10.3390/microorganisms8040495





