Assessment of Diversity of Culturable Marine Yeasts Associated with Corals and Zoanthids in the Gulf of Thailand, South China Sea
Abstract
1. Introduction
2. Materials and Methods
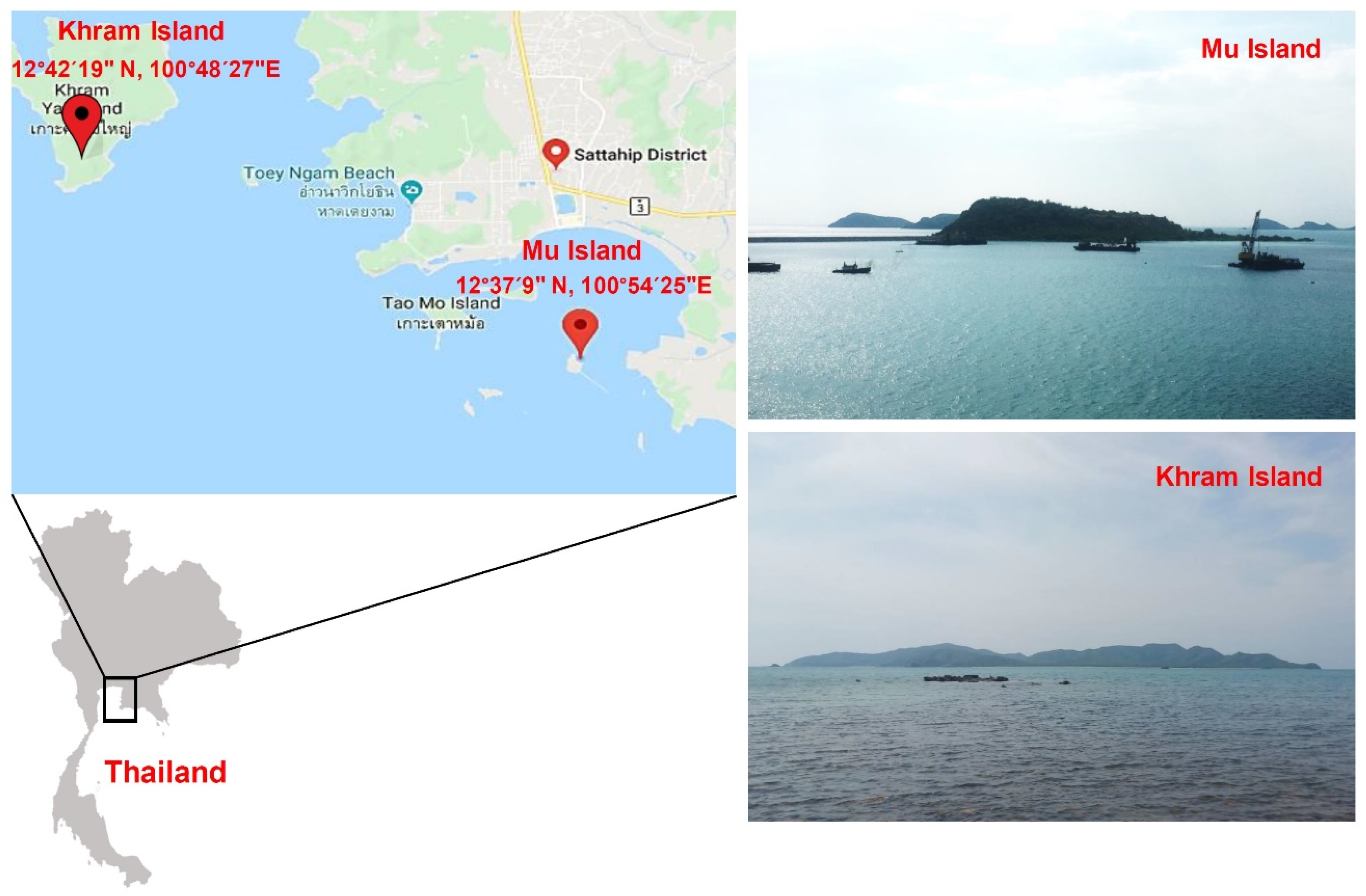
2.1. Collection and Identification of Samples
2.2. Marine Yeast Isolation
2.3. Yeast Identification and Phylogenetic Analysis
2.4. Biodiversity Analysis
3. Results and Discussion
3.1. Collection and Identification of Corals and Zoanthids
3.2. Marine Yeast Isolation and Identification
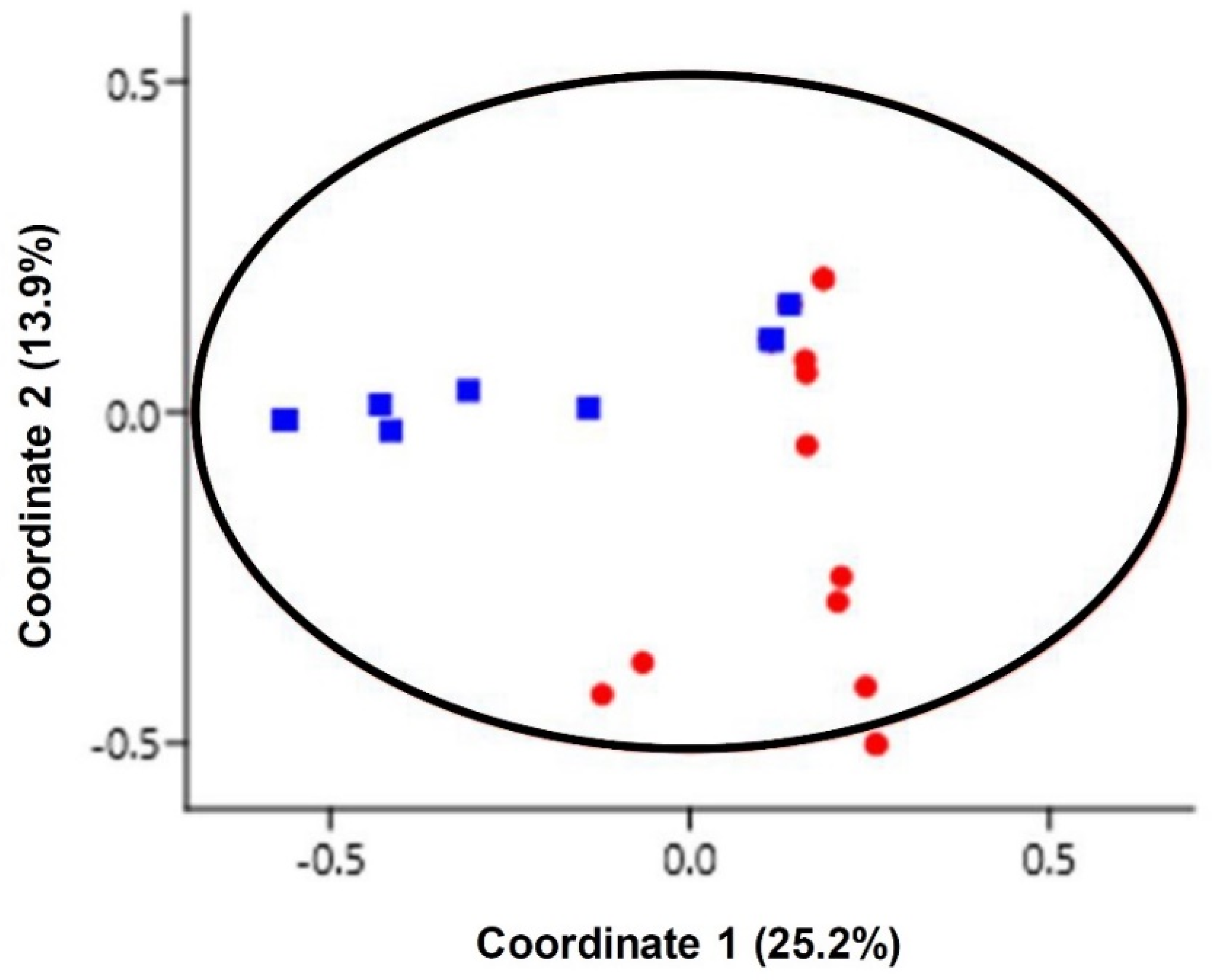
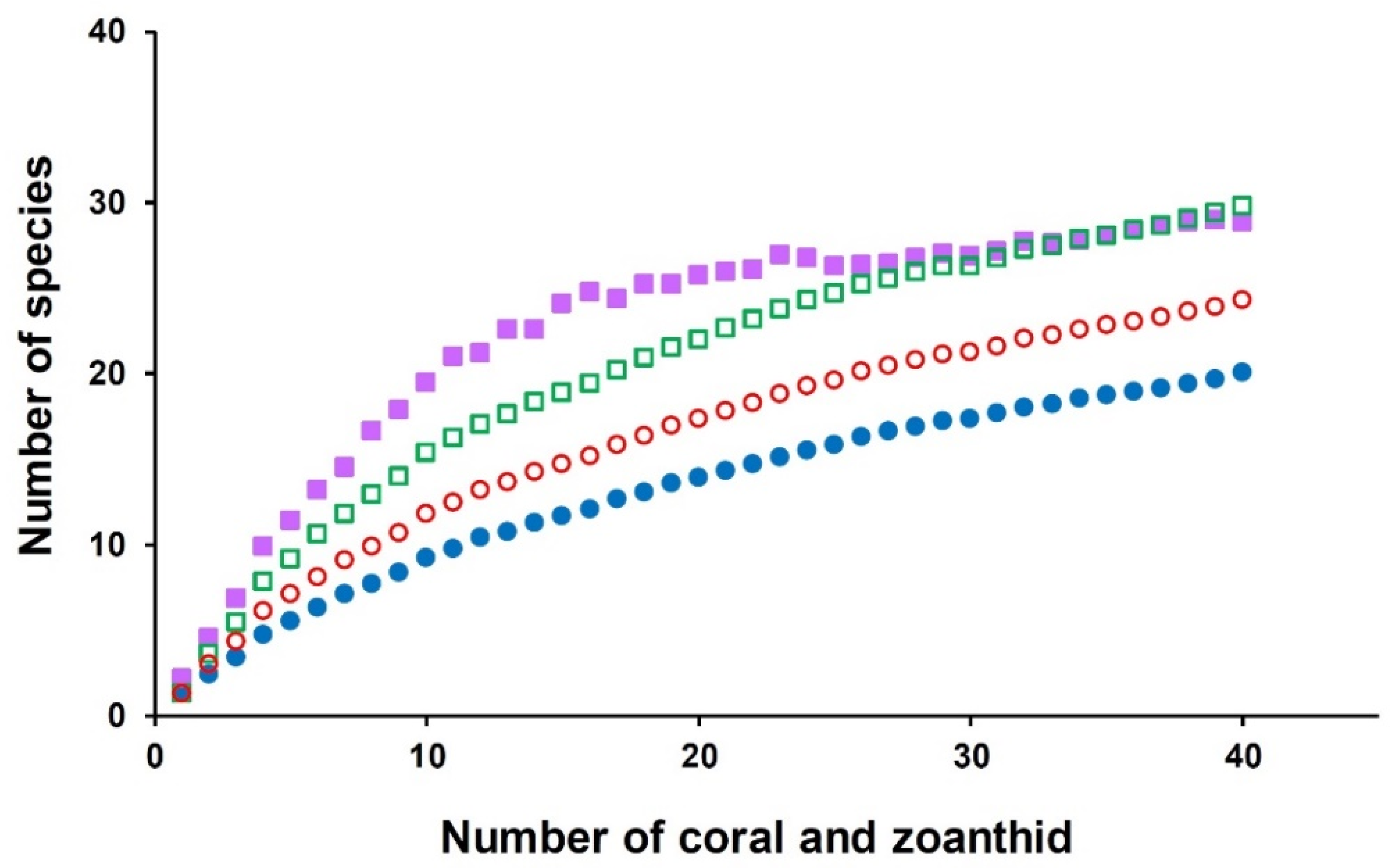
3.3. Yeast Diversity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chi, Z.M.; Liu, G.; Zhao, S.; Li, J.; Peng, Y. Marine yeasts as biocontrol agents and producers of bio-products. Appl. Microbiol. Biotechnol. 2010, 86, 1227–1241. [Google Scholar] [CrossRef] [PubMed]
- Kutty, S.N.; Philip, R. Marine yeasts—A review. Yeast 2008, 25, 465–483. [Google Scholar] [CrossRef] [PubMed]
- Zaky, A.S.; Tucker, G.A.; Daw, Z.Y.; Du, C. Marine yeast isolation and industrial application. FEMS Yeast Res. 2014, 14, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hua, M.; Song, C.; Chi, Z. Occurrence and Diversity of Marine Yeasts in Antarctica Environments. J. Ocean Univ. China 2012, 11, 70–74. [Google Scholar] [CrossRef]
- Chi, Z.; Liu, G.L.; Lu, Y.; Jiang, H.; Chi, Z.M. Bio-products produced by marine yeasts and their potential applications. Bioresour. Technol. 2016, 202, 244–252. [Google Scholar] [CrossRef]
- Naim, M.A.; Smidt, H.; Sipkema, D. Fungi found in Mediterranean and North Sea sponges: How specific are they? PeerJ 2017, 5, e3722. [Google Scholar] [CrossRef][Green Version]
- Paulino, G.V.B.; Félix, C.R.; Broetto, L.; Landell, M.F. Diversity of culturable yeasts associated with zoanthids from Brazilian reef and its relation with anthropogenic disturbance. Mar. Pollut. Bull. 2017, 123, 253–260. [Google Scholar] [CrossRef]
- Li, Z. Sponge and coral microbiomes. In Symbiotic Microbiomes of Coral Reefs Sponges and Corals; Li, Z., Ed.; Spinger Nature: Berlin/Heidelberg, Germany, 2019; pp. 17–28. [Google Scholar]
- Glynn, P.W.; Enochs, l.C. Invertebrates and their roles in coral reef ecosystems. In Coral Reefs: An Ecosystem in Transition; Dubinsky, Z., Stambler, N., Eds.; Srpinger Science Business Media: Berlin/Heidelberg, Germany, 2011; pp. 273–325. [Google Scholar]
- Orlić, S. Microbial diversity of sponge/coral microbiome. In Symbiotic Microbiomes of Coral Reefs Sponges and Corals; Li, Z., Ed.; Spinger Nature: Berlin/Heidelberg, Germany, 2019; pp. 29–41. [Google Scholar]
- Reimer, J.D.; Ono, S.; Sinniger, F.; Tsukahara, J. Distribution of zooxanthellate zoanthid species (Zoantharia: Anthozoa: Hexacorallia) in southern Japan limited by cold temperatures. Galaxea J. Coral Reef Stud. 2008, 10, 57–67. [Google Scholar] [CrossRef][Green Version]
- Roberts, T.E.; Maloney, J.M.; Sweatman, H.P.A.; Bridge, T.C.L. Benthic community composition on submerged reefs in the central Great Barrier Reef. Coral Reefs 2015, 34, 569–580. [Google Scholar] [CrossRef]
- Dumalagan, E.E.; Cabaitan, P.C.; Bridge, T.C.L.; Go, K.T.; Quimpo, T.J.R.; Olavides, R.D.D.; Munar, J.C.; Villanoy, C.L.; Siringan, F.P. Spatial variability in benthic assemblage composition in shallow and upper mesophotic coral ecosystems in the Philippines. Mar. Environ. Res. 2019, 150, 104772. [Google Scholar] [CrossRef]
- Reimer, J.D. Preliminary survey of zooxanthellate zoanthid diversity (Hexacorallia: Zoantharia) from southern Shikoku, Japan. Kuroshio Biosph. 2007, 3, 1–16. [Google Scholar]
- Reimer, J.D.; Albinsky, D.; Yang, S.Y.; Lorion, J. Zoanthid (Cnidaria: Anthozoa: Hexacorallia: Zoantharia) species of coral reefs in Palau. Mar. Biodiv. 2014, 44, 37–44. [Google Scholar] [CrossRef]
- Chiou, S.F.; Kuo, J.; Wongd, T.Y.; Fan, T.Y.; Tew, K.S.; Liu, J.K. Analysis of the coral associated bacterial community structures in healthy and diseased corals from off-shore of southern Taiwan. J. Environ. Sci. Health B 2010, 45, 408–415. [Google Scholar] [CrossRef]
- Meron, D.; Hazanov, L.; Fine, M.; Banin, E. Effect of ocean acidification on the coral microbial community. In Beneficial Microorganisms in Multicellular Life Forms; Rosenberg, E., Gophna, U., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 163–173. [Google Scholar]
- Hong, M.J.; Yu, Y.T.; Chen, C.A.; Chiang, P.W.; Tang, S.L. Influence of species specificity and other factors on bacteria associated with the coral Stylophora pistillata in Taiwan. Appl. Environ. Microbiol. 2009, 75, 7797–7806. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rohwer, F.; Seguritan, V.; Azam, F.; Knowlton, N. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 2002, 243, 1–10. [Google Scholar] [CrossRef]
- Chen, Y.H.; Kuo, J.; Sung, P.J.; Chang, Y.C.; Lu, M.C.; Wong, T.Y.; Liu, J.K.; Weng, C.F.; Twan, W.H.; Kuo, F.W. Isolation of marine bacteria with antimicrobial activities from cultured and field-collected soft corals. World J. Microbiol. Biotechnol. 2012, 28, 3269–3279. [Google Scholar] [CrossRef]
- Burgaud, G.; Arzur, D.; Durand, L.; Cambon-Bonavita, M.A.; Barbier, G. Marine culturable yeasts in deep-sea hydrothermal vents: Species richness and association with fauna. FEMS Microbiol. Ecol. 2010, 73, 121–133. [Google Scholar] [CrossRef]
- Limtong, S.; Kaewwichian, R. The diversity of culturable yeasts in the phylloplane of rice in Thailand. Ann. Microbiol. 2015, 65, 667–675. [Google Scholar] [CrossRef]
- Khunnamwong, P.; Jindamorakot, S.; Limtong, S. Endophytic yeasts diversity in leaf tissue of rice, corn and sugarcane cultivated in Thailand assessed by a culture-dependent approach. Fungal Biol. 2018. [Google Scholar] [CrossRef]
- Srisuk, N.; Nutaratat, P.; Surussawadee, J.; Limtong, S. Yeast Communities in Sugarcane Phylloplane. Microbiology 2019, 88, 353–369. [Google Scholar] [CrossRef]
- Jaiboon, K.; Lertwattanasakul, N.; Limtong, S. Yeasts from peat in a tropical peat swamp forest in Thailand and their ability to produce ethanol, indole-3-acetic acid and extracellular enzymes. Mycol. Prog. 2016, 15, 755–770. [Google Scholar] [CrossRef]
- Boonmak, C.; Khunnamwong, P.; Limtong, S. Yeast communities of primary and secondary peat swamp forests in southern Thailand. Antonie Van Leeuwenhoek 2019, 113, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Limtong, S.; Yongmanitchai, W.; Kawasaki, H.; Seki, T. Candida phangngensis sp. nov., an anamorphic yeast species in the Yarrowia clade, isolated from water in mangrove forests in Phang-Nga Province, Thailand. Int. J. Syst. Evol. Microbiol. 2008, 58, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Am-In, S.; Limtong, S.; Yongmanitchai, W.; Jindamorakot, S. Candida andamanensis sp. nov., Candida laemsonensis sp. nov., and Candida ranongensis sp. nov., three anamorphic yeast species isolated from estuarine waters in a mangrove forest in Ranong Province, Thailand. Int. J. Syst. Evol. Microbiol. 2011, 61, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Hoondee, P.; Wattanagonniyom, T.; Weeraphan, T.; Tanasupawat, S.; Savarajara, A. Occurrence of oleaginous yeast from mangrove forest in Thailand. World J. Microbiol. Biotechnol. 2019, 35, 108. [Google Scholar] [CrossRef] [PubMed]
- Marine Knowledge Hub. Available online: www.mkh.in.th (accessed on 29 March 2010).
- Pootakham, W.; Mhuantong, W.; Yoocha, T.; Putchim, L.; Sonthirod, C.; Naktang, C.; Thongtham, N.; Tangphatsornruang, S. High resolution profiling of coral-associated bacterial communities using full-length 16S rRNA sequence data from PacBio SMRT sequencing system. Sci. Rep. 2017, 7, 2774. [Google Scholar] [CrossRef]
- Somboonna, N.; Wilantho, A.; Monanunsap, S.; Chavanich, S.; Tangphatsornruang, S.; Tongsima, S. Microbial communities in the reef water at Kham Island, lower Gulf of Thailand. PeerJ 2017, 5, e3625. [Google Scholar] [CrossRef]
- Somboonna, N.; Wilantho, A.; Rerngsamran, P.; Tongsima, S. Marine Bacterial Diversity in Coastal Sichang Island, the Upper Gulf of Thailand, in 2011 Wet Season. Front. Mar. Sci. 2019, 6, 308. [Google Scholar] [CrossRef]
- Veron, J. Corals of the World; Australian Institute of Marine Science: Townsville, Australia, 2000; Volume 1, 463p.
- Fabricius, K.E.; Alderslade, P. Soft Corals and Sea Fans: A Comprehensive Guide to the Tropical Shallow-Water Genera of the Central-West Pacific, the Indian Ocean and the Red Sea; Australian Institute of Marine Science: Melbourne, Australia, 2001; 272p.
- Reimer, J.D. Key to field identification of shallow water brachycnemic zoanthids (Order Zoantharia: Suborder Brachycnemina) present in Okinawa. Galaxea 2010, 12, 23–29. [Google Scholar] [CrossRef]
- Limtong, S.; Yongmanitchai, W.; Tun, M.M.; Kawasaki, H.; Seki, T. Kazachstania siamensis sp. nov., an ascomycetous yeast species from forest soil in Thailand. Int. J. Syst. Evol. Microbiol. 2007, 57, 419–422. [Google Scholar] [CrossRef]
- Ruiz-Barba, J.L.; Maldonado, A.; Jiménez-Díaz, R. Small-scale total DNA extraction from bacteria and yeast for PCR applications. Anal. Biochem. 2005, 347, 333–335. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Fell, J.W.; Boekhout, T.; Fonseca, A.; Scorzetti, G.; Statzell-Tallman, A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 2000, 50, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Colwell, R. EstimateS: Statistical Estimation of Species Richness and Shared Species from Samples. Version 8.2. User’s Guide and Application. 2006. Available online: http://viceroy.eeb.uconn.edu/EstimateS (accessed on 26 January 2020).
- Cheablam, O.; Shrestha, R.P. Climate change trends and its impact on tourism resources in Mu Ko Surin marine national park, Thailand. Asia Pac. J. Tour. Res. 2015, 20, 435–454. [Google Scholar] [CrossRef]
- Aubrecht, C.; Elvidge, C.D.; Longcore, T.; Rich, C.; Safran, J.; Strong, A.E.; Eakin, C.M.; Baugh, K.E.; Tuttle, B.T.; Howard, A.T.; et al. A global inventory of coral reef stressors based on satellite observed nighttime lights. Geocarlo Int. 2008, 23, 467–479. [Google Scholar] [CrossRef]
- Godinho, V.M.; de Paula, M.T.R.; Silva, D.A.S.; Paresque, K.; Martins, A.P.; Colepicolo, P.; Rosa, C.A.; Rosa, L.H. Diversity and distribution of hidden cultivable fungi associated with marine animals of Antarctica. Fungal Biol. 2019, 123, 507–516. [Google Scholar] [CrossRef]
- Vaca, I.; Faúndez, C.; Maza, F.; Paillavil, B.; Hernández, V.; Acosta, F.; Levicán, G.; Martínez, C.; Chávez, R. Cultivable psychrotolerant yeasts associated with Antarctic marine sponges. World J. Microbiol. Biotechnol. 2013, 29, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Pang, K.L.; Luo, Z.H. High fungal diversity and abundance recovered in the deep-sea sediments of the Pacific Ocean. Microb. Ecol. 2014, 68, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Gadanho, M.; Sampaio, J.P. Occurrence and diversity of yeasts in the Mid-Atlantic Ridge hydrothermal fields near the Azores archipelago. Microb. Ecol. 2005, 50, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Li, B.; Zheng, C.; Wang, G. Molecular detection of fungal communities in the Hawaiian marine sponges Suberites zeteki and Mycale armata. Appl. Environ. Microbiol. 2008, 74, 6091–6101. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Yanagida, F.; Chen, L.Y. Isolation of marine yeasts from coastal waters of northeastern Taiwan. Aquat. Biol. 2009, 8, 55–60. [Google Scholar] [CrossRef][Green Version]
- Papadakis, J.S.; Mavridou, A.; Richardson, S.C.; Lamprini, M.; Marcelou, U. Bather-related microbial and yeast populations in sand and seawater. Water Res. 1997, 31, 799–804. [Google Scholar] [CrossRef]
- Fell, J.W.; Statzell-Tallman, A.; Scorzetti, G.; Gutiérrez, M.H. Five new species of yeasts from fresh water and marine habitats in the Florida Everglades. Antonie Van Leeuwenhoek 2011, 99, 533–549. [Google Scholar] [CrossRef]
- Kunthiphun, S.; Chokreansukchai1, P.; Hondee, P.; Tanasupawat, S.; Savarajara, A. Diversity and characterization of cultivable oleaginous yeasts isolated from mangrove forests. World J. Microbiol. Biotechnol. 2018, 34, 125. [Google Scholar] [CrossRef]
- Coelho, M.A.; Almeida, J.M.; Martins, I.M.; da Silva, A.J.; Sampaio, J.P. The dynamics of the yeast community of the Tagus river estuary: Testing the hypothesis of the multiple origins of estuarine yeasts. Antonie Van Leeuwenhoek 2010, 98, 331–342. [Google Scholar] [CrossRef]
- Evangelista, S.R.; Miguel, M.G.; Silva, C.F.; Pinheiro, A.C.; Schwan, R.F. Microbiological diversity associated with the spontaneous wet method of coffee fermentation. Int. J. Food Microbiol. 2015, 210, 102–112. [Google Scholar] [CrossRef]
- Limtong, S.; Kaewwichian, R.; Yongmanitchai, W.; Kawasaki, H. Diversity of culturable yeasts in phylloplane of sugarcane in Thailand and their capability to produce indole-3-acetic acid. World J. Microbiol. Biotechnol. 2014, 30, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.W.F.; Dayo-Owoyemi, I.; Nobre, F.S.; Pagnocca, F.C.; Chaud, L.C.S.; Pessoa, A.; Felipe, M.G.A.; Sette, L.D. Taxonomic assessment and enzymes production by yeasts isolated from marine and terrestrial Antarctic samples. Extremophiles 2013, 17, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Cadete, R.M.; Melo, M.A.; Dussan, K.J.; Rodrigues, R.C.; da Silva, S.S.; Zilli, J.E.; Vital, M.J.S.; Gomes, F.C.O.; Lachance, M.A.; Rosa, C.A. Diversity and physiological characterization of D-xylosefermenting yeasts isolated from the Brazilian Amazonian forest. PLoS ONE 2012, 7, e43135. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.; de Souza, A.C.; Magalhatilde, K.T.; Dias, D.R.; Silva, C.F.; Schwan, R.F. Yeasts diversity in Brazilian Cerrado soils: Study of the enzymatic activities. Afr. J. Microbiol. Res 2013, 7, 4176–4190. [Google Scholar] [CrossRef]
- Burgaud, G.; Coton, M.; Jacques, N.; Debaets, S.; Maciel, N.O.; Rosa, C.A.; Gadanho, M.; Sampaio, J.P.; Casaregola, S. Yamadazyma barbieri f.a. sp. nov., an ascomycetous anamorphic yeast isolated from a Mid-Atlantic Ridge hydrothermal site (_2300 m) and marine coastal waters. Int. J. Syst. Evol. Microbiol. 2016, 66, 3600–3606. [Google Scholar] [CrossRef]
- Statzell-Tallman, A.; Belloch, C.; Fell, J.W. Kwoniella mangroviensis gen. nov., sp. nov. a tremellaceous yeast from mangrove habitats in the Florida Everglades and Bahamas. FEMS Yeast Res. 2008, 8, 103–113. [Google Scholar] [CrossRef]
- Gadanho, M.; Almeida, J.M.F.; Sampaio, J.P. Assessment of yeast diversity in a marine environment in the south of Portugal by microsatellite-primed PCR. Antonie Van Leeuwenhoek 2003, 84, 217–227. [Google Scholar] [CrossRef]
- Nagano, Y.; Nagahama, T.; Hatada, Y.; Nonoura, T.; Takami, H.; Miyazaki, J.; Takai, K.; Horikoshi, K. Fungal diversity in deep-sea sediments—The presence of novel fungal groups. Fungal Ecol. 2010, 3, 316–325. [Google Scholar] [CrossRef]
- Yang, S.P.; Wu, Z.H.; Jian, J.C. Distribution of marine red yeasts in shrimps and the environments of shrimp culture. Curr. Microbiol. 2011, 62, 1638–1642. [Google Scholar] [CrossRef]
- Loureiro, S.T.A.; de Queiroz-Cavalcanti, M.A.; Neves, R.P.; de Oliveira-Passavante, J.Z. Yeasts isolated from sand and sea water in beaches of Olinda, Pernambuco state, Brazil. Braz. J. Microbiol. 2005, 36, 1–8. [Google Scholar] [CrossRef][Green Version]
- Sampaio, A.; Sampaio, J.P.; Lĕao, C. Dynamics of yeast populations recovered from decaying leaves in a nonpolluted stream: A 2-year study on the effects of leaf litter type and decomposition time. FEMS Yeast Res. 2007, 7, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Gadanho, M.; Libkind, D.; Sampaio, J.P. Yeast diversity in the extreme acidic environments of the Iberian Pyrite Belt. Microb. Ecol. 2006, 52, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Santiago, I.F.; Rosa, C.A.; Rosa, L.H. Endophytic symbiont yeasts associated with the Antarctic angiosperms Deschampsia antarctica and Colobanthus quitensis. Polar Biol. 2017, 40, 177–183. [Google Scholar] [CrossRef]
- Nagahama, T.; Hamamoto, M.; Nakase, T.; Takami, H.; Horikoshi, K. Distribution and identification of red yeasts in deep-sea environments around the northwest Pacific Ocean. Antonie Van Leeuwenhoek 2001, 80, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Raghukumar, C.; Meena, R.M.; Verma, P.; Shouche, Y. Fungal diversity in deep sea sediments revealed by culture-dependent and culture-independent approaches. Fungal Ecol. 2012, 5, 543–553. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Tang, G.L.; Xu, X.Y.; Nong, X.H.; Qi, S.H. Insights into deep-sea sediment fungal communities from the East Indian Ocean using targeted environmental sequencing combined with traditional cultivation. PLoS ONE 2014, 9, e109118. [Google Scholar] [CrossRef]
- Piñar, G.; Dalnodar, D.; Voitl, C.; Reschreiter, H.; Sterflinger, K. Biodeterioration risk threatens the 3100 year old staircase of Hallstatt (Austria): Possible involvement of halophilic microorganisms. PLoS ONE 2016, 11, e0148279. [Google Scholar]
- Rohwer, F.; Breitbart, M.; Jara, J.; Azam, F.; Knowlton, N. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 2001, 20, 85–91. [Google Scholar]
| Collection Date | Physical Property of Seawater | Total Samples | Sample Code | Coral/Zoanthid | No. of Strain | Strain DMKU | GenBank Accession no. | Yeast Species | ||
|---|---|---|---|---|---|---|---|---|---|---|
| pH | Temp. (°C) | Salinity (PSU) | ||||||||
| Sampling sites S1 | ||||||||||
| 6 March 2016 | 8.17 | 29.4 | 33.2 | 2 | S1 | Cladiella sp. | 0 | ND | ||
| SZ1 | Unknown zoanthid | 4 | 319-1 | LC415303 | Meyerozyma guilliermondii | |||||
| 319-2 | LC415304 | Candida tropicalis | ||||||||
| C319-1 | LC415305 | Wickerhamomyces anomalus | ||||||||
| C319-2 | LC415306 | Candida metapsilosis | ||||||||
| 31 July 2016 | 8.37 | 30.8 | 32.7 | 2 | R1 | Unknown hard coral | 0 | ND | ||
| R2 | Unknown hard coral | 0 | ND | |||||||
| 21 January 2017 | 8.37 | 19.7 | 31.7 | 23 | W1 | Sinularia sp. | 4 | J7-1 | LC431005 | Papiliotrema laurentii |
| JC7-2 | LC431006 | Rhodotorula diobovata | ||||||||
| JC7-4 | LC431007 | Naganishia liquefaciens | ||||||||
| JC7-5 | LC431008 | Candida parapsilosis | ||||||||
| W2 | Pavona sp. | 1 | JC8-1 | LC431009 | Papiliotrema laurentii | |||||
| W3 | Klyxum sp. | 1 | JC9-1 | LC431010 | Cystobasidium calyptogenae | |||||
| W4 | Cladiella sp. | 0 | ND | |||||||
| W5 | Cladiella sp. | 1 | JC14-1 | LC431017 | Candida parapsilosis | |||||
| W6 | Sarcophyton sp. | 1 | JC15-1 | LC431018 | Rhodotorula mucilaginosa | |||||
| W7 | Klyxum sp. | 1 | J18-1 | LC431020 | Candida spencermartinsiae | |||||
| W8 | Cladiella sp. | 0 | ND | |||||||
| W9 | Sarcophyton sp. | 0 | ND | |||||||
| W10 | Acropora sp. | 1 | JC22-1 | LC497427 | Candida parapsilosis | |||||
| W11 | Sinularia sp. | 0 | ND | |||||||
| W12 | Sarcophyton sp. | 1 | JC24-1 | LC511611 | Sterigmatomyces halophilus | |||||
| W13 | Pavona decussata | 2 | JC25-1 | LC497428 | Papiliotrema laurentii | |||||
| JC25-2 | LC497429 | Candida tropicalis | ||||||||
| W14 | Pocillopora damicornis | 0 | ND | |||||||
| W15 | Acropora sp. | 0 | ND | |||||||
| W16 | Cladiella sp. | 5 | J33-2 | LC497434 | Rhodotorula mucilaginosa | |||||
| J33-3 | LC497435 | Papiliotrema laurentii | ||||||||
| JC33-1 | LC497436 | Rhodotorula diobovata | ||||||||
| JC33-2 | LC511612 | Rhodosporidiobolus fluvialis | ||||||||
| JC33-4 | LC511613 | Cystobasidium calyptogenae | ||||||||
| W17 | Klyxum sp. | 2 | JC35-1 | LC497442 | Rhodotorula diobovata | |||||
| JC35-2 | LC511614 | Cystobasidium calyptogenae | ||||||||
| W18 | Cladiella sp. | 1 | J37-1 | LC497437 | Potential new species closest to Rhodotorula toruloides | |||||
| W19 | Klyxum sp. | 0 | ND | |||||||
| W20 | Cladiella sp. | 1 | JC46-1 | LC511615 | Papiliotrema laurentii | |||||
| WZ1 | Unknown zoanthid | 4 | J29-5 | LC497430 | Meyerozyma guilliermondii | |||||
| JC29-2 | LC497431 | Kodamaea ohmeri | ||||||||
| JC29-6 | LC497432 | Papiliotrema laurentii | ||||||||
| JC29-7 | LC497433 | Meyerozyma caribbica | ||||||||
| WZ2 | Unknown zoanthid | 4 | JC34-1 | LC497439 | Papiliotrema laurentii | |||||
| JC34-2 | LC497440 | Rhodotorula diobovata | ||||||||
| JC34-5 | LC511616 | Rhodosporidiobolus fluvialis | ||||||||
| JC34-7 | LC497441 | Rhodotorula mucilaginosa | ||||||||
| WZ3 | Unknown zoanthid | 0 | ND | |||||||
| Sampling site S2 | ||||||||||
| 26 March 2016 | 7.94 | 27.1 | 31.9 | 4 | S2 | Sinularia sp. | 0 | ND | ||
| S3 | Sarcophyton sp. | 1 | C322-1 | LC415308 | Filobasidium uniguttulatum | |||||
| S4 | Pocillopora damicornis | 2 | 3222-1 | LC415320 | Rhodotorula mucilaginosa | |||||
| 3222-2 | LC511617 | Naganishia liquefaciens | ||||||||
| S5 | Sarcophyton sp. | 1 | 3236-1 | LC511618 | Rhodotorula mucilaginosa | |||||
| 30 July 2016 | 8.13 | 28.6 | 31.9 | 4 | R3 | Cladiella sp. | 1 | 729-1 | LC430111 | Rhodotorula mucilaginosa |
| R4 | Cladiella sp. | 1 | 7231-1 | LC430581 | Rhodotorula mucilaginosa | |||||
| R5 | Unknown coral | 3 | 7235-1 | LC430589 | Rhodotorula mucilaginosa | |||||
| C7235-3 | LC430590 | Candida zeylanoides | ||||||||
| C7235-4 | LC430591 | Vishniacozyma victoriae | ||||||||
| R6 | Cladiella sp. | 1 | 7241-1 | LC430599 | Rhodotorula mucilaginosa | |||||
| 18 February 2017 | 8.40 | 20.5 | 31.2 | 5 | W21 | Sarcophyton sp. | 4 | FC12-1 | LC511619 | Meyerozyma caribbica |
| FC12-2 | LC511620 | Rhodotorula mucilaginosa | ||||||||
| FC12-3 | LC511621 | Candida parapsilosis | ||||||||
| FC12-4 | LC511622 | Candida tropicalis | ||||||||
| W22 | Cladiella sp. | 0 | ND | |||||||
| WZ4 | Unknown zoanthid | 0 | ND | |||||||
| WZ5 | Unknown zoanthid | 2 | F9-1 | LC511623 | Rhodotorula mucilaginosa | |||||
| FC9-1 | LC511624 | Rhodotorula toruloides | ||||||||
| WZ6 | Unknown zoanthid | 0 | ND | |||||||
| Phylum | Subphylum | Family | Genus | Species | No. of Strain | Total | Frequency of Occurrence a (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Mu | Khram | |||||||||
| 2016 | 2017 | 2016 | 2017 | |||||||
| Ascomycota (16 strains) | Saccharomycotina | Debaryomycetaceae | Meyerozyma | M. caribbica | 0 | 1 | 0 | 1 | 2 | 5.0 |
| M. guilliermondii | 1 | 1 | 0 | 0 | 2 | 5.0 | ||||
| Candida | C. tropicalis | 1 | 1 | 0 | 1 | 3 | 7.5 | |||
| C. parapsilosis | 0 | 3 | 0 | 1 | 4 | 10.0 | ||||
| C. zeylanoides | 0 | 0 | 1 | 0 | 1 | 2.5 | ||||
| C. spencermartinsiae | 0 | 1 | 0 | 0 | 1 | 2.5 | ||||
| C. metapsilosis | 1 | 0 | 0 | 0 | 1 | 2.5 | ||||
| Metschnikowiaceae | Kodamaea | K. omeri | 0 | 1 | 0 | 0 | 1 | 2.5 | ||
| Phaffomycetaceae | Wickerhamomyces | W. anomalus | 1 | 0 | 0 | 0 | 1 | 2.5 | ||
| Basidiomycota (34 strains) | Agaricomycotina | Bulleribasidiaceae | Vishniacozyma | V. victoriae | 0 | 0 | 1 | 0 | 1 | 2.5 |
| Filobasidiaceae | Filobasidium | F. uniguttulatum | 0 | 0 | 1 | 0 | 1 | 2.5 | ||
| Naganishia | N. liquefaciens | 0 | 1 | 1 | 0 | 2 | 5.0 | |||
| Rhynchogastremataeae | Papiliotrema | P. laurentii | 0 | 7 | 0 | 0 | 7 | 17.5 | ||
| Pucciniomycotina | Agaricostilbaceae | Sterigmatomyces | S. halophilus | 0 | 1 | 0 | 0 | 1 | 2.5 | |
| Cystobasidiaceae | Cystobasidium | Cys. calyptogenae | 0 | 3 | 0 | 0 | 3 | 7.5 | ||
| Sporidiobolaceae | Rhodotorula | R. diobovata | 0 | 4 | 0 | 0 | 4 | 10.0 | ||
| R. mucilaginosa | 0 | 3 | 6 | 2 | 11 | 27.5 | ||||
| Potential new species closest to R.toruloides | 0 | 1 | 0 | 0 | 1 | 2.5 | ||||
| R. toruloides | 0 | 0 | 0 | 1 | 1 | 2.5 | ||||
| Rhodosporidiobolus | Rh. fluvialis | 0 | 2 | 0 | 0 | 2 | 5.0 | |||
| Total number of yeast strain | 4 | 30 | 10 | 6 | 50 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaewkrajay, C.; Chanmethakul, T.; Limtong, S. Assessment of Diversity of Culturable Marine Yeasts Associated with Corals and Zoanthids in the Gulf of Thailand, South China Sea. Microorganisms 2020, 8, 474. https://doi.org/10.3390/microorganisms8040474
Kaewkrajay C, Chanmethakul T, Limtong S. Assessment of Diversity of Culturable Marine Yeasts Associated with Corals and Zoanthids in the Gulf of Thailand, South China Sea. Microorganisms. 2020; 8(4):474. https://doi.org/10.3390/microorganisms8040474
Chicago/Turabian StyleKaewkrajay, Chutima, Thanongsak Chanmethakul, and Savitree Limtong. 2020. "Assessment of Diversity of Culturable Marine Yeasts Associated with Corals and Zoanthids in the Gulf of Thailand, South China Sea" Microorganisms 8, no. 4: 474. https://doi.org/10.3390/microorganisms8040474
APA StyleKaewkrajay, C., Chanmethakul, T., & Limtong, S. (2020). Assessment of Diversity of Culturable Marine Yeasts Associated with Corals and Zoanthids in the Gulf of Thailand, South China Sea. Microorganisms, 8(4), 474. https://doi.org/10.3390/microorganisms8040474





