The Limits and Avoidance of Biases in Metagenomic Analyses of Human Fecal Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. DNA Extraction According to the G’NOME Protocol
2.3. DNA Extraction Using the PROMEGA Kit
2.4. DNA Extraction Using the QIAGEN Kit
2.5. Quantification and Quality Control of Genomic DNA
2.6. 16S rRNA and Whole Metagenome Sequencing
2.7. Bionformatics Analysis
2.8. Genus Name Correction
2.9. Statistical Analysis
2.10. Data Availability
3. Results
3.1. Comparison of Three DNA Extraction Methods
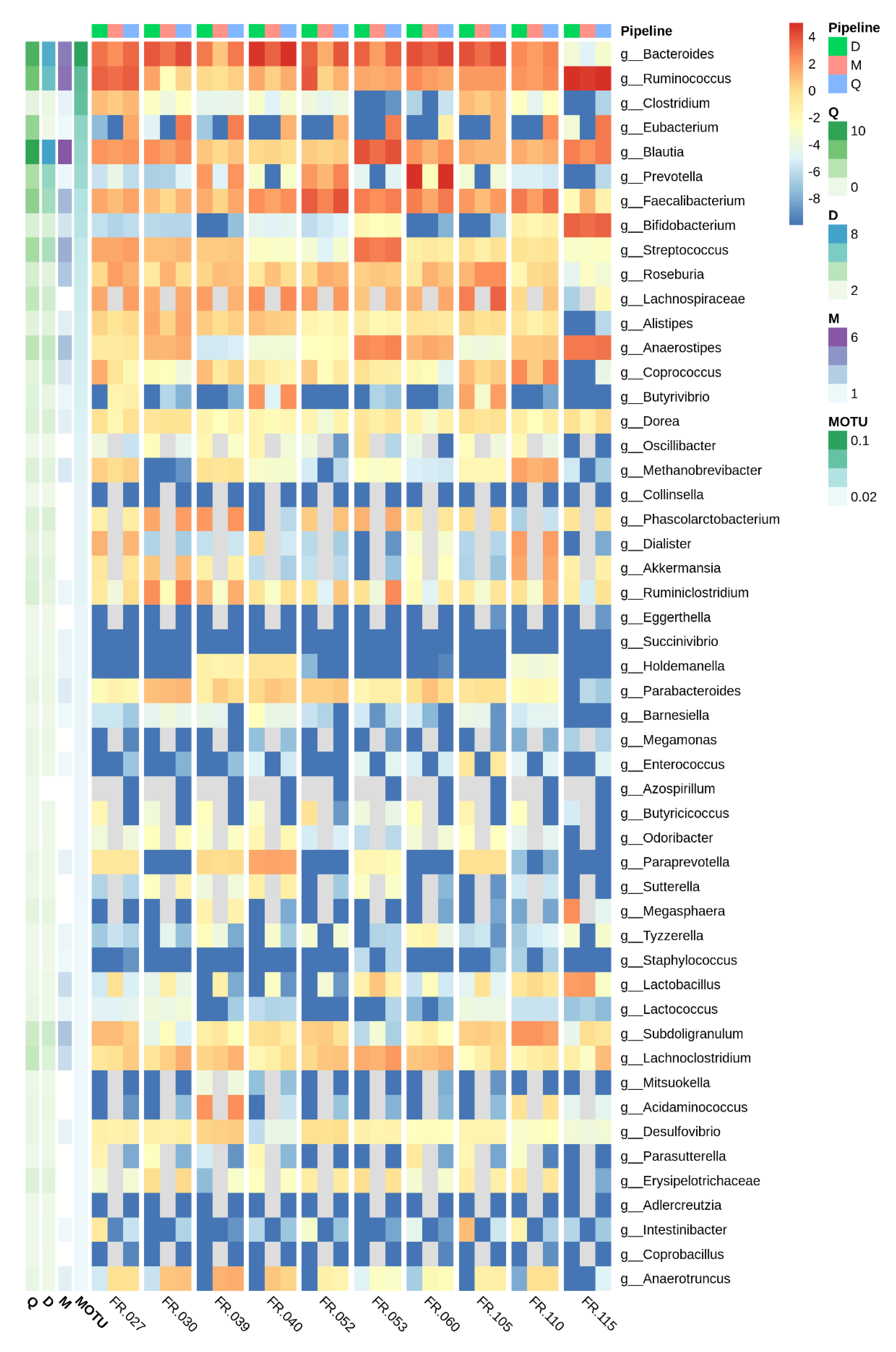
3.2. 16S rRNA and Taxonomical Assignments
3.3. 16S rRNA Taxonomic Assignments
3.3.1. Low Abundance Reads
3.3.2. Abundant Reads
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, A.C.; Hoffmann, C.; Mota, J.F. The human gut microbiota: Metabolism and perspective in obesity. Gut Microbes 2018, 9, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallianou, N.G.; Stratigou, T.; Tsagarakis, S. Microbiome and diabetes: Where are we now? Diabetes Res. Clin. Pract. 2018, 146, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Dos Reis, S.A.; Da Conceição, L.L.; Peluzio, M.D.C.G. Intestinal microbiota and colorectal cancer: Changes in the intestinal microenvironment and their relation to the disease. J. Med. Microbiol. 2019, 68, 1391–1407. [Google Scholar] [CrossRef]
- Sobhani, I.; Bergsten, E.; Couffin, S.; Amiot, A.; Nebbad, B.; Barau, C.; De’Angelis, N.; Rabot, S.; Canoui-Poitrine, F.; Mestivier, D.; et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc. Natl. Acad. Sci. USA 2019, 116, 24285–24295. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [Green Version]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N.; et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef]
- Yu, J.; Feng, Q.; Wong, S.H.; Zhang, D.; Liang, Q.Y.; Qin, Y.; Tang, L.; Zhao, H.; Stenvang, J.; Li, Y.; et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017, 66, 70–78. [Google Scholar] [CrossRef]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogtmann, E.; Hua, X.; Zeller, G.; Sunagawa, S.; Voigt, A.Y.; Hercog, R.; Goedert, J.J.; Shi, J.; Bork, P.; Sinha, R. Colorectal Cancer and the Human Gut Microbiome: Reproducibility with Whole-Genome Shotgun Sequencing. PLoS ONE 2016, 11, e0155362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, J.P.; Edwards, D.J.; Harwich, M.D.; Rivera, M.C.; Fettweis, J.M.; Serrano, M.G.; Reris, R.A.; Sheth, N.U.; Huang, B.; Girerd, P.; et al. The truth about metagenomics: Quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015, 15, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furet, J.-P.; Firmesse, O.; Gourmelon, M.; Bridonneau, C.; Tap, J.; Mondot, S.; Dorã, J.; Corthier, G. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 2009, 68, 351–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahlroos, T.; Tynkkynen, S. Quantitative strain-specific detection ofLactobacillus rhamnosusGG in human faecal samples by real-time PCR. J. Appl. Microbiol. 2009, 106, 506–514. [Google Scholar] [CrossRef]
- Suzuki, M.T.; Taylor, L.T.; DeLong, E.F. Quantitative Analysis of Small-Subunit rRNA Genes in Mixed Microbial Populations via 5J-Nuclease Assays. Appl. Environ. Microbiol. 2000, 66, 10. [Google Scholar] [CrossRef] [Green Version]
- Manz, W.; Amann, R.; Vancanneyt, M.; Schleifer, K.-H.; Ludwig, W. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 1996, 142, 1097–1106. [Google Scholar] [CrossRef] [Green Version]
- Huijsdens, X.W.; Linskens, R.K.; Mak, M.; Meuwissen, S.G.M.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H.M. Quantification of Bacteria Adherent to Gastrointestinal Mucosa by Real-Time PCR. J. Clin. Microbiol. 2002, 40, 4423–4427. [Google Scholar] [CrossRef] [Green Version]
- Aronesty, E. Ea-Utils: Command-Line Tools for Processing Biological Sequencing Data. Available online: https://github.com/expressionanalysis/ea-utils (accessed on 26 February 2016).
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.-J.; Tappu, R. MEGAN Community Edition—Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Milanese, A.; Mende, D.R.; Paoli, L.; Salazar, G.; Ruscheweyh, H.-J.; Cuenca, M.; Hingamp, P.; Alves, R.; Costea, P.I.; Coelho, L.P.; et al. Microbial abundance, activity and population genomic profiling with mOTUs. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Spakowicz, D.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volant, S.; Lechat, P.; Woringer, P.; Motreff, L.; Campagne, P.; Malabat, C.; Kennedy, S.; Ghozlane, A. SHAMAN: A user-friendly website for metataxonomic analysis from raw reads to statistical analysis. BMC Bioinform. 2020, 21, 345. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [Green Version]
- Quereda, J.J.; Dussurget, O.; Nahori, M.-A.; Ghozlane, A.; Volant, S.; Dillies, M.-A.; Regnault, B.; Kennedy, S.; Mondot, S.; Villoing, B.; et al. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. USA 2016, 113, 5706–5711. [Google Scholar] [CrossRef] [Green Version]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Van Nhieu, J.T.; Furet, J.-P. Microbial Dysbiosis in Colorectal Cancer (CRC) Patients. PLoS ONE 2011, 6, e16393. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.Y.; Song, E.-J.; Kim, S.H.; Lee, J.; Nam, Y.-D. Comparison of DNA extraction methods for human gut microbial community profiling. Syst. Appl. Microbiol. 2018, 41, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, L.M.; Castro, J.C.; Kyrpides, N.C.; Cole, J.R.; Tiedje, J.M.; Konstantinidis, K.T. How Much Do rRNA Gene Surveys Underestimate Extant Bacterial Diversity? Appl. Environ. Microbiol. 2018, 84, e00014-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 2018, 34, 2371–2375. [Google Scholar] [CrossRef] [PubMed]
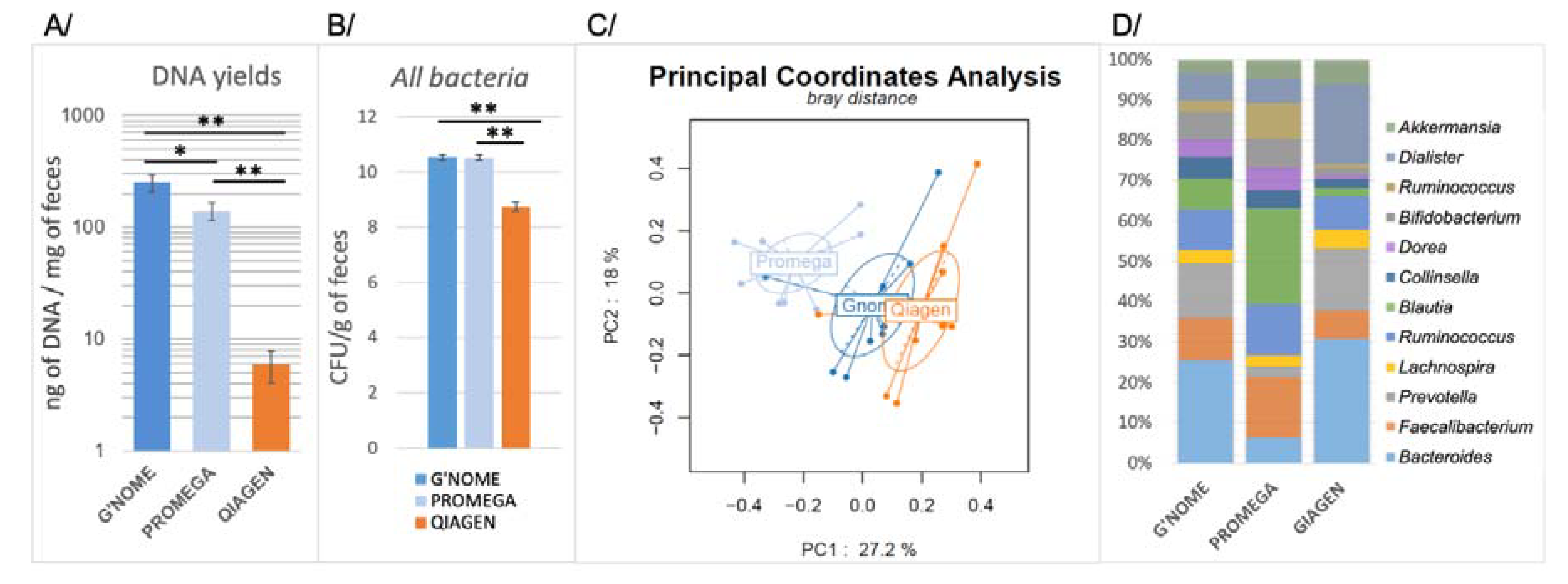

| DNA Extraction Method | Kits and References | Company | Lysis Procedure | Handling Time | DNA Yields (ng/mg) | DNA purity (A260/A280) |
|---|---|---|---|---|---|---|
| G’NOME | G’NOME DNA isolation kit® (#112010600) | MP Biomedicals Santa Ana, CA, USA | BB, CLB, T | 24h | 252.01 ± 44.67 | 1.74 ± 0.02 |
| PROMEGA | Wizard Genomic DNA purification kit® (#A1120) | Promega Madison, WI, USA | L, M, CLB, T | 7h | 139.39 ± 24.65 | 1.69 ± 0.04 |
| QIAGEN | QIAamp DNA Stool Mini Kit® (#12830) | Qiagen Hilden, Germany | CLB, T | 1h | 5.93 ± 1.83 | 2.20 ± 0.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bergsten, E.; Mestivier, D.; Sobhani, I. The Limits and Avoidance of Biases in Metagenomic Analyses of Human Fecal Microbiota. Microorganisms 2020, 8, 1954. https://doi.org/10.3390/microorganisms8121954
Bergsten E, Mestivier D, Sobhani I. The Limits and Avoidance of Biases in Metagenomic Analyses of Human Fecal Microbiota. Microorganisms. 2020; 8(12):1954. https://doi.org/10.3390/microorganisms8121954
Chicago/Turabian StyleBergsten, Emma, Denis Mestivier, and Iradj Sobhani. 2020. "The Limits and Avoidance of Biases in Metagenomic Analyses of Human Fecal Microbiota" Microorganisms 8, no. 12: 1954. https://doi.org/10.3390/microorganisms8121954
APA StyleBergsten, E., Mestivier, D., & Sobhani, I. (2020). The Limits and Avoidance of Biases in Metagenomic Analyses of Human Fecal Microbiota. Microorganisms, 8(12), 1954. https://doi.org/10.3390/microorganisms8121954





