Morphology, Ultrastructure, and Mitochondrial Genome of the Marine Non-Photosynthetic Bicosoecid Cafileria marina Gen. et sp. nov.
Abstract
1. Introduction
2. Material and Methods
2.1. Culture Conditions
2.2. Light and Scanning Electron Microscopy
2.3. Transmission Electron Microscopy
2.4. Confocal Microscopy
2.5. DNA Isolation and rDNA PCR
2.6. Mitochondrial Genome Assembly and Analysis
2.7. Phylogenetic Analyses
3. Results
3.1. Cultivation
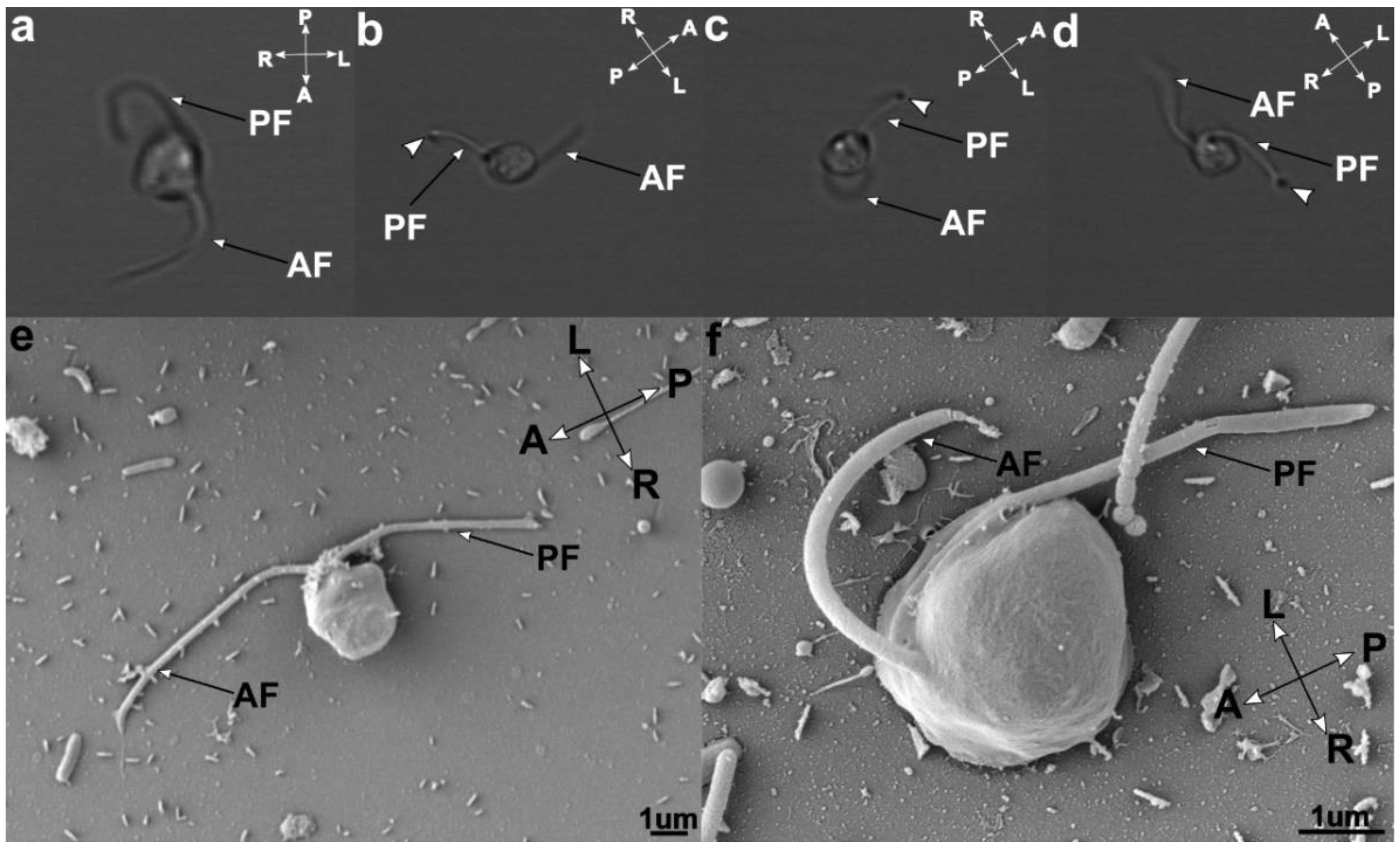
3.2. Cell Morphology
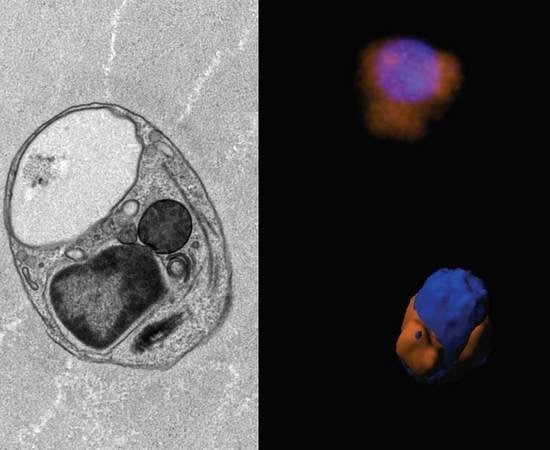
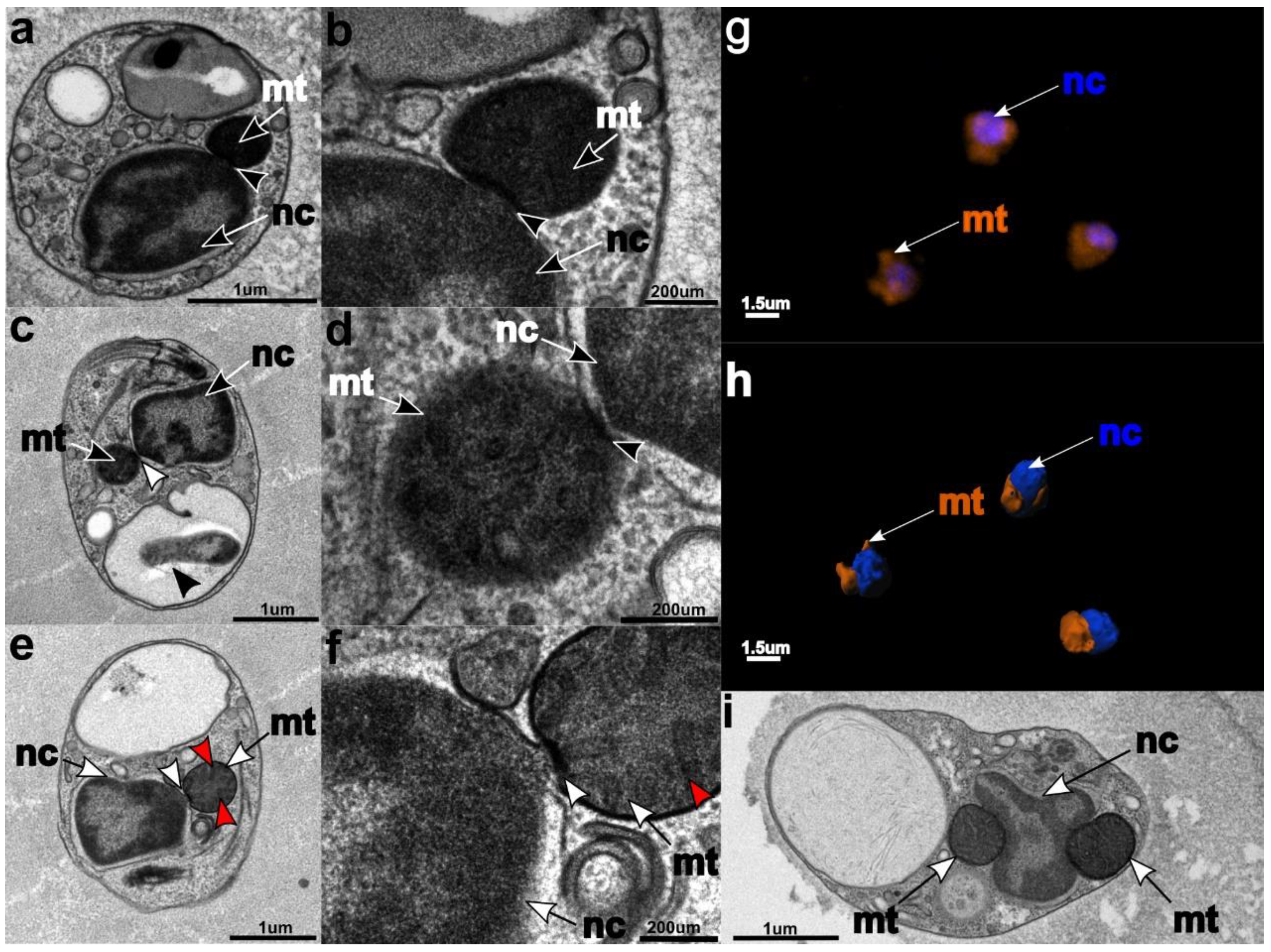
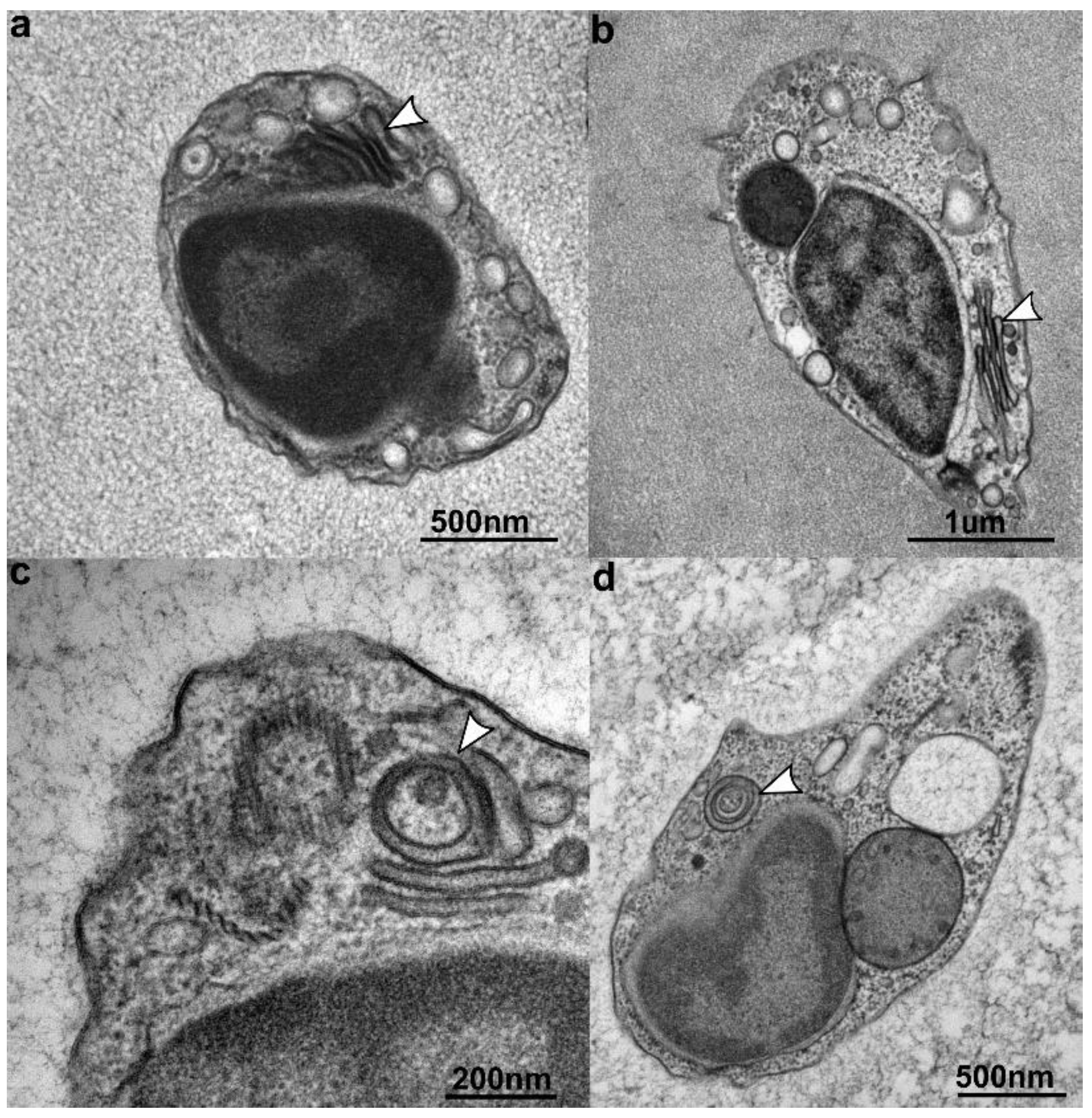
3.3. Ultrastructure
3.4. Flagellar Apparatus
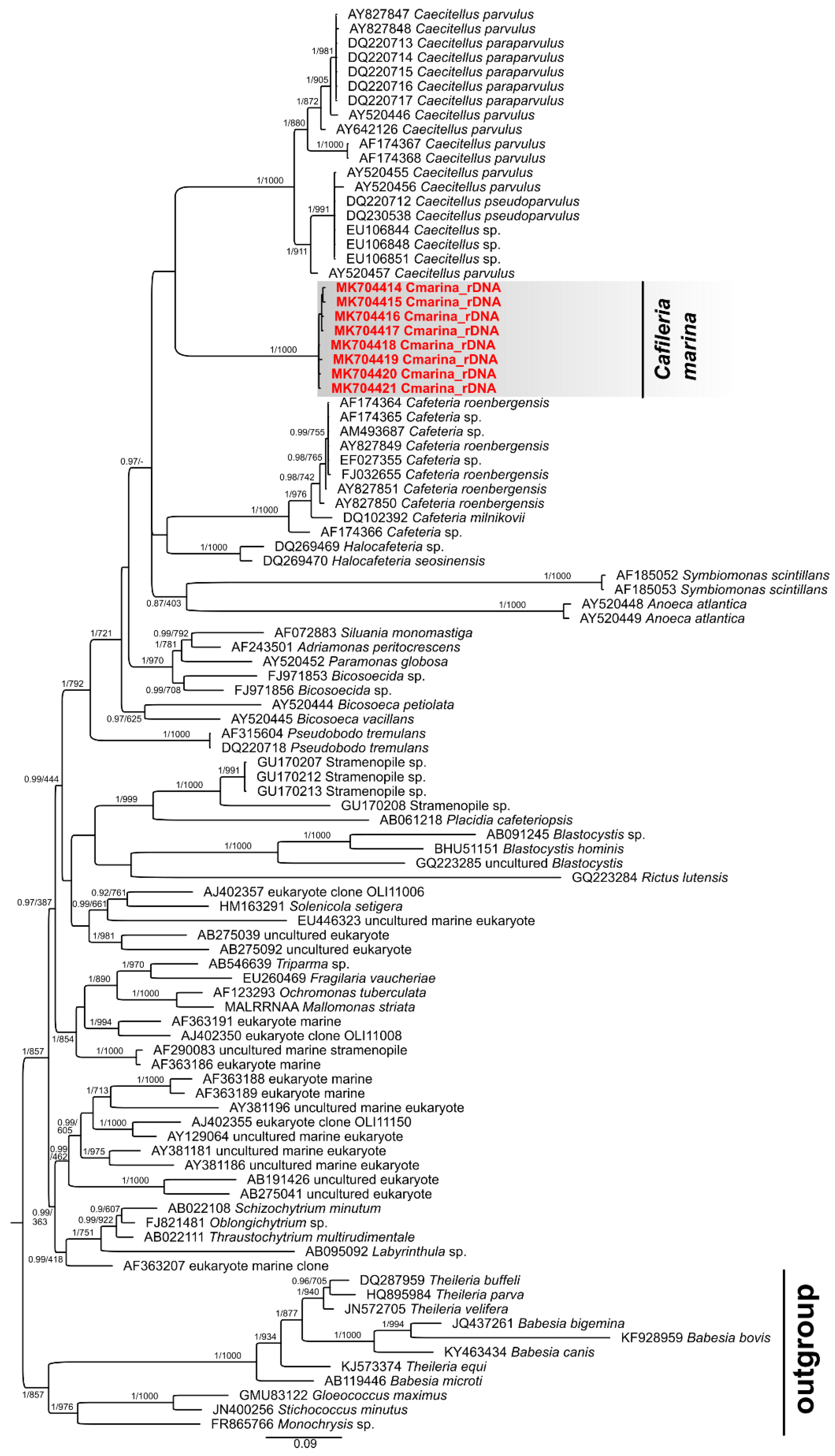
3.5. Phylogenetic Analysis
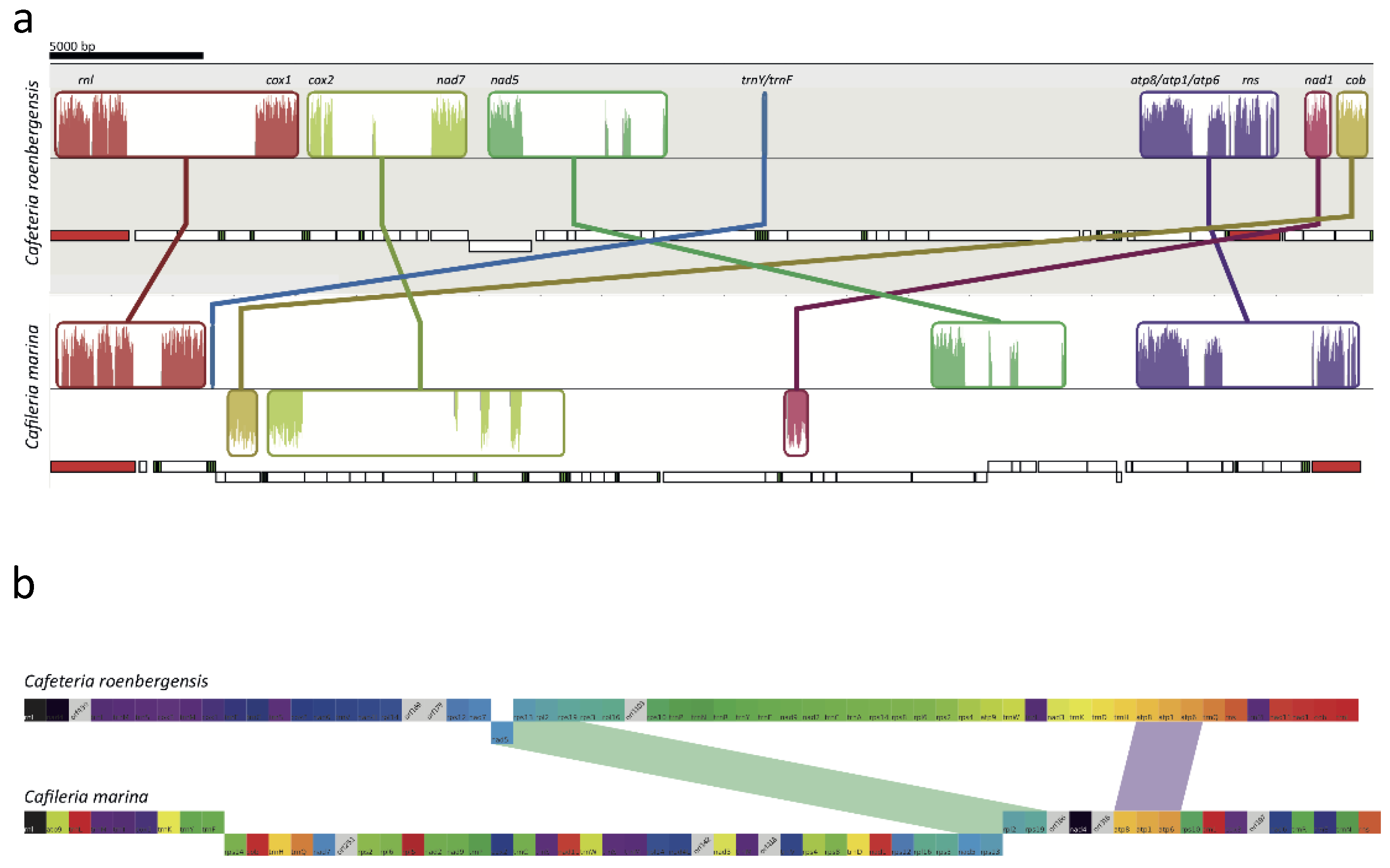
3.6. Mitochondrial Genome
4. Discussion
4.1. Taxonomic Summary
4.2. Cafileria n. gen.
4.3. Type species
4.4. Type locality
4.5. Cafileria marina n. sp.
4.5.1. Description
4.5.2. Etymology
4.5.3. Hapantotype
4.5.4. Assignation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nakano, S.I.; Ishii, N.; Manage, P.M.; Kawabata, Z. Trophic roles of heterotrophic nanoflagellates and ciliates among planktonic organisms in a hypereutrophic pond. Aquat. Microb. Ecol. 1998, 16, 153–161. [Google Scholar] [CrossRef]
- Sanders, R.W.; Porter, K.G.; Bennett, S.J.; Debiase, A.E. Seasonal patterns by flagellates, ciliates, rotifers, and cladocerans in a freshwater community cladocerans planktonic and. Limnol. Oceanogr. 1989, 34, 673–687. [Google Scholar] [CrossRef]
- Kopylov, A.I.; Kosolapov, D.B.; Romanenko, A.V.; Degermendzhy, A.G. Structure of planktonic microbial food web in a brackish stratified Siberian lake. Aquat. Ecol. 2002, 36, 179–204. [Google Scholar] [CrossRef]
- Saccà, A.; Borrego, C.M.; Renda, R.; Triadó-Margarit, X.; Bruni, V.; Guglielmo, L. Predation impact of ciliated and flagellated protozoa during a summer bloom of brown sulfur bacteria in a meromictic coastal lake. FEMS Microbiol. Ecol. 2009, 70, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Yubuki, N.; Leander, B.S.; Silberman, J.D. Ultrastructure and molecular phylogenetic position of a novel phagotrophic stramenopile from low oxygen environments: Rictus lutensis gen. et sp. nov. (Bicosoecida, incertae sedis). Protist 2010, 161, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Scheckenbach, F.; Wylezich, C.; Weitere, M.; Hausmann, K.; Arndt, H. Molecular identity of strains of heterotrophic flagellates isolated from surface waters and deep-sea sediments of the South Atlantic based on SSU rDNA. Aquat. Microb. Ecol. 2005, 38, 239–247. [Google Scholar] [CrossRef]
- Park, J.S.; Cho, B.C.; Simpson, A.G.B. Halocafeteria seosinensis gen. et sp. nov. (Bicosoecida), a halophilic bacterivorous nanoflagellate isolated from a solar saltern. Extremophiles 2006, 10, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.I. Mixotrophy in planktonic protists: An overview. Freshw. Biol. 2000, 45, 219–226. [Google Scholar] [CrossRef]
- Massana, R.; Terrado, R.; Forn, I.; Lovejoy, C.; Pedrós-Alió, C. Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ. Microbiol. 2006, 8, 1515–1522. [Google Scholar] [CrossRef]
- Moriya, M.; Nakayama, T.; Inouye, I. Ultrastructure and 18S rDNA sequence analysis of Wobblia lunata gen. et sp. nov., a new heterotrophic flagellate (Stramenopiles, Incertae sedis). Protist 2000, 151, 41–55. [Google Scholar] [CrossRef]
- Sekiguchi, H.; Moriya, M.; Nakayama, T.; Inouye, I. Vestigial chloroplasts in heterotrophic stramenopiles Pteridomonas danica and Ciliophrys infusionum (Dictyochophyceae). Protist 2002, 153, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.A. Biology and systematics of heterokont and haptophyte algae. Am. J. Bot. 2004, 91, 1508–1522. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T.; Chao, E.E.-Y. Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista). J. Mol. Evol. 2006, 62, 388–420. [Google Scholar] [CrossRef] [PubMed]
- Riisberg, I.; Orr, R.J.S.; Kluge, R.; Shalchian-Tabrizi, K.; Bowers, H.A.; Patil, V.; Edvardsen, B.; Jakobsen, K.S. Seven Gene Phylogeny of Heterokonts. Protist 2009, 160, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Kamikawa, R.; Moog, D.; Zauner, S.; Tanifuji, G.; Ishida, K.I.; Miyashita, H.; Mayama, S.; Hashimoto, T.; Maier, U.G.; Archibald, J.M.; et al. A non-photosynthetic diatom reveals early steps of reductive evolution in plastids. Mol. Biol. Evol. 2017, 34, 2355–2366. [Google Scholar] [CrossRef] [PubMed]
- Kamikawa, R.; Azuma, T.; Ishii, K.I.; Matsuno, Y.; Miyashita, H. Diversity of organellar genomes in non-photosynthetic diatoms. Protist 2018, 169, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Olefeld, J.L.; Majda, S.; Albach, D.C.; Marks, S.; Boenigk, J. Genome size of chrysophytes varies with cell size and nutritional mode. Org. Divers Evol. 2018, 18, 163–173. [Google Scholar] [CrossRef]
- Rottberger, J.; Gruber, A.; Boenigk, J.; Kroth, P.G. Influence of nutrients and light on autotrophic, mixotrophic and heterotrophic freshwater chrysophytes. Aquat. Microb. Ecol. 2013, 71, 179–191. [Google Scholar] [CrossRef]
- Dorrell, R.G.; Azuma, T.; Nomura, M.; Audren de Kerdrel, G.; Paoli, L.; Yang, S.; Bowler, C.; Ishii, K.-I.; Miyashita, H.; Gile, G.H.; et al. Principles of plastid reductive evolution illuminated by nonphotosynthetic chrysophytes. Proc. Natl. Acad. Sci. USA 2019, 16, 6914–6923. [Google Scholar] [CrossRef] [PubMed]
- Ševčíková, T.; Horák, A.; Klimeš, V.; Zbránková, V.; Demir-Hilton, E.; Sudek, S.; Jenkins, J.; Schmutz, J.; Přibyl, P.; Fousek, J.; et al. Updating algal evolutionary relationships through plastid genome sequencing: Did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci. Rep. 2015, 5, 10134. [Google Scholar] [CrossRef] [PubMed]
- Archibald, J.M. Genomic perspectives on the birth and spread of plastids. Proc. Natl. Acad. Sci. USA 2015, 112, 10147–10153. [Google Scholar] [CrossRef] [PubMed]
- Bodył, A. Did some red alga-derived plastids evolve via kleptoplastidy? A hypothesis. Biol. Rev. 2018, 93, 201–222. [Google Scholar] [CrossRef] [PubMed]
- Burki, F. The Convoluted Evolution of Eukaryotes With Complex Plastids. In Advances in Botanical Research, 1st ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2017; pp. 1–30. [Google Scholar]
- Oborník, M. The Birth of Red Complex Plastids: One, Three, or Four Times? Trends Parasitol. 2018, 34, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Baurain, D.; Brinkmann, H.; Petersen, J.; Rodríguez-Ezpeleta, N.; Stechmann, A.; Demoulin, V.; Roger, A.J.; Burger, G.; Lang, B.F.; Philippe, H. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol. Biol. Evol. 2010, 27, 1698–1709. [Google Scholar] [CrossRef]
- Derelle, R.; López-garcía, P.; Timpano, H.; Moreira, D. A phylogenomic framework to study the diversity and evolution of stramenopiles (=heterokonts). Mol. Biol. Evol. 2016, 33, 2890–2898. [Google Scholar] [CrossRef]
- Bouwmeester, K.; Van Poppel, P.M.J.A.; Govers, F. Genome Biology Cracks Enigmas of Oomycete Plant Pathogens. Annu. Plant Rev. Online 2009, 34, 102–133. [Google Scholar]
- Del Campo, J.; Sieracki, M.E.; Molestina, R.; Keeling, P.; Massana, R.; Ruiz-Trillo, I. The others: Our biased perspective of eukaryotic genomes. Trends Ecol. Evol. 2014, 29, 252–259. [Google Scholar] [CrossRef]
- Del Campo, J.; Massana, R. Emerging diversity within chrysophytes, choanoflagellates and bicosoecids based on molecular surveys. Protist 2011, 162, 435–448. [Google Scholar] [CrossRef]
- Shiratori, T.; Thakur, R.; Ishida, K.-I. Pseudophyllomitus vesiculosus (Larsen and Patterson 1990) Lee, 2002, a poorly studied phagotrophic biflagellate is the first characterized member of Stramenopile environmental clade MAST-6. Protist 2017, 168, 439–451. [Google Scholar] [CrossRef]
- Aleoshin, V.V.; Mylnikov, A.P.; Mirzaeva, G.S.; Mikhailov, K.V.; Karpov, S.A. Heterokont predator Develorapax marinus gen. et sp. nov.—A model of the ochrophyte ancestor. Front. Microbiol. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Seeleuthner, Y.; Mondy, S.; Lombard, V.; Carradec, Q.; Pelletier, E.; Wessner, M.; Leconte, J.; Mangot, J.-F.; Poulain, J.; Labadie, K.; et al. Single-cell genomics of multiple uncultured stramenopiles reveals underestimated functional diversity across oceans. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Moestrup, Ø. Current status of chrysophyte ‘splinter groups’: Synurophytes, pedinellids, silicoflagellates. In Chrysophyte algae: Ecology, Phylogeny, Development; Sandgren, C., Smol, J.P., Kristiansen, J., Eds.; Cambridge University Press: Cambridge, UK, 1995; pp. 75–91. [Google Scholar]
- Preisig, H.R. A modern concept of chrysophyte classification. In Chrysophyte algae: Ecology, Phylogeny, Development; Sandgren, C., Smol, J.P., Kristiansen, J., Eds.; Cambridge University Press: Cambridge, UK, 1995; pp. 47–74. [Google Scholar]
- O’Kelly, C.J.; Patterson, D.J. The flagellar apparatus of Cafeteria roenbergensis Fenchel & Patterson, 1988 (Bicosoecales = Bicosoecida). Eur. J. Protistol. 1996, 32, 216–226. [Google Scholar]
- Karpov, S.A.; Kersanach, R.; Williams, D.M. Ultrastructure and 18S rRNA gene sequence of a small heterotrophic flagellate Siluania monomastiga gen. et sp. nov. (Bicosoecida). Eur. J. Protistol. 1998, 34, 415–425. [Google Scholar] [CrossRef]
- Harder, C.B.; Ekelund, F.; Karpov, S.A. Ultrastructure and Phylogenetic Position of Regin rotiferus and Otto terricolus Genera et Species Novae (Bicosoecida, Heterokonta/Stramenopiles). Protist 2014, 165, 144–160. [Google Scholar] [CrossRef] [PubMed]
- Guillard, R.R.L.; Ryther, J.H. Studies on marine planktonic diatoms I. Cyclotella nana (Hustedt) and Detonula confervacea (Cleve) Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef]
- Guillard, R.R.L. Culture of phytoplankton for feeding marine invertebrates. In Culture of Marine Invertebrate Animals; Smith, W.L., Chanley, M.H., Eds.; Plenum Press: New York, NY, USA, 1975; pp. 29–60. [Google Scholar]
- Moore, R.B.; Oborník, M.; Janouškovec, J.; Chrudimský, T.; Vancová, M.; Green, D.H.; Wright, S.W.; Davies, N.M.; Bolch, C.J.S.; Heimann, K.; et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature 2008, 451, 959–963. [Google Scholar] [CrossRef]
- Oborník, M.; Vancová, M.; Lai, D.H.; Janouškovec, J.; Keeling, P.J.; Lukeš, J. Morphology and ultrastructure of multiple life cycle stages of the photosynthetic relative of apicomplexa, Chromera velia. Protist 2011, 162, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Laetsch, D.R.; Blaxter, M.L. BlobTools: Interrogation of genome assemblies. F1000Research 2017, 1287, 1–16. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Peabody, D.S. Translation Initiation at Non-AUG Triplets in Mammalian Cells. J. Biol. Chem. 1969, 264, 5031–5035. [Google Scholar]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.I.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J. Tracer V1.5. Available online: http//beast.bio.ed.ac.uk/Tracer 2009 (accessed on 5 August 2019).
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4.3. Mol. Evol. Phylogenetics Epidemiol. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 5 August 2019).
- Campelo, F.; van Galen, J.; Turacchio, G.; Parashuraman, S.; Kozlov, M.M.; García-Parajo, M.F.; Malhotra, V. Sphingomyelin metabolism controls the shape and function of the golgi cisternae. eLife 2017, 6, e24603. [Google Scholar] [CrossRef]
- Tachikawa, M.; Mochizuki, A. Golgi apparatus self-organizes into the characteristic shape via postmitotic reassembly dynamics. Proc. Natl. Acad. Sci. USA 2017, 114, 5177–5182. [Google Scholar] [CrossRef]
- Gruber, A.; Rocap, G.; Kroth, P.G.; Armbrust, E.V.; Mock, T. Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 2015, 81, 519–528. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Nielsen, H.; Von Heijne, G.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Ševčíková, T.; Klimeš, V.; Zbránková, V.; Strnad, H.; Hroudová, M.; Vlček, Č.; Eliáš, M. A comparative analysis of mitochondrial genomes in eustigmatophyte algae. Genome Biol. Evol. 2016, 8, 705–722. [Google Scholar] [CrossRef]
- Griessmann, K. Uber marine Flagellaten. Arch. Protistenk 1913, 32, 1–78. [Google Scholar]
- Larsen, J.; Patterson, D.J. Some flagellates (Protista) from tropical marine sediments. J. Nat. Hist. 1990, 24, 801–937. [Google Scholar] [CrossRef]
- O’Kelly, C.J.; Nerad, T.A. Kinetid architecture and bicosoecid affinities of the marine heterotrophic nanoflagellate Caecitellus parvulus (Griessmann, 1913) Patterson et al., 1993. Eur. J. Protistol. 1998, 34, 369–375. [Google Scholar]
- Andersen, R.A. Synurophyceae classis nov., a new class of algae. Am. J. Bot. 1987, 74, 337–353. [Google Scholar] [CrossRef]
- Andersen, R.A. The cytoskeleton of chromophyte algae. Protoplasma 1991, 164, 143–159. [Google Scholar] [CrossRef]
- Dzeja, P.P.; Bortolon, R.; Perez-Terzic, C.; Holmuhamedov, E.L.; Terzic, A. Energetic communication between mitochondria and nucleus directed by catalyzed phosphotransfer. Proc. Natl. Acad. Sci. USA 2002, 99, 10156–10161. [Google Scholar] [CrossRef]
- Al-Mehdi, A.-B.; Pastukh, V.M.; Swiger, B.M.; Reed, D.J.; Patel, M.R.; Bardwell, G.C.; Pastukh, V.V.; Alexeyev, M.F.; Gillespie, M.N. Perinuclear Mitochondrial Clustering Creates an Oxidant-Rich Nuclear Domain Required for Hypoxia-Induced Transcription. Sci. Signal. 2012, 5, 1–20. [Google Scholar] [CrossRef]
- Picard, M. Mitochondrial synapses: Intracellular communication and signal integration. Trends Neurosci. 2015, 38, 468–474. [Google Scholar] [CrossRef]
- Prachař, J. Intimate contacts of mitochondria with nuclear envelope as a potential energy gateway for nucleo-cytoplasmic mRNA transport. Gen. Physiol. Biophys. 2003, 22, 525–534. [Google Scholar]
- Delprat, B.; Rieusset, J.; Delettre, C. Defective Endoplasmic Reticulum–Mitochondria Connection Is a Hallmark of Wolfram Syndrome. Contact 2019, 2, 251525641984740. [Google Scholar] [CrossRef]
- Rowland, A.A.; Voeltz, G.K. Endoplasmic reticulum-mitochondria contacts: Function of the junction. Nat. Rev. Mol. Cell Biol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- Elbaz, Y.; Schuldiner, M. Staying in touch: The molecular era of organelle contact sites. Trends Biochem. Sci. 2011, 36, 616–623. [Google Scholar] [CrossRef]
- Friedman, J.R.; Lackner, L.L.; West, M.; DiBenedetto, J.R.; Nunnari, J.; Voeltz, G.K. ER tubules mark sites of mitochondrial division. Science 2011, 334, 358–362. [Google Scholar] [CrossRef]
- Salazar-Roa, M.; Malumbres, M. Fueling the Cell Division Cycle. Trends Cell Biol. 2017, 27, 69–81. [Google Scholar] [CrossRef]
- Hancock, K.; Jahduk, S.L. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J. Biol. Chem. 1990, 265, 19208–19215. [Google Scholar]
- Tan, T.H.P.; Pach, R.; Crausaz, A.; Ivens, A.; Schneider, A. tRNAs in Trypanosoma brucei: Genomic organization, expression, and mitochondrial import. Mol. Cell. Biol. 2002, 22, 3707–3717. [Google Scholar] [CrossRef] [PubMed]
- Zabezhinsky, D.; Slobodin, B.; Rapaport, D.; Gerst, J.E. An essential role for COPI in mRNA localization to mitochondria and mitochondrial function. Cell Rep. 2016, 15, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Michaud, M.; Maréchal-Drouard, L.; Duchêne, A.M. Targeting of cytosolic mRNA to mitochondria: Naked RNA can bind to the mitochondrial surface. Biochimie 2014, 100, 159–166. [Google Scholar] [CrossRef]
- Fogarty, N.M.E.; Ferguson-Smith, A.C.; Burton, G.J. Syncytial knots (Tenney-parker changes) in the human placenta: Evidence of loss of transcriptional activity and oxidative damage. Am. J. Pathol. 2013, 183, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Weaver, D.; Shirihai, O.; Hajnóczky, G. Mitochondrial kiss-and-run: Interplay between mitochondrial motility and fusion-fission dynamics. EMBO J. 2009, 28, 3074–3089. [Google Scholar] [CrossRef] [PubMed]
- Giorgi, C.; Missiroli, S.; Patergnani, S.; Duszynski, J.; Wieckowski, M.R.; Pinton, P. Mitochondria-Associated Membranes: Composition, Molecular Mechanisms, and Physiopathological Implications. Antioxid. Redox Signal. 2015, 22, 995–1019. [Google Scholar] [CrossRef] [PubMed]
- Campelo, F.; Arnarez, C.; Marrink, S.J.; Kozlov, M.M. Helfrich model of membrane bending: From Gibbs theory of liquid interfaces to membranes as thick anisotropic elastic layers. Adv. Colloid. Interface Sci. 2014, 208, 25–33. [Google Scholar] [CrossRef] [PubMed]
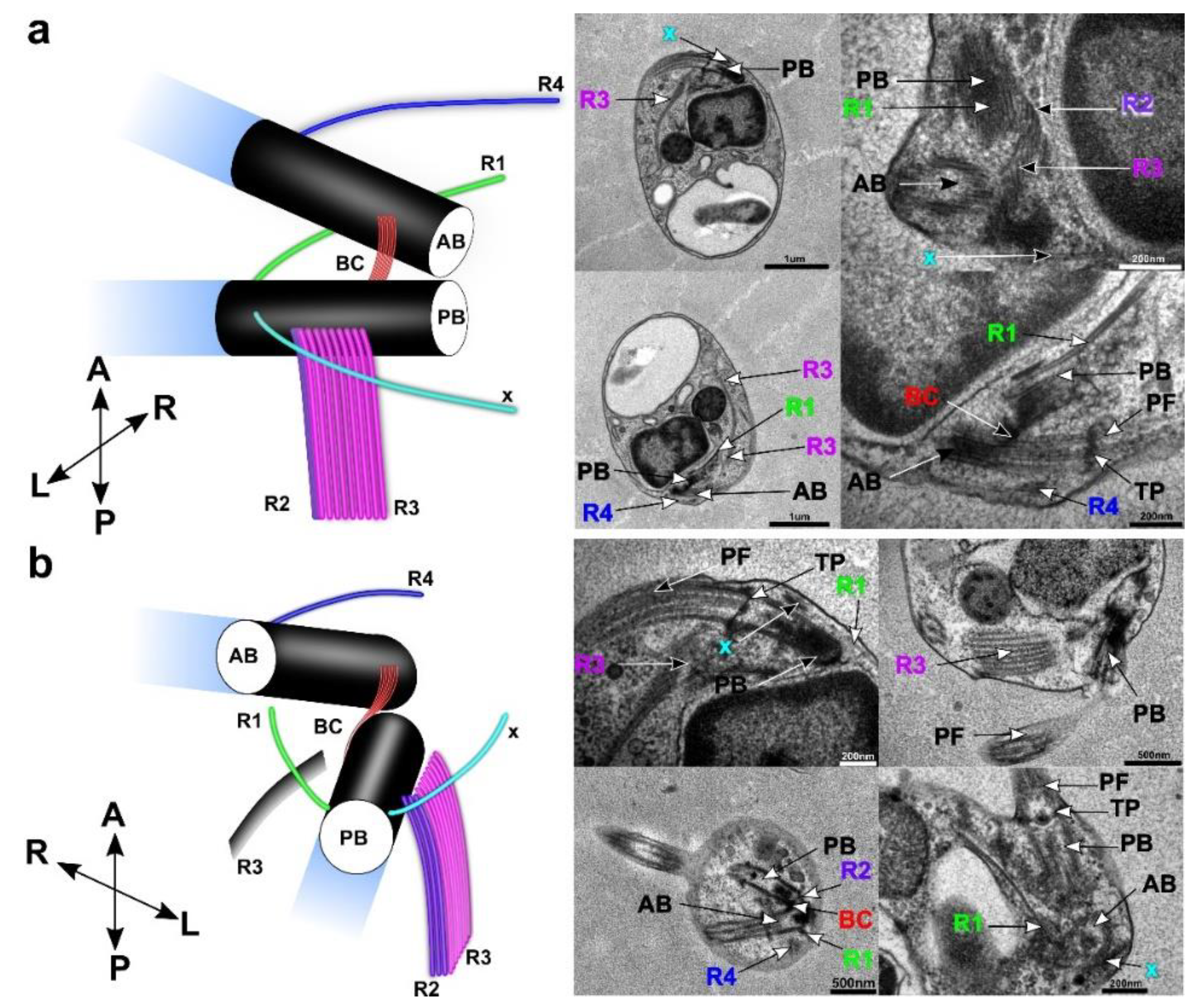
| Ultrastructure of the Flagellar Apparatus | R1 | R2 | R3 | R4 | Additional Microtubular Structures | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Present | Number of Microtubules | Original Name in the Publication | Present | Number of Microtubules | Original Name in the Publication | Present | Number of Microtubules | Original Name in the Publication | Present | Number of Microtubules | Original Name in the Publication | ||
| Species and publication | |||||||||||||
| Halocafeteria seosinensis | Not confirmed | + | 8 | Not confirmed | Not confirmed | ||||||||
| Park et al., 2006 [7] | |||||||||||||
| Cafeteria roenbergensis | + | 2 | R4 | + | 3 | abc | + | 8 | + | 2 | R1 | x | |
| O’Kelly and Patterson 1996 [35] | Secondary cytoskeletal microtubules | ||||||||||||
| Rictus lutensis | + | 2 | R4 | + | 3 | abc | + | ~4–50 | R2 | + | 2 | R3 | x |
| Yubuki et al., 2010 [5] | S | ||||||||||||
| Caecitellus parvulus | + | 2 | R1 | + | 3 | abc | + | ~8–24 | R2 | + | 2 | R1 | x |
| O’Kelly and Nerad 1998 [64] | |||||||||||||
| Cafileria marina | + | 1 | + | 3 | + | 8 | + | 1 | x | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jirsová, D.; Füssy, Z.; Richtová, J.; Gruber, A.; Oborník, M. Morphology, Ultrastructure, and Mitochondrial Genome of the Marine Non-Photosynthetic Bicosoecid Cafileria marina Gen. et sp. nov. Microorganisms 2019, 7, 240. https://doi.org/10.3390/microorganisms7080240
Jirsová D, Füssy Z, Richtová J, Gruber A, Oborník M. Morphology, Ultrastructure, and Mitochondrial Genome of the Marine Non-Photosynthetic Bicosoecid Cafileria marina Gen. et sp. nov. Microorganisms. 2019; 7(8):240. https://doi.org/10.3390/microorganisms7080240
Chicago/Turabian StyleJirsová, Dagmar, Zoltán Füssy, Jitka Richtová, Ansgar Gruber, and Miroslav Oborník. 2019. "Morphology, Ultrastructure, and Mitochondrial Genome of the Marine Non-Photosynthetic Bicosoecid Cafileria marina Gen. et sp. nov." Microorganisms 7, no. 8: 240. https://doi.org/10.3390/microorganisms7080240
APA StyleJirsová, D., Füssy, Z., Richtová, J., Gruber, A., & Oborník, M. (2019). Morphology, Ultrastructure, and Mitochondrial Genome of the Marine Non-Photosynthetic Bicosoecid Cafileria marina Gen. et sp. nov. Microorganisms, 7(8), 240. https://doi.org/10.3390/microorganisms7080240