Monitoring Biofilm Formation and Microbial Interactions that May Occur During a Salmonella Contamination Incident across the Network of a Water Bottling Plant
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and DNA Manipulations
2.2. Plasmid Constructions
2.3. Water Sample Collection and Sampling
2.4. DNA Extraction and Bacterial Identification
2.5. Evaluation of Biofilm Forming Ability
2.6. A Study of Monospecies and Multispecies-Biofilm Formation and the Virulence of Salmonella Typhimurium with Fluorescence Microscopy
2.7. Data Analysis
3. Results and Discussion
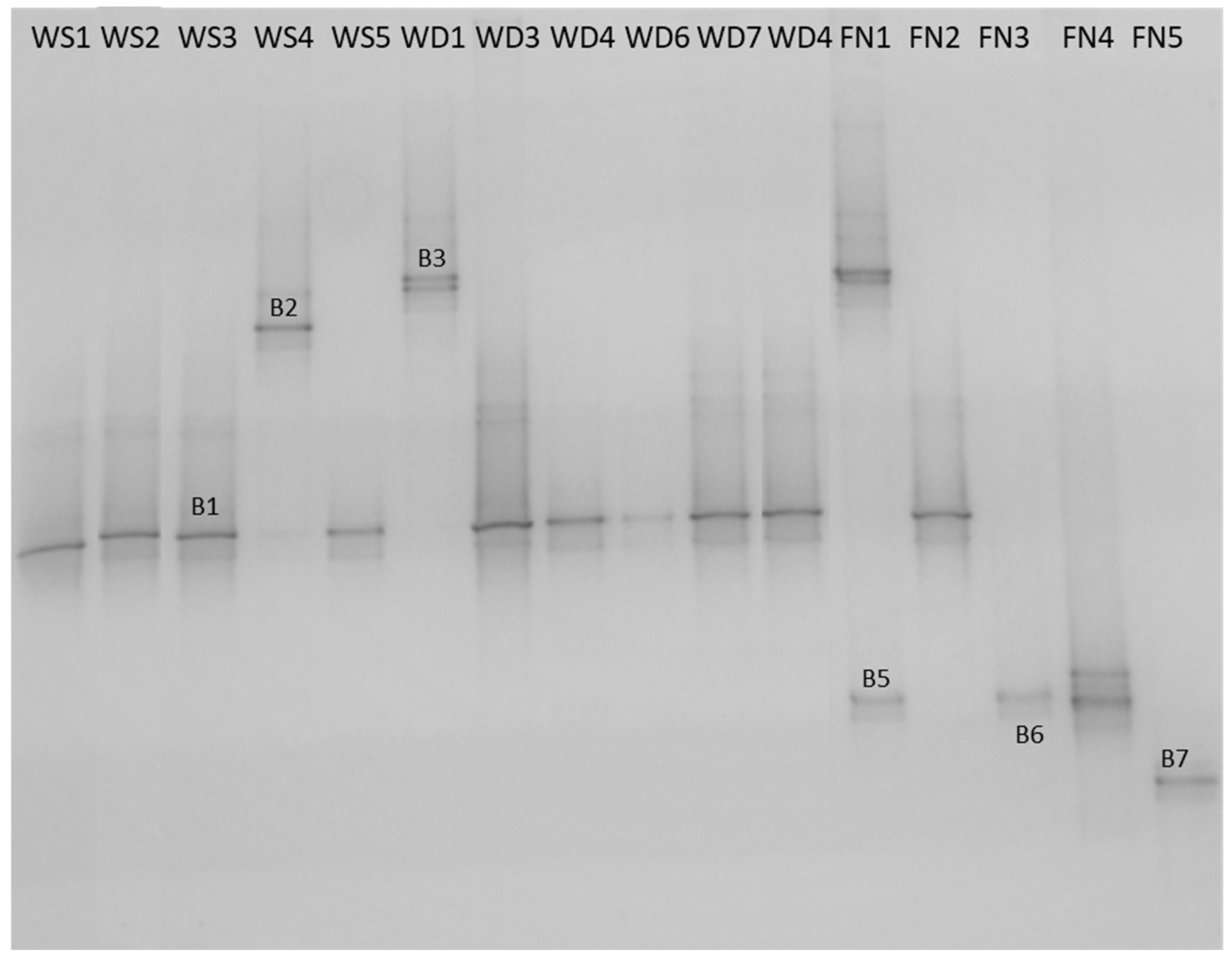
3.1. Water Sample Collection and Bacterial Identification
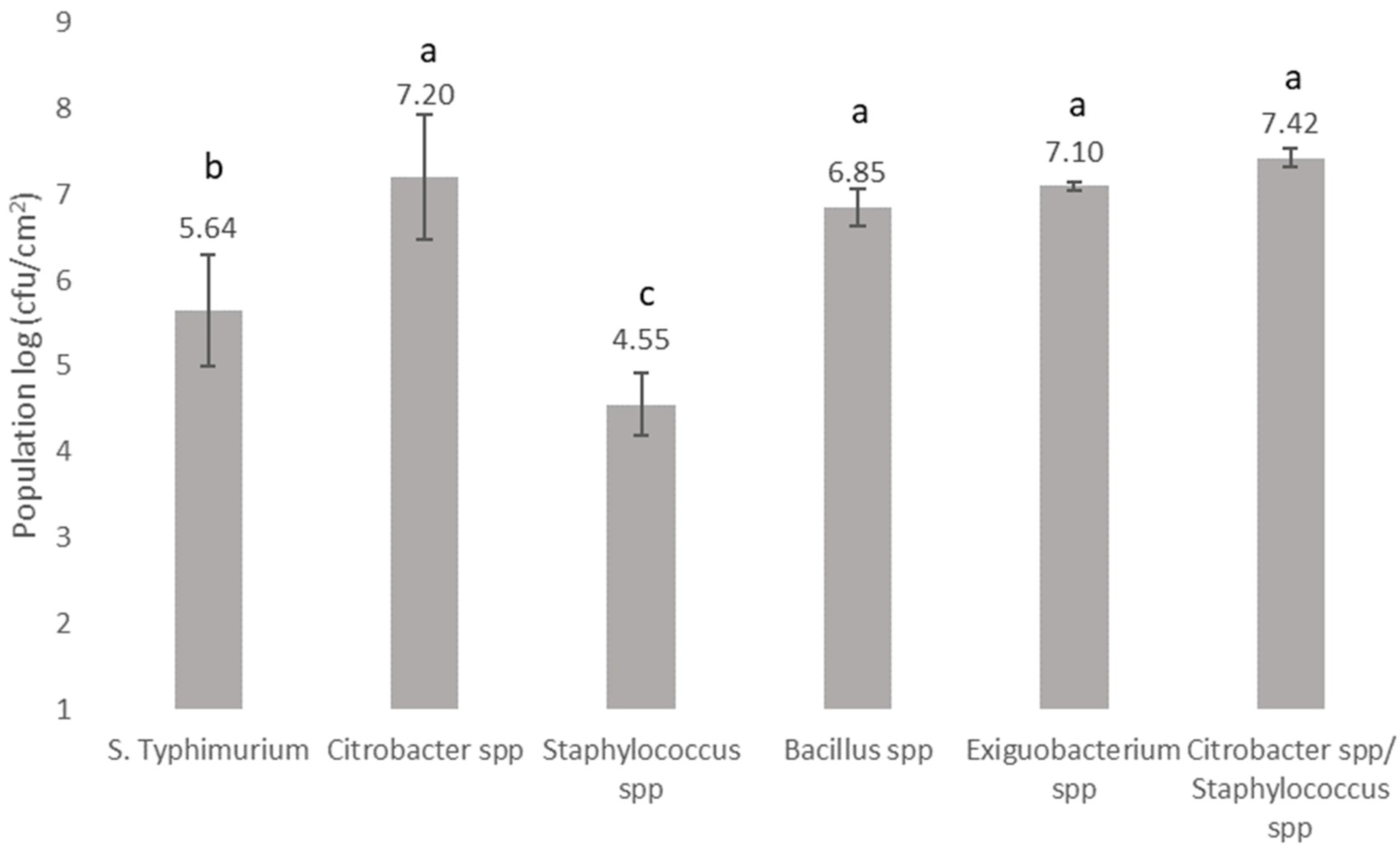
3.2. Biofilm Forming Ability and Microbial Interactions of Bacteria Recovered from the Bottling Plant and Salmonella on Stainless Steel Surfaces
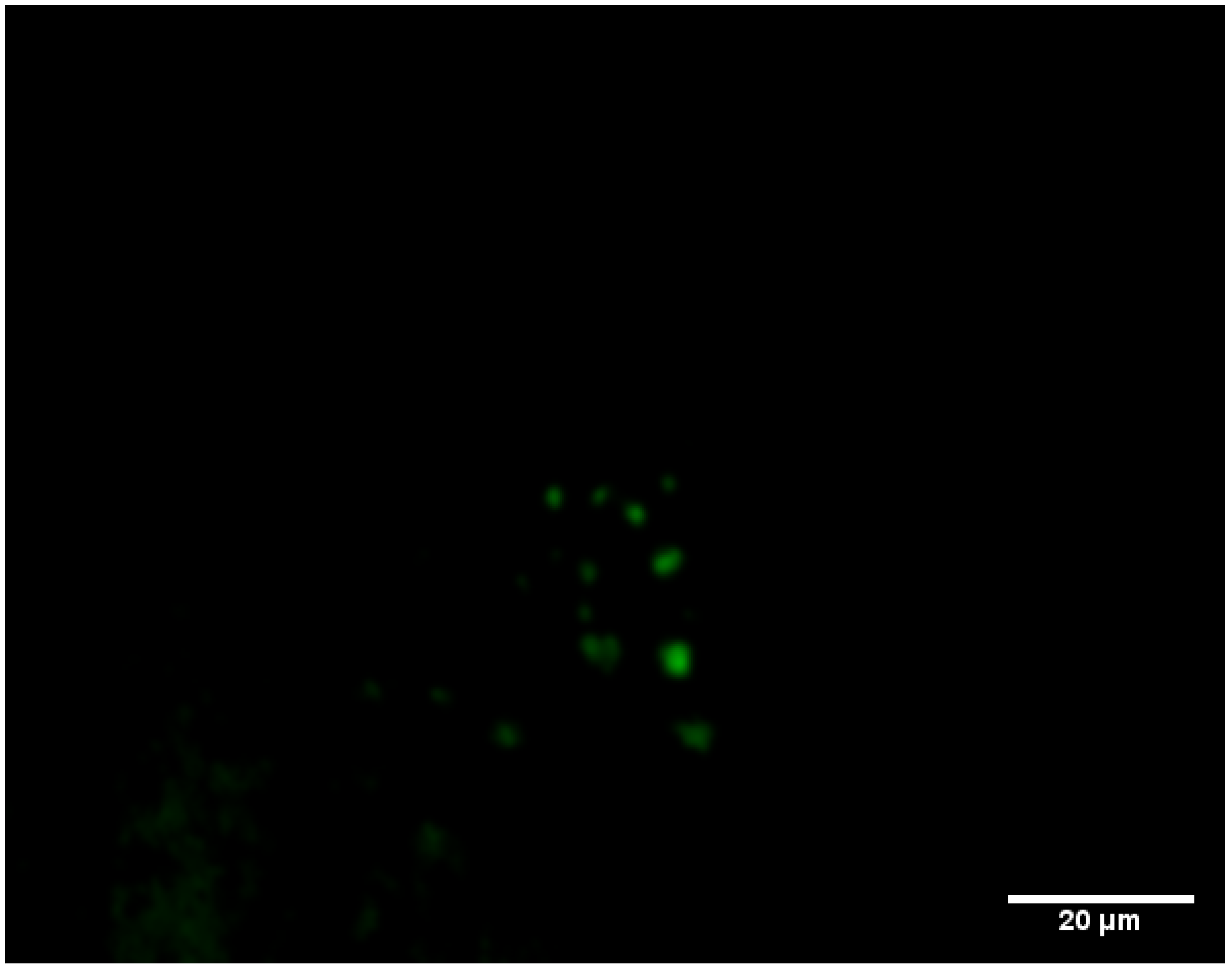
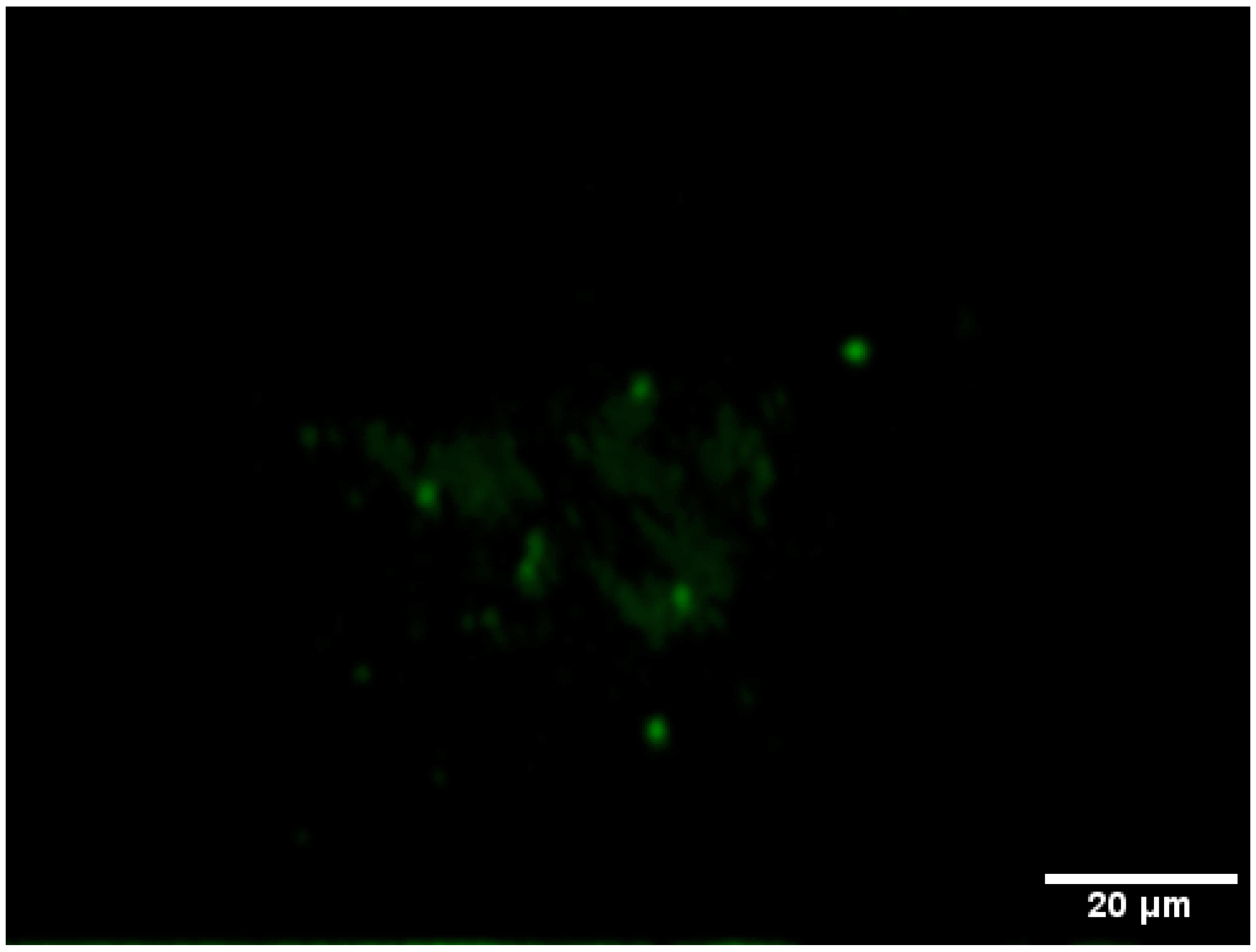
3.3. Monitoring the Biofilm Formation and Virulence of Salmonella in Monospecies and Multispecies Communities by Fluorescent-Based Bioreporters
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Quantitative Microbial Risk Assessment: Application for Water Safety Management; WHO Library Cataloguing-in-Publication Data; WHO: Geneva, Switzerland, 2016.
- Levantesi, C.; Bonadonna, L.; Briancesco, R.; Grohmann, E.; Toze, S.; Tandoi, V. Salmonella in surface and drinking water: Occurrence and water-mediated transmission. Food Res. Int. 2012, 45, 587–602. [Google Scholar] [CrossRef]
- EFSA. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 2015, 13, 3991. [Google Scholar] [CrossRef]
- White, A.P.; Surette, M.G. Comparative genetics of the rdar morphotype in Salmonella. J. Bacteriol. 2006, 188, 8395–8406. [Google Scholar] [CrossRef] [PubMed]
- Giaouris, E.D.; Nychas, G.J.E. The adherence of Salmonella Enteritidis PT4 to stainless steel: The importance of the air-liquid interface and nutrient availability. Food Microbiol. 2006, 23, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, K.; Oliveira, T.; Teixeira, P.; Azeredo, J.; Henriques, M.; Oliveira, R. Comparison of the adhesion ability of different Salmonella Enteritidis serotypes to materials used in kitchens. J. Food Protect. 2006, 69, 2352–2356. [Google Scholar] [CrossRef]
- Chia, T.W.R.; Goulter, R.M.; McMeekin, T.; Dykes, G.A.; Fegan, N. Attachment of different Salmonella serovars to materials commonly used in a poultry processing plant. Food Microbiol. 2009, 26, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Marin, C.; Hernandiz, A.; Lainez, M. Biofilm development capacity of Salmonella strains isolated in poultry risk factors and their resistance against disinfectants. Poultry Sci. 2009, 88, 424–431. [Google Scholar] [CrossRef]
- Rodrigues, D.; Teixeira, P.; Oliveira, R.; Azeredo, J. Salmonella enterica Enteritidis biofilm formation and viability on regular and triclosan-impregnated bench cover materials. J. Food Protect. 2011, 74, 32–37. [Google Scholar] [CrossRef]
- Giaouris, E.; Chorianopoulos, N.; Nychas, G.J.E. Effect of temperature, pH, and water activity on biofilm formation by Salmonella enterica Enteritidis PT4 on stainless steel surfaces as indicated by the bead vortexing method and conductance measurements. J. Food Prot. 2005, 68, 2149–2154. [Google Scholar] [CrossRef]
- Zijnge, V.; van Leeuwen, M.B.M.; Degener, J.E.; Abbas, F.; Thurnheer, T.; Gmur, R.; Harmsen, H.J.M. Oral biofilm architecture on natural teeth. PLoS ONE 2010, 5, e9321. [Google Scholar] [CrossRef]
- Hermans, K.; Nguyen, T.L.; Roberfroid, S.; Schoofs, G.; Verhoeven, T.; De Coster, D.; Vanderleyden, J.; De Keersmaecker, S.C. Gene expression analysis of monospecies Salmonella Typhimurium biofilms using differential fluorescence induction. J. Microbiol. Meth. 2011, 84, 467–478. [Google Scholar] [CrossRef]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef]
- Koutsoumanis, K.P.; Aspridou, Z. Individual cell heterogeneity in predictive Food Microbiology: Challenges in predicting a ‘noisy’ world. Int. J. Food Microb. 2016, 240, 3–10. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Fryer, R.M.; Randall, J.; Yoshida, T.; Hsiao, L.L.; Blumenstock, J.; Jensen, K.E.; Dimofte, T.; Jensen, R.V.; Gullans, S.R. Global analysis of gene expression: Methods, interpretation, and pitfalls. Exp. Nephrol. 2002, 10, 64–74. [Google Scholar] [CrossRef]
- Casassola, A.; Brammer, S.P.; Chaves, M.S.; Martinelli, J.A.; Grando, M.F.; Denardin, N.D. Gene expression: A review on methods for the study of defense-related gene differential expression in plants. Am. J. Plant Sci. 2013, 4, 64–73. [Google Scholar] [CrossRef]
- Van de Meer, J.R.; Belkin, S. Where microbiology meets microengineering: Design and applications of reporter bacteria. Nat. Rev. 2010, 8, 511–522. [Google Scholar] [CrossRef]
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; De Keersmaecker, S.C.J. Salmonella biofilms: An overview on occurrence, structure, regulation and eradication. Food Res. Int. 2012, 45, 502–531. [Google Scholar] [CrossRef]
- Bhavsar, A.P.; Brown, N.F.; Stoepel, J.; Wiermer, M.; Martin, D.D.O.; Hsu, K.J.; Imami, K.; Ross, C.J.; Hayden, M.R.; Foster, L.J.; et al. The Salmonella Type III Effector sspH2 Specifically exploits the NLR co-chaperone activity of SGT1 to subvert immunity. PLoS Pathog. 2013, 9, e1003518. [Google Scholar] [CrossRef]
- Bogomolnaya, L.M.; Aldrich, L.; Ragoza, Y.; Talamantes, M.; Andrews, K.D.; McClelland, M.; Andrews-Polymenis, H.L. Identification of novel factors involved in modulating motility of Salmonella enterica serotype Typhimurium. PLoS ONE 2014, 9, e111513. [Google Scholar] [CrossRef]
- Hallez, R.; Letesson, J.J.; Vandenhaute, J.; De Bolle, X. Gateway-based destination vectors for functional analyses of bacterial ORFeomes: Application to the Min system in Brucella abortus. Appl. Environ. Microbiol. 2007, 73, 1375–1379. [Google Scholar] [CrossRef]
- Venieri, D.; Vantarakis, A.; Komninou, G.; Papapetropoulou, M. Microbiological evaluation of bottled non-carbonated (“still) water from domestic brands in Greece. Int. J. Food Microbiol. 2005, 107, 68–72. [Google Scholar] [CrossRef]
- Riepl, M.; Schauer, S.; Knetsch, S.; Holzhammer, E.; Farnleitner, A.H.; Sommer, R.; Kirschner, A.K.T. Applicability of solid-phase cytometry and epifluorescence microscopy for rapid assessment of the microbiological quality of dialysis water. Nephrol. Dial. Transplant. 2011, 26, 3640–3645. [Google Scholar] [CrossRef]
- Ercolini, D.; Russo, F.; Torrieri, E.; Masi, P.; Villani, F. Changes in the spoilage related microbiota of beef during refrigerated storage under different packaging conditions. Appl. Environ. Microbiol. 2006, 72, 4663–4671. [Google Scholar] [CrossRef]
- Muyzer, G.; De Weal, E.C.; Uitterlinden, A. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Kostaki, M.; Chorianopoulos, N.; Braxou, E.; Nychas, G.J.E.; Giaouris, E. Differential biofilm formation and chemical disinfection resistance of sessile cells of Listeria monocytogenes strains under monospecies and dual-species (with Salmonella enterica) conditions. Appl. Environ. Microbiol. 2012, 78, 2586–2595. [Google Scholar] [CrossRef]
- Cabral, J.P. Water microbiology. Bacterial pathogens and water. Int. J. Environ. Res. Public Health 2010, 7, 3657–3703. [Google Scholar] [CrossRef]
- Fiello, M.; Mikell, J.R.; Moore, M.T.; Cooper, C.M. Variability in the characterization of total coliforms, fecal coliforms and Escherichia coli in recreational water supplies of North Mississippi, USA. Bull. Environ. Contam. Toxicol. 2014, 93, 133–137. [Google Scholar] [CrossRef]
- Pindi Pavan Kumar, P.; Raghuveer, Y.; Shiva Shanker, A. Identification of opportunistic pathogenic bacteria in drinking water samples of different rural health centers and their clinical impacts on humans. Biomed. Res. Int. 2013, 34, 8250. [Google Scholar]
- Badger, J.L.; Stins, M.F.; Kim, K.S. Citrobacter freundii invades and replicates in human brain microvascular endothelial cells. Inf. Immun. 1999, 67, 4208–4215. [Google Scholar]
- Percival, S.L.; Chalmers, R.M.; Embrey, M.; Hunter, P.R.; Sellwood, J.; WynJones, P. Microbiology of Waterborne Diseases; Elsevier Academic Press: San Diego, CA, USA, 2004. [Google Scholar]
- Jayasekara, N.Y.; Heard, G.M.; Cox, J.M.; Fleet, G.H. Populations of pseudomonads and related bacteria associated with bottled non-carbonated mineral water. Food Microbiol. 1998, 15, 167–176. [Google Scholar] [CrossRef]
- Palleroni, N.J. Pseudomonas. In Topley & Wilson’s Microbiology and Microbial Infections; Mahy, B.W.J., Ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2010. [Google Scholar]
- Kim, S.H.; Wei, C.I. Molecular characterization of biofilm formation and attachment of Salmonella enterica serovar Typhimurium DT104 on food contact surfaces. J. Food Protect. 2009, 72, 1841–1847. [Google Scholar] [CrossRef]
- Vantarakis, A.; Smaili, M.; Detorakis, I.; Vantarakis, G.; Papapetropoulou, M. Diachronic long-term surveillance of bacteriological quality of bottled water in Greece (1995–2010). Food Control 2013, 33, 63–67. [Google Scholar] [CrossRef]
- Chorianopoulos, N.G.; Giaouris, E.D.; Kourkoutas, Y.; Nychas, G.J. Inhibition of the early stage of Salmonella enterica serovar Enteritidis biofilm development on stainless steel by cell-free supernatant of a Hafnia alvei culture. Appl. Environ. Microbiol. 2010, 76, 2018–2022. [Google Scholar] [CrossRef]
- Malheiros, P.; Passos, C.; Casarin, L.; Serraglio, L.; Tondo, E. Evaluation of growth and transfer of Staphylococcus aureus from poultry meat to surfaces of stainless steel and polyethylene and their disinfection. Food Control 2010, 21, 298–301. [Google Scholar] [CrossRef]
- Michu, E.; Cervinkova, D.; Babak, V.; Kyrova, K.; Jaglic, Z. Biofilm formation on stainless steel by Staphylococcus epidermidis in milk and influence of glucose and sodium chloride on the development of ica-mediated biofilms. Int. Dairy J. 2011, 21, 179–184. [Google Scholar] [CrossRef]
- Faille, C.; Bénézech, T.; Midelet-Bourdin, G.; Lequette, Y.; Clarisse, M.; Ronse, G.; Ronse, A.; Slomianny, C. Sporulation of Bacillus spp. within biofilms: A potential source of contamination in food processing environments. Food Microbiol. 2014, 40, 64–74. [Google Scholar] [CrossRef]
- Nam, H.; Seo, H.S.; Bang, J.; Kim, H.; Beuchat, L.R.; Ryu, J.H. Efficacy of gaseous chlorine dioxide in inactivating Bacillus cereus spores attached to and in a biofilm on stainless steel. Int. J. Food Microbiol. 2014, 188, 122–127. [Google Scholar] [CrossRef]
- Gunduz, G.T.; Tuncel, G. Biofilm formation in an ice cream plant. Anton. Leeuw. 2006, 89, 329–336. [Google Scholar] [CrossRef]
- Pereira, A.L.; Silva, T.N.; Gomes, A.C.M.M.; Araújo, A.C.G.; Giugliano, L.G. Diarrhea associated biofilm formed by enteroaggregative Escherichia coli and aggregative Citrobacter freundii: A consortium mediated by putative F pili. BMC Microbiol. 2010, 10, 57. [Google Scholar] [CrossRef]
- Weiler, C.; Ifland, A.; Naumann, A.; Kleta, S.; Noll, M. Incorporation of Listeria monocytogenes strains in raw milk biofilms. Int. J. Food Microbiol. 2013, 161, 61–68. [Google Scholar] [CrossRef]
- Zengler, K.; Palsson, B.O. A road map for the development of community systems (CoSy) biology. Nat. Rev. Microbiol. 2012, 10, 366–372. [Google Scholar] [CrossRef]
- Burmølle, M.; Ren, D.; Bjarnsholt, T.; Sørensen, S.J. Interactions in multispecies biofilms: Do they actually matter? Trends Microbiol. 2014, 22, 84–91. [Google Scholar] [CrossRef]
- Lenz, D.H.; Miller, M.B.; Zhu, J.; Kulkarni, R.V.; Bassler, B.L. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 2005, 58, 1186–1202. [Google Scholar] [CrossRef]
- Heroven, A.K.; Sest, M.; Pisano, F.; Scheb-Wetzel, M.; Steinmann, R.; Bohme, K.; Klein, J.; Münch, R.; Schomburg, D.; Dersch, P. Crp induces switching of the CsrB and CsrC RNAs in Yersinia pseudotuberculosis and links nutritional status to virulence. Front. Cell Infect. Microbiol. 2012, 2, 158. [Google Scholar] [CrossRef]
- Brencic, A.; Lory, S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 2009, 72, 612–632. [Google Scholar] [CrossRef]
- Martínez, C.L.; Vadyvaloo, V. Mechanisms of post-transcriptional gene regulation in bacterial biofilms. Front. Cell Infect. Microbiol. 2014, 4, 38. [Google Scholar] [CrossRef]
- Jonas, K.; Edwards, A.N.; Ahmad, I.; Romeo, T.; Römling, U.; Melefors, O. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ. Microbiol. 2010, 12, 524–540. [Google Scholar] [CrossRef]
- Karampoula, F. Biofilms in Water Industry: CASE study of Salmonella enterica Serovar Typhimurium—Biofilm Formation and Control In Vitro and In Situ. Master’s Thesis, Agricultural University of Athens, Athens, Greece, 2015. [Google Scholar]

| Gene | Gene Function | Primer Name | Primer Sequence (5′ to 3′) a | Restriction Sites | Amplified Region b | Reference |
|---|---|---|---|---|---|---|
| csrA | General regulator: motility, biofilm formation, virulence [19] | csrA-F1 | TTAAACCATGGGAATTGATTGGGATGGTCGCTC | NcoI | −389…+183 | This study |
| csrA-R1 | GTAACTCGAGTGCTGGGATTTTTCAGCCTGG | XhoI | This study | |||
| csgB | Nucleator in the assembly of curli (coiled surface structures) on the cell surface [19] | csgB-F1 | ATCCCCATGGCGGCATAACACACACCAAC | NcoI | −374…+156 | This study |
| csgB-R1 | CTATTACTCGAGCCGACTTGACCAATAATGGC | XhoI | This study | |||
| sspH2 | virulence factor [20] | sspH2-F1 | GCGTTCCATGGTTGCCTGATACGGATGAAAACCG | NcoI | −374…+159 | This study |
| sspH2-R1 | CAAATCCTCGAGATGCACTCCAGCGCTTCAGTC | XhoI | This study | |||
| fliD | Motility regulator [21] | fliD-F1 | GCATACCATGGAACAGTTGCAGCCGTATCGCTG | NcoI | −831…+48 | This study |
| fliD-R1 | TTTTCCTCGAGCCATAGGCGGTTAGCTTTGCCG | XhoI | This study |
| Microorganism | Source/Sample | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water Source | Water Distribution Network | Final Products | ||||||||||||||
| WS1 | WS2 | WS3 | WS4 | WS5 | WD1 | WD3 | WD4 | WD6 | WD7 | WD10 | FN1 | FN2 | FN3 | FN4 | FN5 | |
| HPC (cfu/mL) * | 4 | 5 | 3 | 29 | 5 | 12 | 7 | 9 | 23 | 26 | 4 | 16 | 15 | 30 | 1 | 3 |
| Citrobacter spp. ** | + | + | + | + | + | + | + | + | + | + | ||||||
| Staphylococcus spp. ** | + | |||||||||||||||
| Staphylococcus spp. ** | + | + | + | + | ||||||||||||
| Pseudomonas spp. ** | + | + | ||||||||||||||
| Bacillus spp. ** | + | + | ||||||||||||||
| Exiguobacterium spp. ** | + | |||||||||||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karampoula, F.; Doulgeraki, A.I.; Fotiadis, C.; Tampakaki, A.; Nychas, G.-J.E. Monitoring Biofilm Formation and Microbial Interactions that May Occur During a Salmonella Contamination Incident across the Network of a Water Bottling Plant. Microorganisms 2019, 7, 236. https://doi.org/10.3390/microorganisms7080236
Karampoula F, Doulgeraki AI, Fotiadis C, Tampakaki A, Nychas G-JE. Monitoring Biofilm Formation and Microbial Interactions that May Occur During a Salmonella Contamination Incident across the Network of a Water Bottling Plant. Microorganisms. 2019; 7(8):236. https://doi.org/10.3390/microorganisms7080236
Chicago/Turabian StyleKarampoula, Foteini, Agapi I. Doulgeraki, Christos Fotiadis, Anastasia Tampakaki, and George-John E. Nychas. 2019. "Monitoring Biofilm Formation and Microbial Interactions that May Occur During a Salmonella Contamination Incident across the Network of a Water Bottling Plant" Microorganisms 7, no. 8: 236. https://doi.org/10.3390/microorganisms7080236
APA StyleKarampoula, F., Doulgeraki, A. I., Fotiadis, C., Tampakaki, A., & Nychas, G.-J. E. (2019). Monitoring Biofilm Formation and Microbial Interactions that May Occur During a Salmonella Contamination Incident across the Network of a Water Bottling Plant. Microorganisms, 7(8), 236. https://doi.org/10.3390/microorganisms7080236





