Biomarkers’ Responses to Reductive Dechlorination Rates and Oxygen Stress in Bioaugmentation Culture KB-1TM
Abstract
:1. Introduction
2. Materials and Methods
2.1. Analytical Methods
2.2. Microbial Culture
2.3. Continuous Feed Experiments
2.4. Oxygen Stress Experiment
2.5. Calculation of Respiration Rates
2.6. Nucleic Acid Extraction and Quantification
2.7. Microarray Analysis
2.8. Proteomic Analysis
3. Results and Discussion
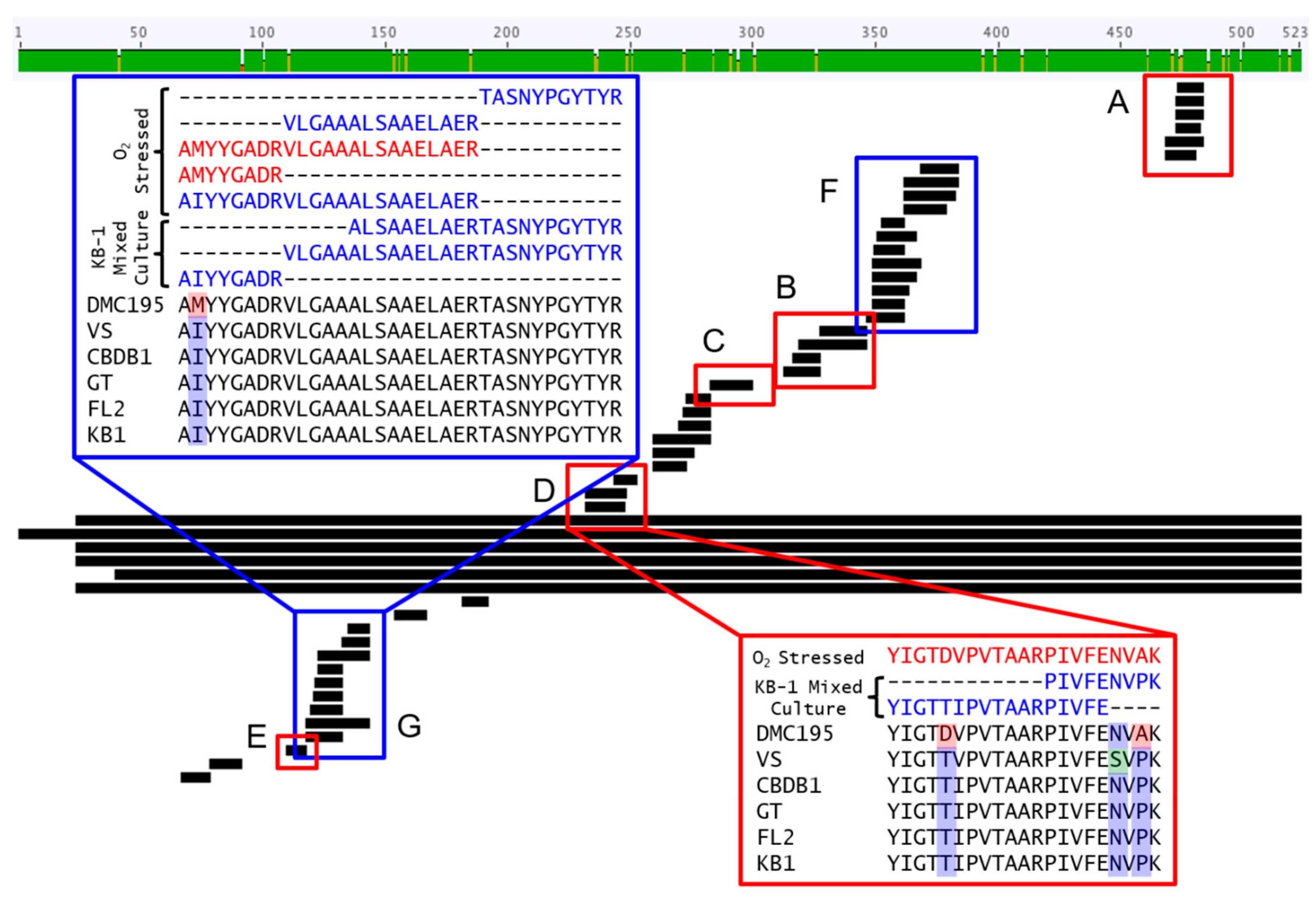
3.1. Proteomic and Transcriptomic Profiling of KB-1TM
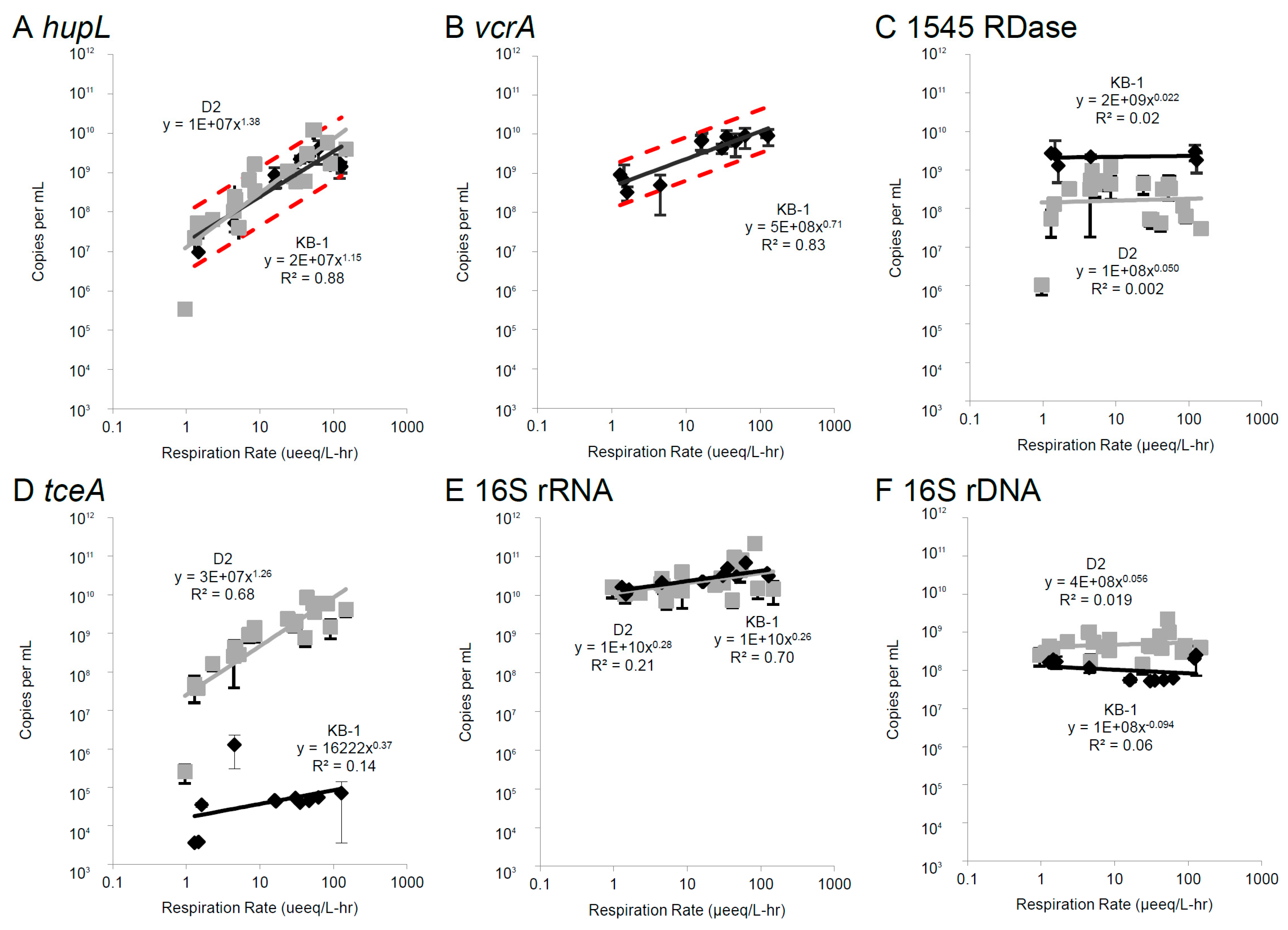
3.2. Correlations Between mRNA Biomarkers and Respiration Rate
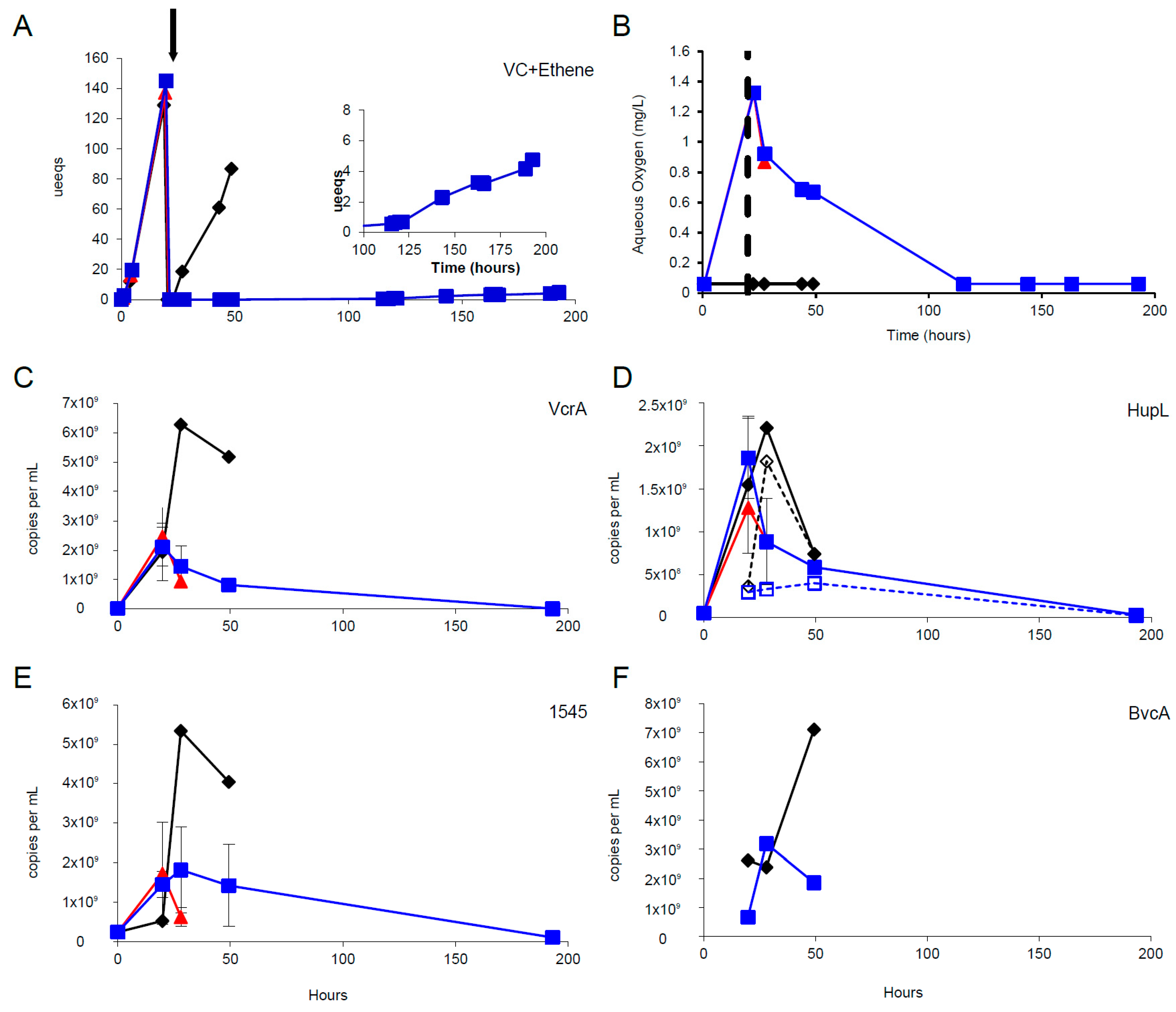
3.3. Oxygen Stress Response
3.4. mRNA Decay Under Oxygen Stress Conditions
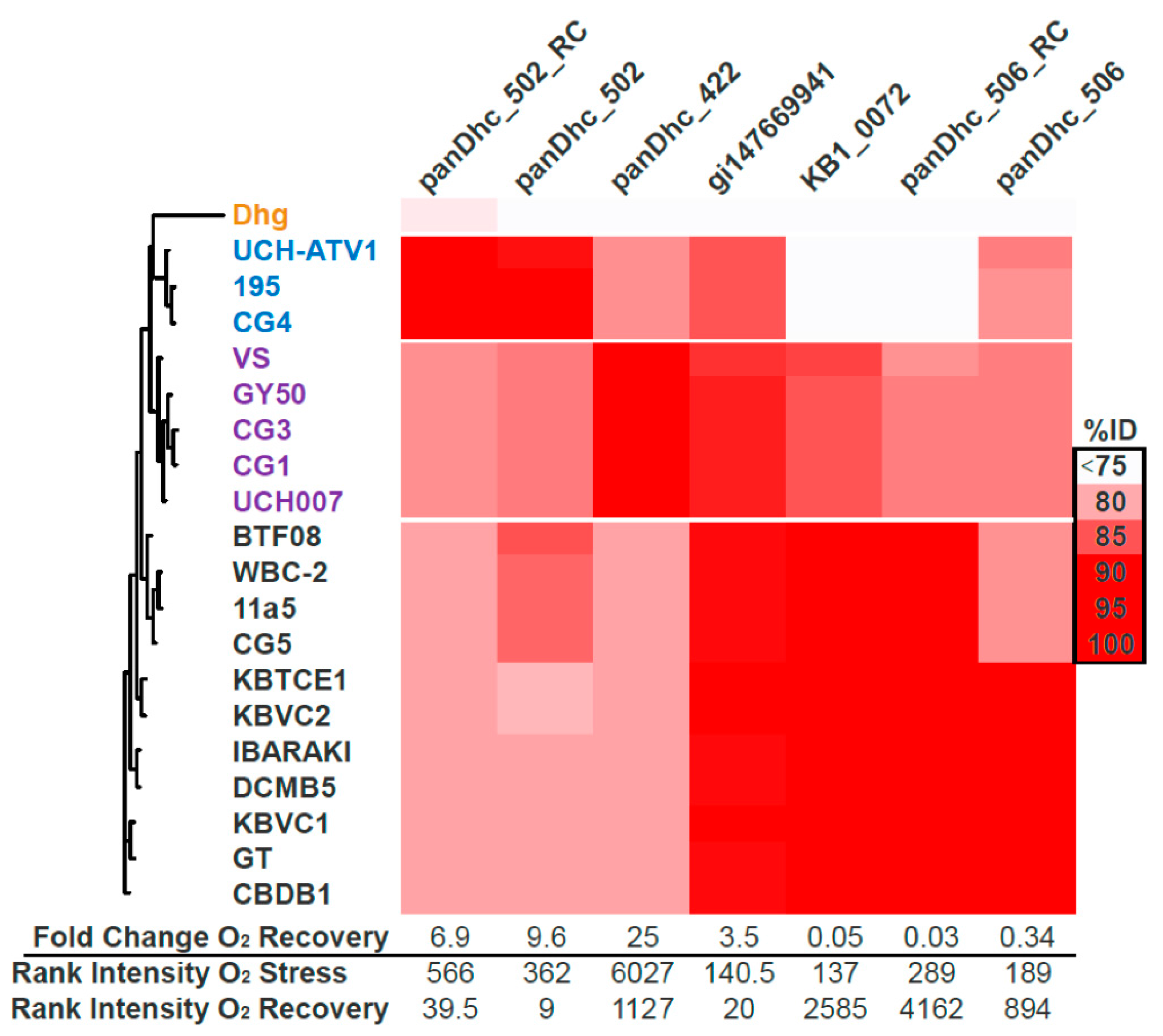
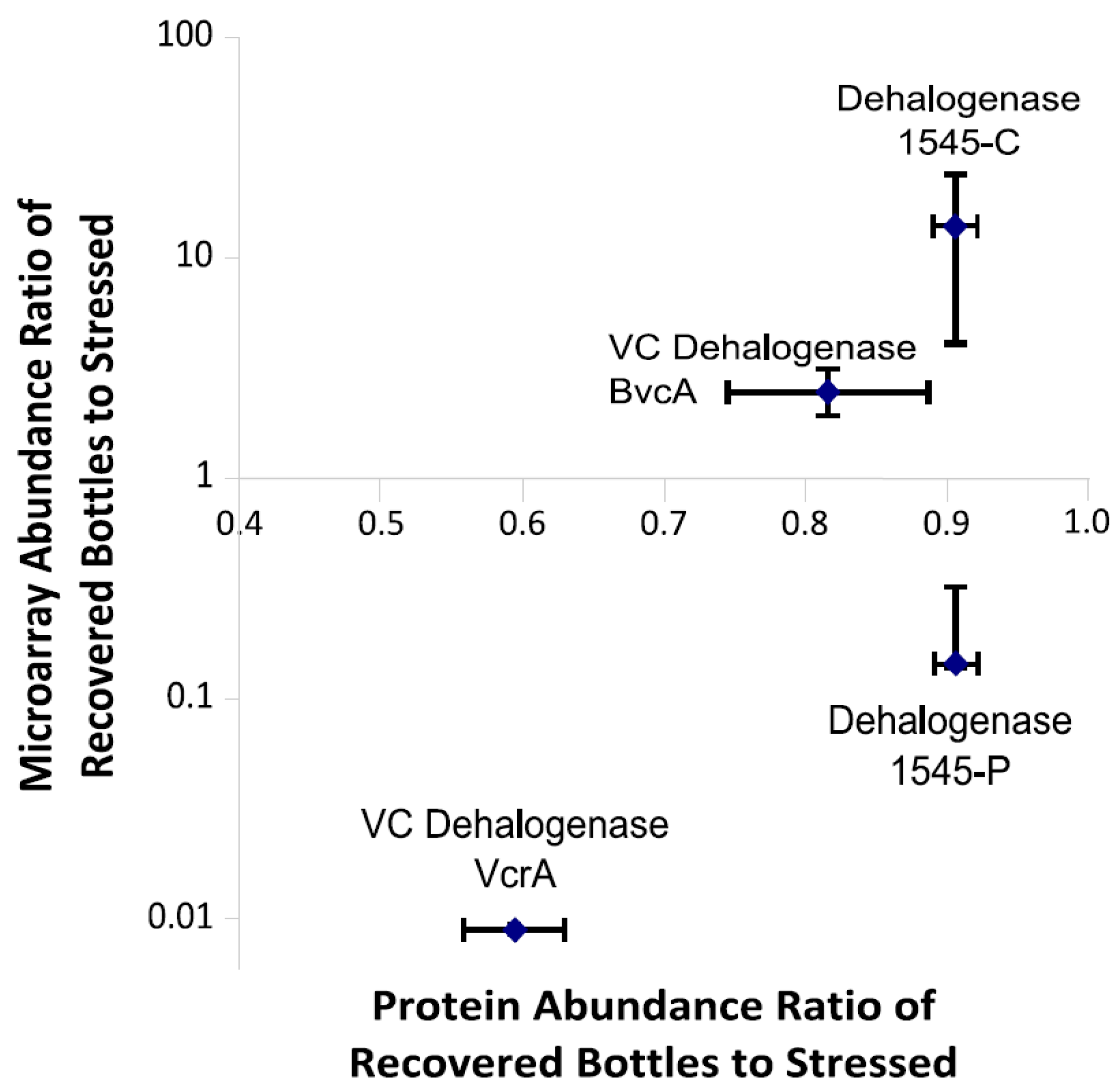
3.5. Microarray Trends in the Oxygen Stress Experiment
3.6. Multiplexed Proteomic Analysis of Oxygen Stress Experiments
3.7. Summary
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hendrickson, E.R.; Payne, J.A.; Young, R.M.; Starr, M.G.; Perry, M.P.; Fahnestock, S.; Ellis, D.E.; Ebersole, R.C. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 2002, 68, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Maymo-Gatell, X.; Chien, Y.; Gossett, J.M.; Zinder, S.H. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 1997, 276, 1568–1571. [Google Scholar] [CrossRef] [PubMed]
- Maymo-Gatell, X.; Anguish, T.; Zinder, S.H. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by Dehalococcoides ethenogenes 195. Appl. Environ. Microbiol. 1999, 65, 3108–3113. [Google Scholar] [PubMed]
- Fennell, D.E.; Nijenhuis, I.; Wilson, S.F.; Zinder, S.H.; Häggblom, M.M. Dehalococcoides ethenogenes Strain 195 Reductively Dechlorinates Diverse Chlorinated Aromatic Pollutants. Environ. Sci. Technol. 2004, 38, 2075–2081. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Sung, Y.; Krajmalnik-Brown, R.; Ritalahti, K.M.; Loffler, F.E. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 2005, 7, 1442–1450. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.; Ritalahti, K.M.; Apkarian, R.P.; Loffler, F.E. Quantitative PCR Confirms Purity of Strain GT, a Novel Trichloroethene-to-Ethene-Respiring Dehalococcoides Isolate. Appl. Environ. Microbiol. 2006, 72, 1980–1987. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.A.; Rosner, B.M.; Von Abendroth, G.; Meshulam-Simon, G.; McCarty, P.L.; Spormann, A.M. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 2004, 70, 4880–4888. [Google Scholar] [CrossRef] [PubMed]
- Cupples, A.M.; Spormann, A.M.; McCarty, P.L. Comparative evaluation of chloroethene dechlorination to ethene by Dehalococcoides-like microorganisms. Environ. Sci. Technol. 2004, 38, 4768–4774. [Google Scholar] [CrossRef] [PubMed]
- Duhamel, M.; Mo, K.; Edwards, E. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl. Environ. Microbiol. 2004, 70, 5538–5545. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Ritalahti, K.M.; Yang, K.-L.; Koenigsberg, S.S.; Loffler, F.E. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 2003, 424, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Molenda, O.; Tang, S.; Edwards, E.A. Identity and Substrate Specificity of Reductive Dehalogenases Expressed in Dehalococcoides-Containing Enrichment Cultures Maintained on Different Chlorinated Ethenes. Appl. Environ. Microbiol. 2015, 81, 4626–4633. [Google Scholar] [CrossRef] [PubMed]
- Committee on In Situ Bioremediation, National Research Council. In Situ Bioremediation: When does It Work; The National Academies Press: Washington, DC, USA, 1993; ISBN 0-309-04896-6. [Google Scholar]
- Rahm, B.G.; Richardson, R.E. Gene Transcripts as Quantitative Bioindicators of Tetrachloroethene, Trichloroethene, and cis-1,2-Dichloroethene Dehalorespiration Rates. Environ. Sci. Technol. 2008, 42, 5099–5105. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.K.H.; Macbeth, T.W.; Sorenson, K.S.; Deeb, R.A.; Alvarez-Cohen, L. Quantifying Genes and Transcripts to Assess the In Situ Physiology of Dehalococcoides spp. in a Trichloroethene-Contaminated Groundwater Site. Appl. Environ. Microbiol. 2008, 74, 2728–2739. [Google Scholar] [CrossRef] [PubMed]
- Rahm, B.G.; Morris, R.M.; Richardson, R.E. Temporal Expression of Respiratory Genes in an Enrichment Culture Containing Dehalococcoides ethenogenes. Appl. Environ. Microbiol. 2006, 72, 5486–5491. [Google Scholar] [CrossRef] [PubMed]
- Rahm, B.G.; Richardson, R.E. Correlation of respiratory gene expression levels and pseudo-steady-state PCE respiration rates in Dehalococcoides ethenogenes. Environ. Sci. Technol. 2008, 42, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Waller, A.S.; Krajmalnik-Brown, R.; Loffler, F.E.; Edwards, E.A. Multiple Reductive-Dehalogenase-Homologous genes are simultaneously transcribed during Dechlorination by Dehalococcoides-Containing Cultures. Appl. Environ. Microbiol. 2005, 71, 8257–8264. [Google Scholar] [CrossRef] [PubMed]
- Behrens, S.; Azizian, M.F.; McMurdie, P.J.; Sabalowsky, A.; Dolan, M.E.; Semprini, L.; Spormann, A.M. Monitoring abundance and expression of Dehalococcoides species chloroethene-reductive dehalogenases in a tetrachloroethene-dechlorinating flow column. Appl. Environ. Microbiol. 2008, 74, 5695–5703. [Google Scholar] [CrossRef] [PubMed]
- Fung, J.M.; Morris, R.M.; Adrian, L.; Zinder, S.H. Expression of reductive dehalogenase Genes in Dehalococcoides ethenogenes Strain 195 Growing on Tetrachloroethene, Trichloroethene, or 2,3-Dichlorophenol. Appl. Environ. Microbiol. 2007, 73, 4439–4445. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.R.; Lee, P.K.H.; Holmes, V.F.; Fortin, A.C.; Alvarez-Cohen, L. Transcriptional expression of the tceA gene in a Dehalococcoides-containing microbial enrichment. Appl. Environ. Microbiol. 2005, 71, 7145–7151. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.R.; Lee, P.K.H.; Holmes, V.F.; Alvarez-Cohen, L. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 2005, 71, 3866–3871. [Google Scholar] [CrossRef] [PubMed]
- Rowe, A.R.; Heavner, G.L.; Mansfeldt, C.B.; Werner, J.J.; Richardson, R.E. Relating Chloroethene Respiration Rates in Dehalococcoides to Protein and mRNA Biomarkers. Environ. Sci. Technol. 2012, 46, 9388–9397. [Google Scholar] [CrossRef] [PubMed]
- Kranzioch, I.; Ganz, S.; Tiehm, A. Chloroethene degradation and expression of Dehalococcoides dehalogenase genes in cultures originating from Yangtze sediments. Environ. Sci. Pollut. Res. 2015, 22, 3138–3148. [Google Scholar] [CrossRef] [PubMed]
- Baelum, J.; Chambon, J.C.; Scheutz, C.; Binning, P.J.; Laier, T.; Bjerg, P.L.; Jacobsen, C.S. A conceptual model linking functional gene expression and reductive dechlorination rates of chlorinated ethenes in clay rich groundwater sediment. Water Res. 2013, 47, 2467–2478. [Google Scholar] [CrossRef] [PubMed]
- Mattes, T.E.; Jin, Y.O.; Livermore, J.; Pearl, M.; Liu, X. Abundance and activity of vinyl chloride (VC)-oxidizing bacteria in a dilute groundwater VC plume biostimulated with oxygen and ethene. Appl. Microbiol. Biotechnol. 2015, 99, 9267–9276. [Google Scholar] [CrossRef] [PubMed]
- Scheutz, C.; Durant, N.; Dennis, P.; Hansen, M.H.; Jorgensen, T.; Jakobsen, R.; Bjerg, P.L. Concurrent ethene generation and growth of Dehalococcoides containing vinyl chloride reductive dehalogenase genes during an enhanced reductive dechlorination field demonstration. Environ. Sci. Technol. 2008, 42, 9302–9309. [Google Scholar] [CrossRef] [PubMed]
- Perez de Mora, A.; Lacourt, A.; McMaster, M.L.; Liang, X.; Dworatzek, S.; Edwards, E.A. Chlorinated electron acceptor availability selects for specific Dehalococcoides populations in dechlorinating enrichment cultures and in groundwater. bioRxiv 2017. [Google Scholar] [CrossRef]
- Morris, R.M.; Fung, J.M.; Rahm, B.G.; Zhang, S.; Freedman, D.L.; Zinder, S.H.; Richardson, R.E. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl. Environ. Microbiol. 2007, 73, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Kublik, A.; Deobald, D.; Hartwig, S.; Schiffmann, C.L.; Andrades, A.; von Bergen, M.; Sawers, R.G.; Adrian, L. Identification of a multi-protein reductive dehalogenase complex in Dehalococcoides mccartyi strain CBDB1 suggests a protein-dependent respiratory electron transport chain obviating quinone involvement: Respiratory reductive dehalogenase protein complex. Environ. Microbiol. 2016, 18, 3044–3056. [Google Scholar] [CrossRef] [PubMed]
- Fennell, D.E.; Gossett, J.M.; Zinder, S.H. Comparison of butyric acid, ethanol, lactic acid, and propionic acid as hydrogen donors for the reductive dechlorination of tetrachloroethene. Environ. Sci. Technol. 1997, 31, 918–926. [Google Scholar] [CrossRef]
- Fennell, D. Comparison of Alternative Hydrogen Donors for Anaerobic Reductive Dechlorination of Tetrachloroethene. Ph.D. Thesis, Cornell University, Ithaca, NY, USA, 1998. [Google Scholar]
- Gossett, J.M. Sustained aerobic oxidation of vinyl chloride at low oxygen concentrations. Environ. Sci. Technol. 2010, 44, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Duhamel, M.; Wehr, S.D.; Yu, L.; Rizvi, H.; Seepersad, D.; Dworatzek, S.; Cox, E.E.; Edwards, E.A. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-dichloroethene and vinyl chloride. Water Res. 2002, 36, 4193–4202. [Google Scholar] [CrossRef]
- Duhamel, M.; Edwards, E.A. Microbial composition of chlorinated ethene-degrading cultures dominated by Dehalococcoides. FEMS Microbiol. Ecol. 2006, 58, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Duhamel, M.; Edwards, E.A. Growth and Yields of Dechlorinators, Acetogens, and Methanogens during Reductive Dechlorination of Chlorinated Ethenes and Dihaloelimination of 1,2-Dichloroethane. Environ. Sci. Technol. 2007, 41, 2303–2310. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Beiko, R.G.; Rowe, A.R.; Richardson, R.E.; Edwards, E.A. Comparative metagenomics of three Dehalococcoides-containing enrichment cultures: The role of the non-dechlorinating community. BMC Genom. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Mansfeldt, C.B.; Heavner, G.W.; Rowe, A.R.; Hayete, B.; Church, B.W.; Richardson, R.E. Inferring Gene Networks for Strains of Dehalococcoides Highlights Conserved Relationships between Genes Encoding Core Catabolic and Cell-Wall Structural Proteins. PLoS ONE 2016, 11, e0166234. [Google Scholar] [CrossRef] [PubMed]
- Peirson, S.N.; Butler, J.N.; Foster, R.G. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003, 31, 1–7. [Google Scholar] [CrossRef]
- Schefe, J.H.; Lehmann, K.E.; Buschmann, I.R.; Unger, T.; Funke-Kaiser, H. Quantitative real-time RT-PCR data analysis: Current concepts and the novel “gene expression’s C T difference” formula. J. Mol. Med. 2006, 84, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Mansfeldt, C.B.; Rowe, A.R.; Heavner, G.L.W.; Zinder, S.H.; Richardson, R.E. Meta-Analyses of Dehalococcoides mccartyi Strain 195 Transcriptomic Profiles Identify a Respiration Rate-Related Gene Expression Transition Point and Interoperon Recruitment of a Key Oxidoreductase Subunit. Appl. Environ. Microbiol. 2014, 80, 6062–6072. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Salehi, M.; Nuin, P.; Tillier, E.R.; Edwards, E.A. Design and Verification of a Pangenome Microarray Oligonucleotide Probe set for Dehalococcoides spp. Appl. Environ. Microbiol. 2011, 77, 5361–5369. [Google Scholar] [CrossRef] [PubMed]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef]
- Kim, S.; Gupta, N.; Pevzner, P.A. Spectral Probabilities and Generating Functions of Tandem Mass Spectra: A Strike against Decoy Databases. J. Proteome Res. 2008, 7, 3354–3363. [Google Scholar] [CrossRef] [PubMed]
- Monroe, M.E.; Shaw, J.L.; Daly, D.S.; Adkins, J.N.; Smith, R.D. MASIC: A software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput. Biol. Chem. 2008, 32, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.; Schaefer, J.; Kuhn, K.; Kienle, S.; Schwarz, J.; Schmidt, G.; Neumann, T.; Johnstone, R.A.W.; Mohammed, A.K.A.; Hamon, C. Tandem Mass Tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2006, 78, 4235. [Google Scholar] [CrossRef]
- Zybailov, B.; Mosley, A.L.; Sardiu, M.E.; Coleman, M.K.; Florens, L.; Washburn, M.P. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 2006, 5, 2339–2347. [Google Scholar] [CrossRef] [PubMed]
- Werner, J.J.; Ptak, A.C.; Rahm, B.G.; Zhang, S.; Richardson, R.E. Absolute quantification of Dehalococcoides proteins: Enzyme bioindicators of chlorinated ethene dehalorespiration. Environ. Microbiol. 2009, 11, 2687–2697. [Google Scholar] [CrossRef] [PubMed]
- Heavner, G.L.W. Biokinetic Modeling, Laboratory Examination and Field Analysis of DNA, RNA and Protein as Robust Molecular Biomarkers of Chloroethene Reductive Dechlorination in Dehalococcoides Mccartyi. Ph.D. Thesis, Cornell University, Ithaca, NY, USA, 2013. [Google Scholar]
- Heavner, G.L.W.; Rowe, A.R.; Mansfeldt, C.B.; Pan, J.K.; Gossett, J.M.; Richardson, R.E. Molecular Biomarker-based biokinetic modeling of a PCE-Dechlorinating and Methanogenic Mixed Culture. Environ. Sci. Technol. 2013, 47, 3724–3733. [Google Scholar] [CrossRef] [PubMed]
- Marshall, I.P.; Berggren, D.R.; Azizian, M.F.; Burow, L.C.; Semprini, L.; Spormann, A.M. The Hydrogenase Chip: A tiling oligonucleotide DNA microarray technique for characterizing hydrogen-producing and -consuming microbes in microbial communities. ISME J. 2011, 6, 814–826. [Google Scholar] [CrossRef] [PubMed]
- Rowe, A.R. Molecular Biomarkers for Respiration in Dehalococcoides ethenogenes and Methanospirillum hungatei: Comparing Protein and Messenger-RNA Abundance in Anaerobes. Ph.D. Thesis, Cornell University, Ithaca, NY, USA, 2011. [Google Scholar]
- Mansfeldt, C.B. Data Driven Hypothesis Modelling of Dehalococcoides mccartyi: Predicted Biology and Biomarkers of Stress in Two Mixed Microbial Communities. Ph.D. Thesis, Cornell University, Ithaca, NY, USA, 2013. [Google Scholar]
- Wei, K.; Grostern, A.; Chan, W.W.M.; Richardson, R.E.; Edwards, E.A. Electron acceptor interactions between organohalide respiring bacteria: Cross-feeding, competition and inhibition. In Organohalide Respiration; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Mayer-Blackwell, K.; Azizian, M.F.; Green, J.K.; Spormann, A.M.; Semprini, L. Survival of Vinyl Chloride Respiring Dehalococcoides mccartyi under Long-Term Electron Donor Limitation. Environ. Sci. Technol. 2017, 51, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Sutton, N.B.; Atashgahi, S.; Saccenti, E.; Grotenhuis, T.; Smidt, H.; Rijnaarts, H.H.M. Microbial Community Response of an Organohalide Respiring Enrichment Culture to Permanganate Oxidation. PLoS ONE 2015, 10, e0134615. [Google Scholar] [CrossRef] [PubMed]
| Homolog Name | Protein Intensity | Microarray Intensity |
|---|---|---|
| VcrA | 4723 | 106,022 |
| DET1545 | 4684 | 32,998 |
| HupL, NiFe Hydrogenase | 959 | 118,339 |
| BvcA | 923 | 153,080 |
| Geobacter PceA | 94 | - |
| KB1_8 (KB1_1468), KB1_9 (KB1_0060) | 66 | 801 |
| DET0180 | 62 | 1212 |
| KB1_1, KB13109_4 (KB1_0054) | 57 | 2279 |
| TceA | 49 | 1298 |
| KB13241_3 (KB1_1549) | 13 | 76 |
| DET1528 | 8.4 | 182 |
| KB13241_7 (KB1_1589), KB1_7 | 7.7 | 845 |
| DET1519 | 2.2 | 3635 |
| DET1538 | 1.1 | 5536 |
| cbdbA80 | 1.1 | 252 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heavner, G.L.W.; Mansfeldt, C.B.; Debs, G.E.; Hellerstedt, S.T.; Rowe, A.R.; Richardson, R.E. Biomarkers’ Responses to Reductive Dechlorination Rates and Oxygen Stress in Bioaugmentation Culture KB-1TM. Microorganisms 2018, 6, 13. https://doi.org/10.3390/microorganisms6010013
Heavner GLW, Mansfeldt CB, Debs GE, Hellerstedt ST, Rowe AR, Richardson RE. Biomarkers’ Responses to Reductive Dechlorination Rates and Oxygen Stress in Bioaugmentation Culture KB-1TM. Microorganisms. 2018; 6(1):13. https://doi.org/10.3390/microorganisms6010013
Chicago/Turabian StyleHeavner, Gretchen L. W., Cresten B. Mansfeldt, Garrett E. Debs, Sage T. Hellerstedt, Annette R. Rowe, and Ruth E. Richardson. 2018. "Biomarkers’ Responses to Reductive Dechlorination Rates and Oxygen Stress in Bioaugmentation Culture KB-1TM" Microorganisms 6, no. 1: 13. https://doi.org/10.3390/microorganisms6010013
APA StyleHeavner, G. L. W., Mansfeldt, C. B., Debs, G. E., Hellerstedt, S. T., Rowe, A. R., & Richardson, R. E. (2018). Biomarkers’ Responses to Reductive Dechlorination Rates and Oxygen Stress in Bioaugmentation Culture KB-1TM. Microorganisms, 6(1), 13. https://doi.org/10.3390/microorganisms6010013





