Nasal Colonizers from Sows in the Federal District of Brazil Showed a Diverse Phenotypic Resistance Profile
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Sampling, Swine Farms and Sow Health Management
2.3. Biosecurity Data Collection
2.4. Culture and Bacterial Isolation
2.5. Antimicrobial Susceptibility Testing
2.6. DNA Extraction and PCR Assays
2.7. Statistical Analyses
3. Results
3.1. Sampling Sow Data
3.2. Lung Lesion Slaughter House Report and Pathogen Detection by PCR or Bacterial Culture in Nasal Samples from Sows
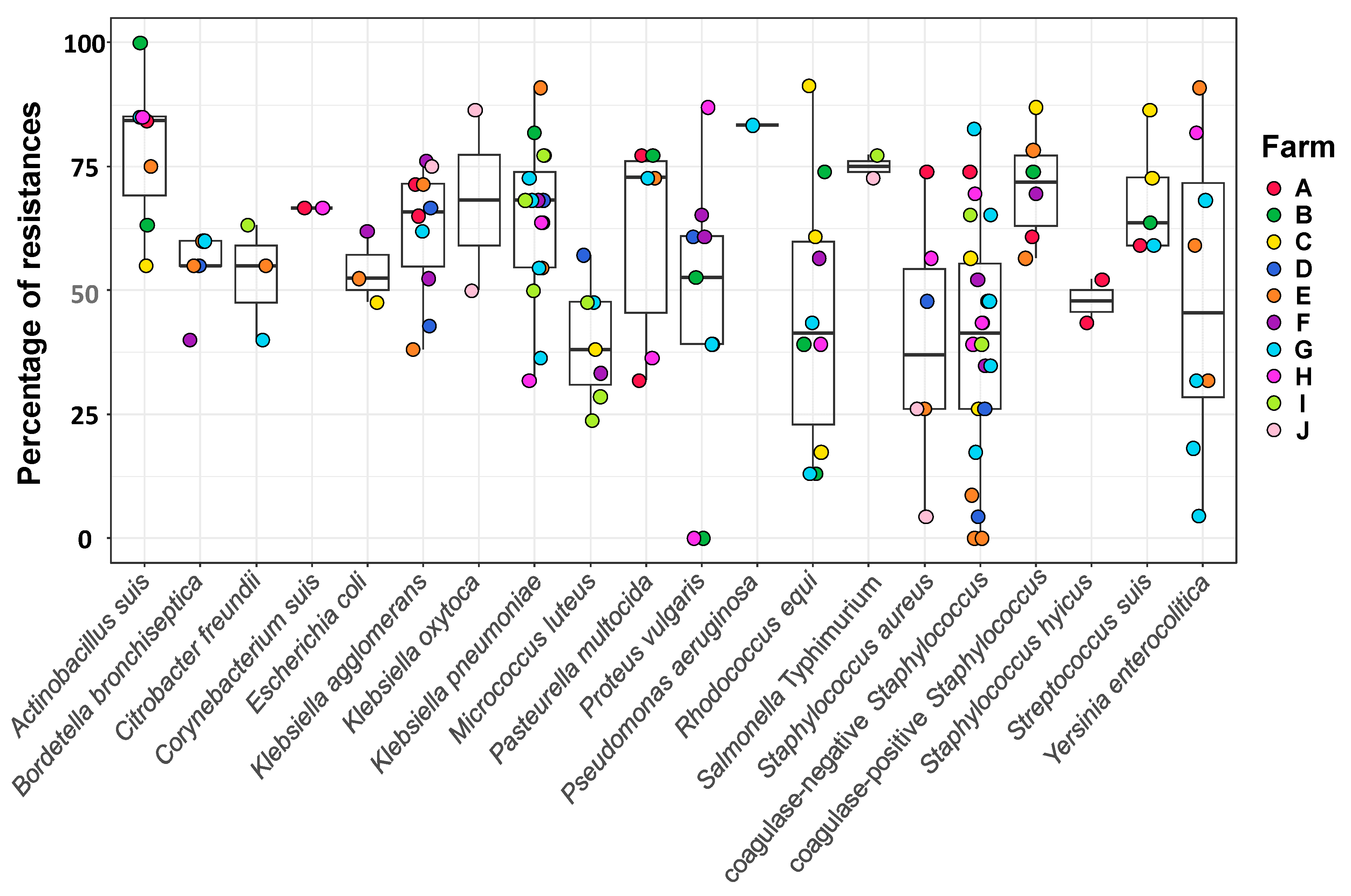
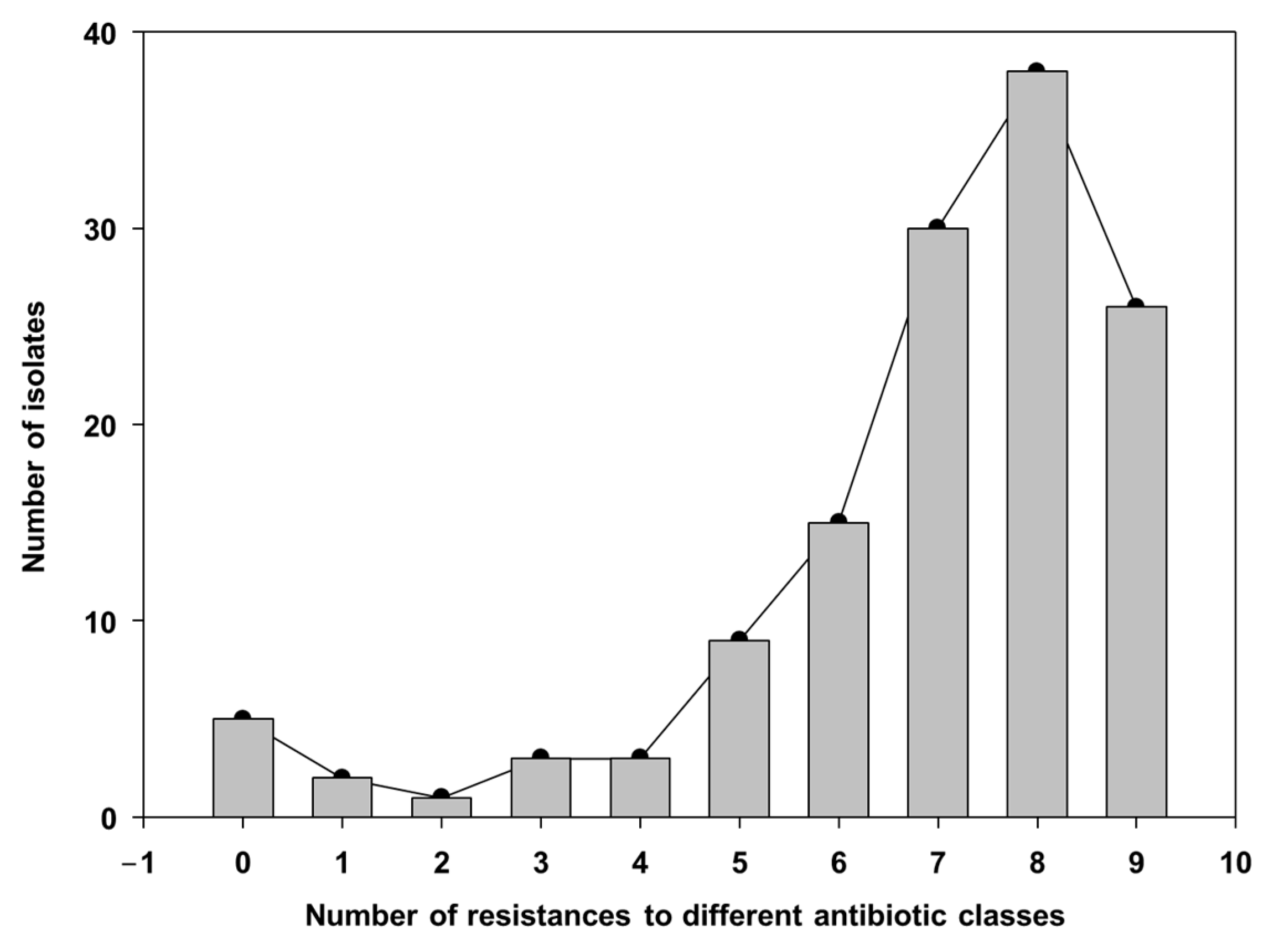
3.3. Resistance Profile of Bacterial Isolates from the Sows’ Nasal Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prescott, J.F. History and current use of antimicrobial drugs in veterinary medicine. Microbiol. Spectr. 2017, 5, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zeon, O.; Kibe, L.W. Antimicrobial Drug Resistance and Antimicrobial Resistant Threats. Physician Assist. Clin. 2023, 8, 411–420. [Google Scholar] [CrossRef]
- Thompson, N.T.; Kitzenberg, D.A.; Kao, D.J. Persister-mediated emergence of antimicrobial resistance in agriculture due to antibiotic growth promoters. AIMS Microbiol. 2023, 9, 738–756. [Google Scholar] [CrossRef]
- Letek, M. Alexander Fleming, The discoverer of the antibiotic effects of penicillin. Front. Young Minds 2020, 8, 159. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D. Selection and evolution of resistance antimicrobial drugs. IUBMB Life 2014, 66, 521–529. [Google Scholar] [CrossRef]
- Collignon, P.J.; McEwen, S.A. One Health—Its importance in helping to better control antimicrobial resistance. Trop. Med. Infect. Dis. 2019, 4, 22. [Google Scholar] [CrossRef]
- Salam, A.; Al-Amin, Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Costa-Hurtado, M.; Barba-Vidal, E.; Maldonado, J.; Aragon, V. Update on Glässer’s disease: How to control the disease under restrictive use of antimicrobials. Vet. Microbiol. 2020, 242, 108595. [Google Scholar] [CrossRef]
- Dutra, M.C.; Moreno, L.Z.; Dias, R.A.; Moreno, A.M. Antimicrobial Use in Brazilian Swine Herds: Assessment of Use and Reduction Examples. Microorganisms 2021, 9, 881. [Google Scholar] [CrossRef]
- Ferdinand, A.S.; Coppo, M.J.C.; Howden, B.P.; Browning, G.F. Tackling antimicrobial resistance by integrating One Health and the Sustainable Development Goals. BMC Glob. Public Health 2023, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Hopman, N.E.M.; Van Dijk, M.A.M.; Broens, E.M.; Wagenaar, J.A.; Heederik, D.J.J.; Van Geijlswijk, I.M. Quantifying antimicrobial use in Dutch companion animals. Front. Vet. Sci. 2019, 6, 158. [Google Scholar] [CrossRef] [PubMed]
- Serpa, M.; Nascimento, J.A.F.B.D.; Alves, M.F.; Guedes, M.I.M.C.; Reis, A.T.; Heinemann, M.B.; Lage, A.P.; Lobato, Z.I.P.; Dorneles, E.M.S. Antimicrobial resistance in bacteria isolated from pigs with respiratory clinical signs in Brazil. Braz. J. Vet. Res. Anim. Sci. 2020, 57, e160956. [Google Scholar] [CrossRef]
- Pirolo, M.; Espinosa-Gongora, C.; Bogaert, D.; Guardabassi, L. The porcine respiratory microbiome: Recent insights and future challenges. Anim. Microbiome 2021, 3, 9. [Google Scholar] [CrossRef]
- ISO/TS 34700:2016; Animal Welfare Management—General Requirements and Guidance for Organizations in the Food Supply Chain. International Organization for Standardization: Geneva, Switzerland, 2016.
- Castle, S.S. Amoxicillin. In xPharm: The Comprehensive Pharmacology Reference; Enna, S.J., Bylund, D.B., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–6. ISBN 9780080552323. [Google Scholar] [CrossRef]
- Christensen, G.; Sørensen, V.; Mousing, J. Disease surveillance as a tool in swine health management. Anim. Health Res. Rev. 2002, 3, 43–47. [Google Scholar] [CrossRef]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. Am. Soc. Microbiol. 2016, 1, 23. Available online: https://asm.org/getattachment/2594ce26-bd44-47f6-8287-0657aa9185ad/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test-Protocol-pdf (accessed on 16 March 2025).
- Brombilla, T.; Ogata, R.A.; Nassar, A.F.C.; Cardoso, M.V.; Ruiz, V.L.A.; Fava, C.D. Effect of bacterial agents of the porcine respiratory disease complex on productive indices and slaughter weight. Ciência Anim. Bras. 2019, 20, e51615. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 25 April 2024).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ggplot2 3.4.0. Tidyverse: [Online]; Available online: https://www.tidyverse.org/blog/2022/11/ggplot2-3-4-0/ (accessed on 4 June 2024).
- Rahn, O. New Principles for the Classification of Bacteria. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. 1937, 96, 273–286. [Google Scholar]
- Luo, Y.; Ren, W.; Smidt, H.; Wright, A.-D.G.; Yu, B.; Schyns, G.; McCormack, U.M.; Cowieson, A.J.; Yu, J.; He, J.; et al. Dynamic Distribution of Gut Microbiota in Pigs at Different Growth Stages: Composition and Contribution. Microbiol. Spectr. 2022, 10, e00688-21. [Google Scholar] [CrossRef]
- Dayao, D.A.E.; Gibson, J.S.; Blackall, P.J.; Turni, C. Antimicrobial resistance in bacteria associated with porcine respiratory disease in Australia. Vet. Microbiol. 2014, 171, 232–235. [Google Scholar] [CrossRef]
- Correa-Fiz, F.; Neila-Ibáñez, C.; López-Soria, S.; Napp, S.; Martinez, B.; Sobrevia, L.; Tibble, S.; Aragon, V.; Migura-Garcia, L. Feed additives for the control of post-weaning Streptococcus suis disease and the effect on the faecal and nasal microbiota. Sci. Rep. 2020, 10, 20354. [Google Scholar] [CrossRef] [PubMed]
- Hadjirin, N.F.; Miller, E.L.; Murray, G.G.R.; Yen, P.L.K.; Phuc, H.D.; Wileman, T.M.; Hernandez-Garcia, J.; Williamson, S.M.; Parkhill, J.; Maskell, D.J.; et al. Large-scale genomic analysis of antimicrobial resistance in the zoonotic pathogen Streptococcus suis. BMC Biol. 2021, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.; Aragon, V.; Brockmeier, S.L.; Gebhart, C.; de Greeff, A.; Kerdsin, A.; O’dea, M.A.; Okura, M.; Saléry, M.; Schultsz, C.; et al. Update on Streptococcus suis research and prevention in the era of antimicrobial restriction: 4th International Workshop on S. suis. Pathogens 2020, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Guitart-Matas, J.; Gonzalez-Escalona, N.; Maguire, M.; Vilaró, A.; Martinez-Urtaza, J.; Fraile, L.; Migura-Garcia, L. Revealing genomic insights of the unexplored porcine pathogen Actinobacillus pleuropneumoniae using whole genome sequencing. Microbiol. Spectr. 2022, 10, e01185-22. [Google Scholar] [CrossRef]
- Espinosa Gongora, C.; Larsen, N.; Schønning, K.; Fredholm, M.; Guardabassi, L. Differential analysis of the nasal microbiome of pig carriers or non-carriers of Staphylococcus aureus. PLoS ONE 2016, 11, e0160331. [Google Scholar] [CrossRef]
- Zhang, C.; Ning, Y.; Zhang, Z.; Song, L.; Qiu, H.; Gao, H. In vitro antimicrobial susceptibility of Streptococcus suis strains isolated from clinically healthy sows in China. Vet. Microbiol. 2008, 131, 386–392. [Google Scholar] [CrossRef]
- Francisco, M.S.; Rossi, C.C.; Brito, M.A.V.P.; Laport, M.S.; Barros, E.M.; Giambiagi-de Marval, M. Characterization of biofilms and antimicrobial resistance of coagulase-negative Staphylococcus species involved with subclinical mastitis. J. Dairy Res. 2021, 88, 179–184. [Google Scholar] [CrossRef]
- Deng, J.; Liu, K.; Wang, K.; Yang, B.; Xu, H.; Wang, J.; Dai, F.; Xiao, X.; Gu, X.; Zhang, L.; et al. The prevalence of coagulase-negative Staphylococcus associated with bovine mastitis in China and its antimicrobial resistance rate: A meta-analysis. J. Dairy Res. 2023, 90, 158–163. [Google Scholar] [CrossRef]
- Hopkins, D.; Poljak, Z.; Farzan, A.; Friendship, R. Factors contributing to mortality during a Streptococcus suis outbreak in nursery pigs. Can. Vet. J. 2018, 59, 623–630. [Google Scholar]
- Murray, G.G.R.; Hossain, A.S.M.M.; Miller, E.L.; Bruchmann, S.; Balmer, A.J.; Matuszewska, M.; Herbert, J.; Hadjirin, N.F.; Mugabi, R.; Li, G.; et al. The emergence and diversification of a zoonotic pathogen from within the microbiota of intensively farmed pigs. Proc. Natl. Acad. Sci. USA 2023, 120, e2307773120. [Google Scholar] [CrossRef]
- Klima, C.L.; Holman, D.B.; Cook, S.R.; Conrad, C.C.; Ralston, B.J.; Allan, N.; Anholt, R.M.; Niu, Y.D.; Stanford, K.; Hannon, S.J.; et al. Multidrug resistance in Pasteurellaceae associated with bovine respiratory disease mortalities in North America from 2011 to 2016. Front. Microbiol. 2020, 11, 606438. [Google Scholar] [CrossRef] [PubMed]
- Torres-Blas, I.; Fernández Aguilar, X.; Cabezón, O.; Aragon, V.; Migura-García, L. Antimicrobial resistance in Pasteurellaceae isolates from Pyrenean Chamois (Rupicapra pyrenaica) and domestic sheep in an alpine ecosystem. Animals 2021, 11, 1686. [Google Scholar] [CrossRef]
- Ministério da Agricultura, Pecuária e Abastecimento. Decreto N° 12.031, de 2024. Diário Oficial da União. Available online: https://www.gov.br/agricultura (accessed on 8 January 2024).
- Caballero-Flores, G.; Pickard, J.M.; Núñez, G. Microbiota-mediated colonization resistance: Mechanisms and regulation. Nat. Rev. Microbiol. 2023, 21, 347–360. [Google Scholar] [CrossRef]
- Blanco-Fuertes, M.; Sibila, M.; Franzo, G.; Obregon-Gutierrez, P.; Illas, F.; Correa-Fiz, F.; Aragón, V. Ceftiofur treatment of sows results in long-term alterations in the nasal microbiota of the offspring that can be ameliorated by inoculation of nasal colonizers. Anim. Microbiome 2023, 5, 53. [Google Scholar] [CrossRef] [PubMed]
- Correa-Fiz, F.; Gonçalves, J.M.S.; Illas, F.; Aragón, V. Antimicrobial removal on piglets promotes health and higher bacterial diversity in the nasal microbiota. Sci. Rep. 2019, 9, 6545. [Google Scholar] [CrossRef]
- Mou, K.T.; Allen, H.K.; Alt, D.P.; Trachselb, J.; Haua, S.J.; Coetzeec, J.F.; Holmand, D.B.; Kellnerb, S.; Lovingb, C.L.; Brockmeier, S.L. Shifts in the nasal microbiota of swine in response to different dosing regimens of oxytetracycline administration. Vet. Microbiol. 2019, 237, 108386. [Google Scholar] [CrossRef]
- Bonillo-Lopez, L.; Obregon-Gutierrez, P.; Huerta, E.; Correa-Fiz, F.; Sibila, M.; Aragon, V. Intensive antibiotic treatment of sows with parenteral crystalline ceftiofur and tulathromycin alters the composition of the nasal microbiota of their offspring. Vet. Res. 2023, 54, 112. [Google Scholar] [CrossRef]
- Mahmmod, Y.S.; Correa-Fiz, F.; Aragon, V. Variations in association of nasal microbiota with virulent and non-virulent strains of Glaesserella (Haemophilus) parasuis in weaning piglets. Vet. Res. 2020, 51, 7. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Stobberingh, E.E.; Savelkoul, P.H.; Wolffs, P.F. The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 2013, 4, 87. [Google Scholar] [CrossRef]
- Scalisi, N.; Kuhnert, P.; Amado, M.E.V.; Overesch, G.; Stärk, K.D.; Ruggli, N.; Jores, J. Seroprevalence of Mycoplasma hyopneumoniae in sows fifteen years after implementation of a control programme for enzootic pneumonia in Switzerland. Vet. Microbiol. 2022, 270, 109455. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). M100-S15 Vol. 25, No. 1. Available online: https://clsi.org/standards/products/microbiology/documents/m100/ (accessed on 16 November 2023).
- Aguilar-Vega, C.; Scoglio, C.; Clavijo, M.J.; Robbins, R.; Karriker, L.; Liu, X.; Martínez-López, B. A tool to enhance antimicrobial stewardship using similarity networks to identify antimicrobial resistance patterns across farms. Sci. Rep. 2023, 13, 2931. [Google Scholar] [CrossRef] [PubMed]

| Farm | Farm Isolation | Swine Herds Distance | Road Distance | Breeders Reposition | Quaran-tine | Vectors Control | Type of Feed | Feed Transport | Vehicle Disinfection | Human Access | Bio Score * |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1 | 0 | 0.5 | 1 | 0.5 | 1 | 1 | 1 | 1 | 1 | 8 |
| B | 0.75 | 1 | 0.75 | 1 | 0 | 0.25 | 1 | 1 | 0.25 | 1 | 7 |
| C | 1 | 0.75 | 0.25 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| D | 0.5 | 1 | 0.75 | 0.5 | 0 | 0.5 | 1 | 1 | 0.75 | 1 | 7 |
| E | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 6 |
| F | 0.5 | 1 | 0.75 | 1 | 0 | 0.5 | 1 | 1 | 0.25 | 1 | 7 |
| G | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0.5 | 0.5 | 7 |
| H | 0.25 | 1 | 0.75 | 1 | 0 | 1 | 1 | 1 | 0.5 | 0.5 | 7 |
| I | 1 | 1 | 0.25 | 1 | 0 | 1 | 1 | 1 | 0.25 | 0.5 | 7 |
| J | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0.5 | 0.5 | 6 |
| Pharmacologic Class | Antimicrobials | Concentration in Disk |
|---|---|---|
| Aminoglycoside | Amikacin (AMI) | 30 µg |
| Gentamicin (GEN) | 10 µg | |
| Neomycin (NEO) | 30 µg | |
| Amphenicol | Florfenicol (FLF) | 30 µg |
| Beta-lactam | Amoxicillin + Clavulanic acid (AMC) | 20 µg |
| Amoxicillin (AMO) | 30 µg | |
| Ampicillin (AMP) | 10 µg | |
| Penicillin | Penicillin (PEN) | 30 µg |
| Cephalosporine | Cephalothin (CFL) | 30 µg |
| Cephalexin (CFE) | 30 µg | |
| Ceftiofur (CFT) | 30 µg | |
| Quinolones | Enrofloxacin (ENO) | 5 µg |
| Marbofloxacin (MBO) | 5 µg | |
| Norfloxacin (NOR) | 10 µg | |
| Lincosamide | Clindamycin (CLI) | 2 µg |
| Macrolide | Erythromycin (ERI) | 15 µg |
| Tylosin (TLS) | 60 µg | |
| Tulathromycin (TUL) | 30 µg | |
| Tetracycline | Tetracycline (TET) | 30 µg |
| Doxycycline (DOX) | 30 µg | |
| Polypeptide | Bacitracin (BC) | 10 µg |
| Sulphonamide | sulfametoxazol (SUL) | 300 µg |
| sulfametoxazol-trimetoprim (SUT) | 25 µg |
| Farm | AMB Agents | Vaccines | Production Type |
|---|---|---|---|
| A | AMO, CLI, TET, ENO, OXY | M. hyopneumoniae, circovirus, P. multocida, S. ser. Typhimurium | One-site-herd: piglet unit production |
| B | AMO, FLF, PEN | M. hyopneumoniae, circovirus, G. parasuis, S. suis. | One-site-herd: piglet unit production |
| C | AMO, FLF | P. multocida, B. bronchiseptica, G. parasuis, S. ser. Typhimurium, E. coli. | Two-site-herd: piglet and gilt and young boar production |
| D | AMO, FLF | G. parasuis | One-site-herd: piglet unit production |
| E | AMO | None | Farrow-to-finish: piglet to hog-finished production |
| F | AMO, FLF, PEN | P. multocida, S. ser. Typhimurium, S. suis. | One-site-herd: piglet unit production |
| G | AMO | None | Farrow-to-finish: piglet to hog-finished production |
| H | AMO, FLF | M. hyopneumoniae, circovirus, P. multocida, S. ser. Typhimurium, S. suis | One-site-herd: piglet unit production |
| I | AMO, FLF, TYL | M. hyopneumoniae, circovirus, P. multocida, G. parasuis, S. ser. Typhimurium | One-site-herd: piglet unit production |
| J | AMO, CLI | P. multocida, G. parasuis | Farrow-to-finish: piglet to hog-finished production |
| Farm | PCR | Culture |
|---|---|---|
| A | P. multocida | A. suis; P. multocida; S. suis. |
| B | P. multocida; G. parasuis | A. suis; P. multocida; S. suis. |
| C | G. parasuis | A. suis; S. suis |
| D | P. multocida | B. bronchiseptica |
| E | P. multocida | A. suis; B. bronchiseptica; P. multocida; Yersinia enterocolitica |
| F | G. parasuis | B. bronchiseptica |
| G | P. multocida | A. suis; B. bronchiseptica; P. multocida; S. suis; Y. enterocolitica |
| H | P. multocida | A. suis; P. multocida; Y. enterocolitica |
| I | G. parasuis | A. suis; S. suis |
| J | A. pleuropneumoniae | S. ser. Typhimurium; S. aureus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rigueira, L.L.; de Sant’Ana, F.J.F.; Dallago, B.S.L.; de Faria, R.S.A.; Rodrigues, M.M.; Obregon-Gutierrez, P.; Aragon, V.; Perecmanis, S. Nasal Colonizers from Sows in the Federal District of Brazil Showed a Diverse Phenotypic Resistance Profile. Microorganisms 2025, 13, 1354. https://doi.org/10.3390/microorganisms13061354
Rigueira LL, de Sant’Ana FJF, Dallago BSL, de Faria RSA, Rodrigues MM, Obregon-Gutierrez P, Aragon V, Perecmanis S. Nasal Colonizers from Sows in the Federal District of Brazil Showed a Diverse Phenotypic Resistance Profile. Microorganisms. 2025; 13(6):1354. https://doi.org/10.3390/microorganisms13061354
Chicago/Turabian StyleRigueira, Luciana Lana, Fabiano José Ferreira de Sant’Ana, Bruno Stéfano Lima Dallago, Rômulo Salignac Araújo de Faria, Maurício Macedo Rodrigues, Pau Obregon-Gutierrez, Virginia Aragon, and Simone Perecmanis. 2025. "Nasal Colonizers from Sows in the Federal District of Brazil Showed a Diverse Phenotypic Resistance Profile" Microorganisms 13, no. 6: 1354. https://doi.org/10.3390/microorganisms13061354
APA StyleRigueira, L. L., de Sant’Ana, F. J. F., Dallago, B. S. L., de Faria, R. S. A., Rodrigues, M. M., Obregon-Gutierrez, P., Aragon, V., & Perecmanis, S. (2025). Nasal Colonizers from Sows in the Federal District of Brazil Showed a Diverse Phenotypic Resistance Profile. Microorganisms, 13(6), 1354. https://doi.org/10.3390/microorganisms13061354








