Abstract
The rapid spread of tet(X) genes capable of inactivating tigecycline represents a critical challenge to global public health. This study aims to explore the distribution, genetic diversity, and transferability of tet(X) genes in Myroides, a genus of Gram-negative bacteria increasingly implicated in multidrug-resistant (MDR) bacterial infections. From 2021 to 2024, 646 samples of chicken, sheep, soil, and water were randomly collected, yielding nine chicken-derived tet(X)-positive Myroides sp. strains in Shandong, China. All of them were MDR to tetracycline, ceftazidime, gentamicin, amikacin, colistin, ciprofloxacin, gatifloxacin, and trimethoprim-sulfamethoxazole, with elevated minimum inhibitory concentrations (MICs) for tigecycline, florfenicol, and macrolides, but exhibited susceptibility to meropenem (100%), ampicillin-sulbactam (66.7%), and cefotaxime (33.3%). A genomic analysis of the isolates and 86 public tet(X)-positive Myroides genomes revealed the widespread distribution of tet(X) and macrolide-inactivating estT genes across 12 Myroides species, including 7 novel species. Eight tet(X) and eight estT variants were identified, half of which were novel. The phylogenetic analysis highlighted interspecies transmission risks, with ISCR2-mediated transposons of tet(X6) and estT-2 across Myroides, Riemerella, Empedobacter, Providencia, Acinetobacter, and Proteus species. These findings illuminate the genomic diversity driving antibiotic resistance in understudied bacterial taxa, with implications for global One Health strategies.
1. Introduction
Myroides, a genus of non-motile, non-fermenting, aerobic, and Gram-negative bacteria belonging to Flavobacteriaceae, comprises 17 validly published and correct species [1,2]. Traditionally, these bacteria are considered non-pathogenic, but recent clinical reports have revealed their pathogenicity, particularly in immunocompromised patients [2,3,4,5]. While the infections caused by Myroides spp. of clinical significance remain limited in scope, surveillance studies show increasing detection rates that may be attributable to advances in molecular diagnostic techniques [3]. Notably, clinically relevant Myroides species such as Myroides odoratimimus and Myroides odoratus demonstrate extensive antimicrobial resistance profiles encompassing β-lactam and aminoglycoside antibiotics, thereby exacerbating therapeutic limitations and emerging as a growing concern in nosocomial infection [2,6,7,8].
As a third-generation tetracycline derivative and the first glycylcycline antibacterial agent, tigecycline holds critical status as a last-line therapeutic option against MDR bacterial pathogens in clinical settings [9]. The compound received initial Food and Drug Administration approval in 2005 for management of complicated intra-abdominal infections and acute bacterial skin/skin structure infections, followed by expanded indications for community-acquired bacterial pneumonia in 2009 [10]. Subsequent regulatory approvals were obtained in the European Union (2006) and China (2011), establishing its global therapeutic application framework supported by accumulating clinical evidence [11,12]. Since the first report in 2019, a variety of tigecycline resistance genes tet(X) have been reported in Enterobacteriaceae, Acinetobacter spp., Empedobacter spp., Elizabethkingia spp., Riemerella anatipestifer, and Pseudomonas caeni in China [13,14,15,16,17,18,19,20,21].
Recently, the tet(X) genes were reported in M. odoratimimus, M. odoratus, and Myroides phaeus from inpatient, pig, fish, and soil samples [6,8,13,22,23]. However, the genetic diversity and transferability of tet(X) genes in Myroides spp. remained poorly understood. In this study, we intend to explore the prevalence, antimicrobial susceptibility, phylogenetic relationship, and transmission risk of tet(X)-positive Myroides sp. isolates, together with blast querying in the public database.
2. Materials and Methods
2.1. Sample Collection and Bacterial Isolation
During a surveillance across three Chinese provinces, Shandong, Jiangsu, and Ningxia, 646 non-redundant samples were randomly collected from 2021 to 2024, comprising chicken feces (n = 449), sheep feces (n = 95), and surrounding environmental samples (soil, n = 60; water, n = 42). Specimens were homogenized in 0.9% sterile saline (1:5 w/v for solids; 1:5 v/v for liquids) with vortex mixing, and 100 μL of supernatant was plated on Luria–Bertani (LB) agar plates supplemented with tigecycline (4 mg/L) for selective isolation of tigecycline-resistant strains. Presumptive tet(X)-positive Myroides sp. strains underwent molecular confirmation via PCR amplification of conserved 16S rRNA regions and tet(X)-specific primers, followed by bidirectional Sanger sequencing [24,25].
2.2. Antibiotic Resistance Evaluation
MICs were determined by two-fold Mueller–Hinton (MH) agar dilution according to the Clinical and Laboratory Standards Institute guidelines [26]. Both Myroides spp. and Acinetobacter spp. are non-motile, non-fermenting, aerobic, and Gram-negative bacteria [2,27], and therefore, Acinetobacter spp. breakpoints were used for interpreting MICs of Myroides spp. in this study, which lacked the standardized breakpoints. These antibiotics (Yuanye, Shanghai, China) included tetracycline (dilution range, 0.5–256 mg/L), amikacin (0.5–256 mg/L), gentamicin (0.25–256 mg/L), ciprofloxacin (0.0039–64 mg/L), gatifloxacin (0.0078–16 mg/L), colistin (0.25–256 mg/L), trimethoprim-sulfamethoxazole (0.5/9.5–16/304 mg/L), ampicillin-sulbactam (0.125/0.0625–256/128 mg/L), ceftazidime (0.125–256 mg/L), cefotaxime (0.03125–256 mg/L), meropenem (0.0078–64 mg/L), tigecycline (0.03125–64 mg/L), florfenicol (2–256 mg/L mg/L), tylosin (0.25–512 mg/L), tilmicosin (0.125–256 mg/L), and tildipirosin (0.125–256 mg/L), of which the last five antibiotics lacked resistance breakpoints. For quality standardization, the reference strain Escherichia coli ATCC 25922 was systematically incorporated as the antimicrobial susceptibility testing control organism.
2.3. Whole Genome Sequencing (WGS)
The genomic landscape of tet(X)-harboring Myroides sp. isolates was characterized through integrated sequencing strategies. Primary sequencing was conducted using the Illumina NovaSeq 6000 platform (ANOROAD, Beijing, China) with 2 × 150 bp paired-end sequencing, followed by de novo assembly via SPAdes version 3.15.5 (Russian Academy of Sciences, St. Petersburg, Russia) [28]. M. odoratimimus C26-4 and C34-1 then underwent third-generation sequencing using Oxford Nanopore PromethION (BENAGEN, Wuhan, China) with ultra-long read strategies, achieving circularized genomes through hybrid assembly with Unicycler version 0.5.1 (University of Melbourne, Melbourne, Australia) [29]. Complementary to experimental data, all publicly available WGS data of tet(X)-positive Myroides sp. strains were retrieved from the NCBI database [30]. All the genome assemblies were checked by CheckM version 1.1.6 (University of Queensland, Brisbane, Australia) and Quast version 5.2.0 (Russian Academy of Sciences, St. Petersburg, Russia), which were defined by <350 contigs, >50 kb N50, >95% genome completeness, <2 genome contamination, and <50 genome heterogeneity [31,32].
2.4. Bioinformatics Analyses
Genomic analyses were conducted using IPGA version 1.09 (Chinese Academy of Sciences, Beijing, China) to determine average nucleotide identity (ANI) and construct a core single-nucleotide polymorphism (SNP) phylogeny for tet(X)-positive Myroides spp., with species delineation based on a >95% ANI threshold against LPSN type strains [33,34,35]. All Myroides genomes were further confirmed by in silico DNA–DNA hybridization (isDDH) analyses and classified by a >70% isDDH threshold [35]. Antibiotic resistance genes (ARGs) and virulence factors were identified by >80% identity and >60% coverage thresholds via ABRicate version 1.0.1 (University of Melbourne, Melbourne, Australia), with phylogeny-heatmap integration performed using ggtreeExtra (Southern Medical University, Guangzhou, China) [36,37]. Maximum likelihood trees of tet(X) or estT variants were generated by MEGA-X version 10.1.8 (Pennsylvania State University, State College, PA, USA) in 500 bootstrap replicates and visualized by FigTree version 1.4.4 (University of Edinburgh, Edinburgh, UK), with novel alleles defined by ≥2% amino acid divergence according to international standards for resistance gene naming [18,38,39]. Three-dimensional structural prediction of Tet(X) and EstT variants was performed through SWISS-MODEL using the experimentally resolved Tet(X2)-tigecycline co-crystal structure (PDB accession number: 4A6N) and our previously simulated structure of EstT-1, respectively [18,40]. Genome annotation utilized RAST version 2.0 (Fellowship for Interpretation of Genomes, Burr Ridge, IL, USA), while chromosomal comparisons of tet(X)-bearing regions employed BRIG version 0.95 (University of Queensland, Brisbane, Australia) [41,42]. Genetic contexts of tet(X) or estT genes were achieved through Easyfig version 2.2.5 (University of Queensland, Brisbane, Australia) [43].
2.5. Phenotypic Experiments of Novel Myroides Species
Morphology of the novel Myroides species we isolated was determined on LB agar plates at 35 °C for 24 h, and through Gram-staining [44]. Anaerobic growth was examined on LB agar plates in an anaerobic bag at 35 °C for 24 h. Growth at different NaCl concentrations (0%, 1%, 2%, 3%, 4%, 5%) or pH (4, 5, 6, 7, 8, 9, 10, 11, 12) was performed in LB broth at 35 °C for 24 h. Growth at various temperatures (20 °C, 25 °C, 30 °C, 35 °C, 37 °C, 38 °C, 39 °C, 40 °C, 41 °C, 42 °C) was tested on LB agar plates for 24 h. Bacterial motility was tested in LB medium with 0.4% agar at 35 °C for 24 h. Hemolysis was examined on LB agar plates containing 5% sheep blood at 35 °C for 24 h. Physiological activities on glucose, oxidase, citrate, maltose, arginine dihydrolase, mannitol, xylose, nitrate reduction, DNA, and acetamide were detected by a non-fermenting bacterial identification kit (HuanKai, Guangzhou, China). M. odoratimimus ATCC BAA-634 was used as the reference standard.
2.6. Cloning Expression
The novel tet(X) and estT variants we identified were directionally cloned into the L-arabinose-inducible pBAD24 expression vector with EcoR I/Sal I and Nhe I/Sal I restriction sites (Table S1), respectively, followed by electroporation into chemically competent E. coli JM109 cells. Transformants were selected on LB agar under ampicillin pressure (100 mg/L) and validated through colony PCR and bidirectional Sanger sequencing [45]. For MIC determination, log-phase cultures induced with 0.1% L-arabinose were subjected to two-fold broth microdilution, with tetracyclines (tetracycline/doxycycline/minocycline/tigecycline) or macrolides (tylosin/tilmicosin/tildipirosin) tested [26]. Isogenic control strains harboring tet(X2), tet(X6), tet(X6)-tet(X2), estT-1.2, estT-2, or empty vector were included for phenotypic benchmarking [18,20].
2.7. Transfer of tet(X) Genes
As documented in our prior research [18], the horizontal transfer potential of tet(X)-mediated tigecycline resistance was evaluated through biparental conjugation assays using rifampicin-resistant Acinetobacter baylyi ADP1 and E. coli C600 as recipients. Following 16-h incubation at a donor-to-recipient ratio of 1:3, putative transconjugant colonies were isolated on selective LB agar plates supplemented with tigecycline (2 mg/L) and rifampin (125 mg/L). All candidate colonies subsequently underwent molecular confirmation through tet(X)-specific PCR amplification, followed by species-specific PCR fingerprinting for A. baylyi identification and enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) genotyping for E. coli validation [46,47]. In addition, natural transformation was conducted to explore the transferability of tet(X) into A. baylyi ADP1, E. coli C600, and M. odoratimimus ATCC BAA-634 [48].
3. Results
3.1. Sporadic Detection of tet(X)-Positive MDR Myroides spp.
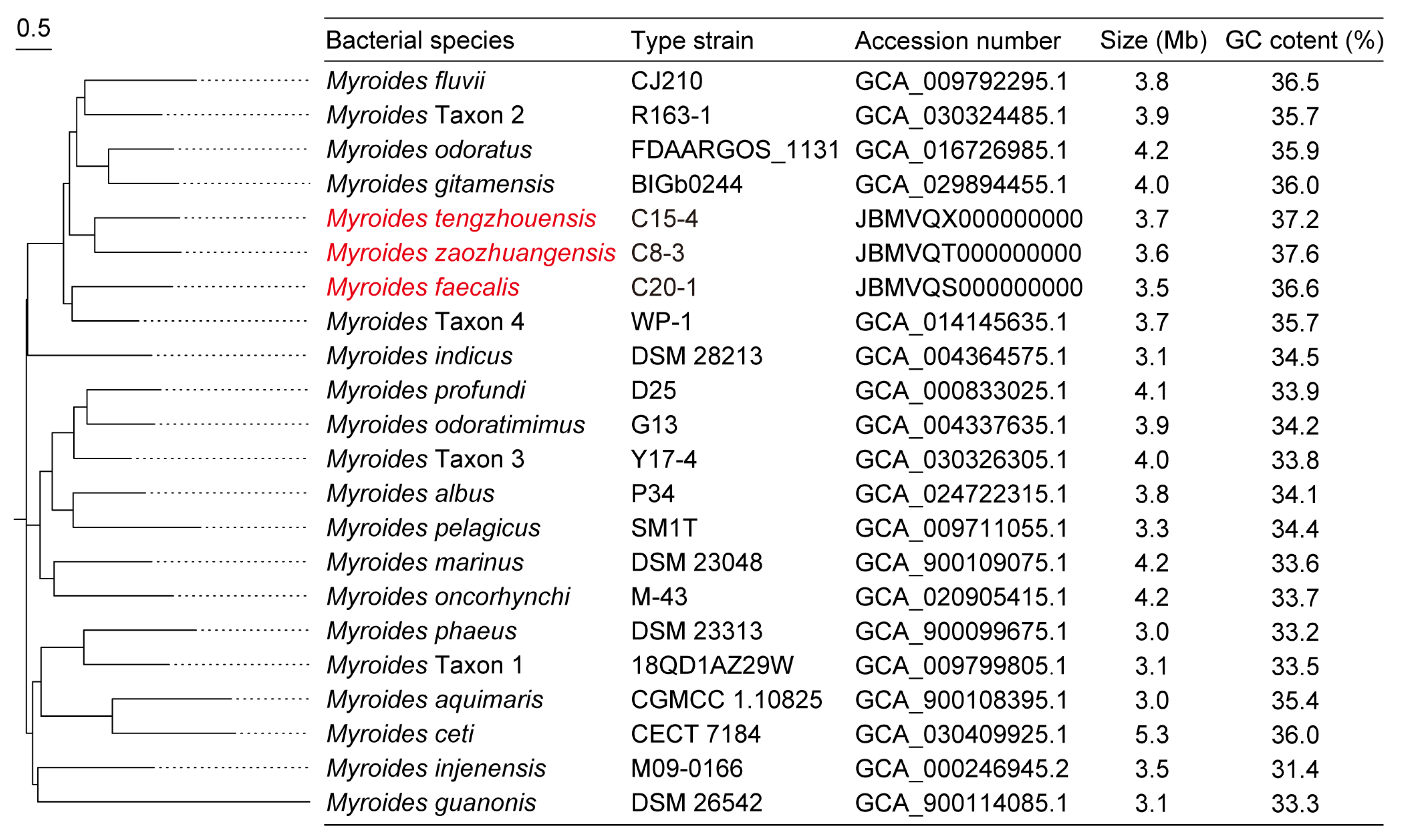
In this study, nine tet(X)-positive Myroides sp. strains were isolated from chicken samples in Shandong, but were negative in sheep, soil, and water samples in Shandong, Jiangsu, and Ningxia, China. These isolates contained five M. odoratimimus and four novel Myroides sp. strains of Myroides tengzhouensis (n = 1), Myroides faecalis (n = 2), and Myroides zaozhuangensis (n = 1), which shared less than 95% intra-species ANI threshold and 70% intra-species isDDH threshold with the reference Myroides species (Figure 1). As shown in Table 1, MICs of 16 antibiotics were tested, of which 12 antibiotics (except florfenicol, tylosin, tilmicosin, and tildipirosin) have been approved for the treatment of human infections in China. All of these strains were MDR to tetracycline, ceftazidime, gentamicin, amikacin, colistin, ciprofloxacin, gatifloxacin, and trimethoprim-sulfamethoxazole but exhibited susceptibility to meropenem (100%), ampicillin-sulbactam (66.7%), and cefotaxime (33.3%). Meanwhile, MIC90 of tigecycline, florfenicol, tylosin, tilmicosin, and tildipirosin were 16 mg/L, 64 mg/L, 512 mg/L, 64 mg/L, and 32 mg/L, respectively.
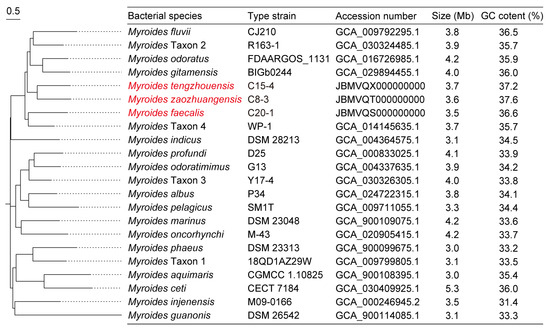
Figure 1.
SNP-based phylogenetic tree of 22 different Myroides species. There are seven novel species named Myroides tengzhouensis, Myroides zaozhuangensis, Myroides faecalis, and Myroides Taxon 1–Taxon 4, respectively, of which the novel Myroides species we isolated are marked in red. Type strains and their GenBank accession numbers, genome sizes, and GC contents are present in parallel. Bar, 0.5 nucleotide substitutions per site.

Table 1.
MICs of tet(X)-positive Myroides sp. isolates.
By querying the public NCBI database (Figure 2), another 86 tet(X)-positive Myroides sp. strains were collected in China (n = 81), Turkey (n = 2), Mexico (n = 2), and the USA (n = 1). In brief, the tet(X) genes were widely distributed in 10 different Myroides species, such as M. odoratimimus (n = 55), Myroides marinus (n = 18), M. phaeus (n = 1), Myroides pelagicus (n = 1), Myroides injenensis (n = 1), and five novel Myroides species of M. zaozhuangensis (n = 2), Myroides Taxon 1 (n = 3), Myroides Taxon 2 (n = 3), Myroides Taxon 3 (n = 1), and Myroides Taxon 4 (n = 1). Source tracing of bacterial strains indicated chicken (n = 33) is the main reservoir of tet(X)-positive Myroides spp., followed by homo sapiens (n = 17), fish (n = 17), fly (n = 11), pig (n = 3), soil (n = 3), cattle (n = 1), and urban (n = 1) samples. Despite the lack of sample sizes and MIC data, these online genomes indicated the sporadic detection and bacterial diversity of Myroides sp. strains carrying tet(X) genes worldwide.

Figure 2.
Phylogeny and ARGs of tet(X)-positive Myroides sp. strains. Bacterial species, geographical location, sampling sources, and GenBank accession numbers are present in parallel. The tet(X)-positive Myroides sp. strains we isolated are marked in red. In addition, the tet(X) and estT genes are colored in brown and cyan, respectively, and the remainder are colored in green. Bar, 0.5 nucleotide substitutions per site.
3.2. Polymorphism of tet(X) and estT
Computational resistance gene profiling of 95 tet(X)-harboring Myroides sp. strains revealed five classes of ARGs for tetracyclines, β-lactams, macrolides, phenicols, and sulfonamides, of which 11.6% carried two tet(X) variants (Figure 2). Among eight tet(X) variants characterized, four were previously annotated ones encompassing tet(X2) (n = 86), tet(X2.2) (n = 4), tet(X6) (n = 4), and tet(X6)-tet(X2) (n = 3). The remaining four novel variants were designated under standardized nomenclature as tet(X2.4) (n = 2), tet(X2.5) (n = 1), tet(X18.2) (n = 1), and tet(X24.2) (n = 5). Particularly, tet(X2) (n = 7), tet(X6) (n = 2), tet(X6)-tet(X2) (n = 3), and tet(X24.2) (n = 4) were identified in our epidemiological investigation. Functional validation of them through heterologous expression in E. coli JM109 demonstrated 2–64-fold MICs against tetracycline, doxycycline, minocycline, and tigecycline compared to an isogenic empty-vector control (Table 2). Homology modeling confirmed Tet(X6), Tet(X6)-Tet(X2), Tet(X18.2), and Tet(X24.2) with a lack of 10 N-terminal amino acids shared six key amino acid residues at S272, M319, T329, N330, I340, and E341, as previously reported [49,50], leading to a high-level resistance phenotype for tetracyclines, while tet(X2), tet(X2.2), tet(X2.4), and tet(X2.5) exhibited a low-level activity (Figure S1).

Table 2.
MICs of the tet(X) and estT clones.
It is noted that 58 (61.1%) out of 95 tet(X)-positive Myroides sp. strains carried estT genes, of which 10.3% carried two estT variants (Figure 2). These variants included estT-1 (n = 3), estT-1.2 (n = 40), estT-1.4 (n = 4), estT-2 (n = 9), and four novel variants (n = 8). According to the gene assignment rule, the novel variants were designated as estT-1.8 (n = 3), estT-1.9 (n = 3), estT-1.10 (n = 1), and estT-2.2 (n = 1). Particularly, estT-1.2 (n = 4), estT-1.8 (n = 3), and estT-2 (n = 6) were identified in our epidemiological investigation. MIC results showed E. coli clones of them that exhibited 2–4-fold increases for 16–atom–containing tylosin, tilmicosin, and tildipirosin of macrolides compared to an isogenic empty-vector control, suggesting a low-level activity (Table 2). Homology modeling of eight EstT proteins was also conducted, with a lack of N-terminal random coil of EstT-1, and it remains to be studied (Figure S2).
3.3. Novel Myroides Species
To date, a total of 22 Myroides species have been reported with different genome sizes but have a similar GC content ranging from 31.4% to 37.6% (Figure 1). The physiological characteristics of novel species M. zaozhuangensis C8-3, M. tengzhouensis C15-4, and M. faecalis C20-1 were analyzed against M. odoratimimus ATCC BAA-634 and described as below (Table S2).
3.3.1. Description of Myroides zaozhuangensis sp. nov.
Myroides zaozhuangensis (zao.zhuang.en’sis. N.L. masc. adj. zaozhuangensis, referring to Zaozhuang, Shandong, China).
Cells are Gram-staining-negative, rod-shaped, aerobic, non-motile, and non-hemolytic. Colonies are light yellow and circular, with a smooth surface and regular margin after 24 h of incubation at LB agar. It survives at pH 5–9, NaCl up to 4%, and temperature up to 41 °C. The tests are negative for glucose, citrate, maltose, mannitol, xylose, and nitrate reduction, but positive for oxidase, arginine dihydrolase, DNA hydrolysis, and acetamide utilization.
The type strain C8-3T (GDMCC 66301) was isolated from a chicken manure sample in 2021 in Shandong, China. The size of whole-genome sequences of the type strain is 3.6 Mb, with a G + C content of 37.6%, which has been deposited in the NCBI database under the GenBank accession number: JBMVQT000000000.
3.3.2. Description of Myroides tengzhouensis sp. nov.
Myroides tengzhouensis (teng.zhou.en’sis. N.L. masc. adj. tengzhouensis, referring to Tengzhou, Shandong, China).
Cells are Gram-staining-negative, rod-shaped, aerobic, non-motile, and non-hemolytic. Colonies are light yellow and circular, with a smooth surface and regular margin after 24 h of incubation on LB agar. It survives at pH 6–8, NaCl up to 3%, and temperature up to 38 °C. The tests are negative for glucose, citrate, maltose, mannitol, xylose, and nitrate reduction, but positive for oxidase, arginine dihydrolase, DNA hydrolysis, and acetamide utilization.
The type strain C15-4T (GDMCC 66292) was isolated from a chicken manure sample in 2021 in Shandong, China. The size of whole-genome sequences of the type strain is 3.7 Mb, with a G + C content of 37.2%, which has been deposited in the NCBI database under the GenBank accession number: JBMVQX000000000.
3.3.3. Description of Myroides faecalis sp. nov.
Myroides faecalis (fae.ca’lis. N.L. masc. adj. faecalis, pertaining to feces).
Cells are Gram-staining-negative, rod-shaped, aerobic, non-motile, and non-hemolytic. Colonies are light yellow and circular, with a smooth surface and regular margin after 24 h of incubation on LB agar. It survives at pH 6–8, NaCl up to 4%, and temperature up to 38 °C. The tests are negative for glucose, citrate, maltose, mannitol, xylose, and nitrate reduction, but positive for oxidase, arginine dihydrolase, DNA hydrolysis, and acetamide utilization.
The type strain C20-1T (GDMCC 66293) was isolated from a chicken manure sample in 2021 in Shandong, China. The size of whole-genome sequences of the type strain is 3.5 Mb, with a G + C content of 36.6%, which has been deposited in the NCBI database under the GenBank accession number: JBMVQS000000000.
3.4. Phylogeny of Myroides spp.
Following SNP analyses, an SNP-based phylogenetic tree of 95 tet(X)-positive Myroides sp. strains was conducted for bacterial relationship (Figure 2). The tree revealed M. marinus, M. phaeus, M. pelagicus, M. injenensis, M. tengzhouensis, M. zaozhuangensis, and Myroides Taxon 1–Taxon 4 formed 10 separate clusters, except the complex of M. odoratimimus and M. faecalis. All the novel Myroides species shared 179-4099 SNPs with the reference strain M. odoratimimus G13 (GenBank accession number: GCA_004337635.1), and there existed a novel bacterial evolutionary branch consisting of M. tengzhouensis, M. zaozhuangensis, Myroides Taxon 2, and Myroides Taxon 4. Scarcely, the clonal transmission risk of tet(X) genes occurred in M. odoratimimus, M. marinus, M. faecalis, M. zaozhuangensis, Myroides Taxon 1, or Myroides Taxon 2, indicating the potential importance of the horizontal transmission route.
3.5. ISCR2-Mediated Transposons of tet(X) and estT
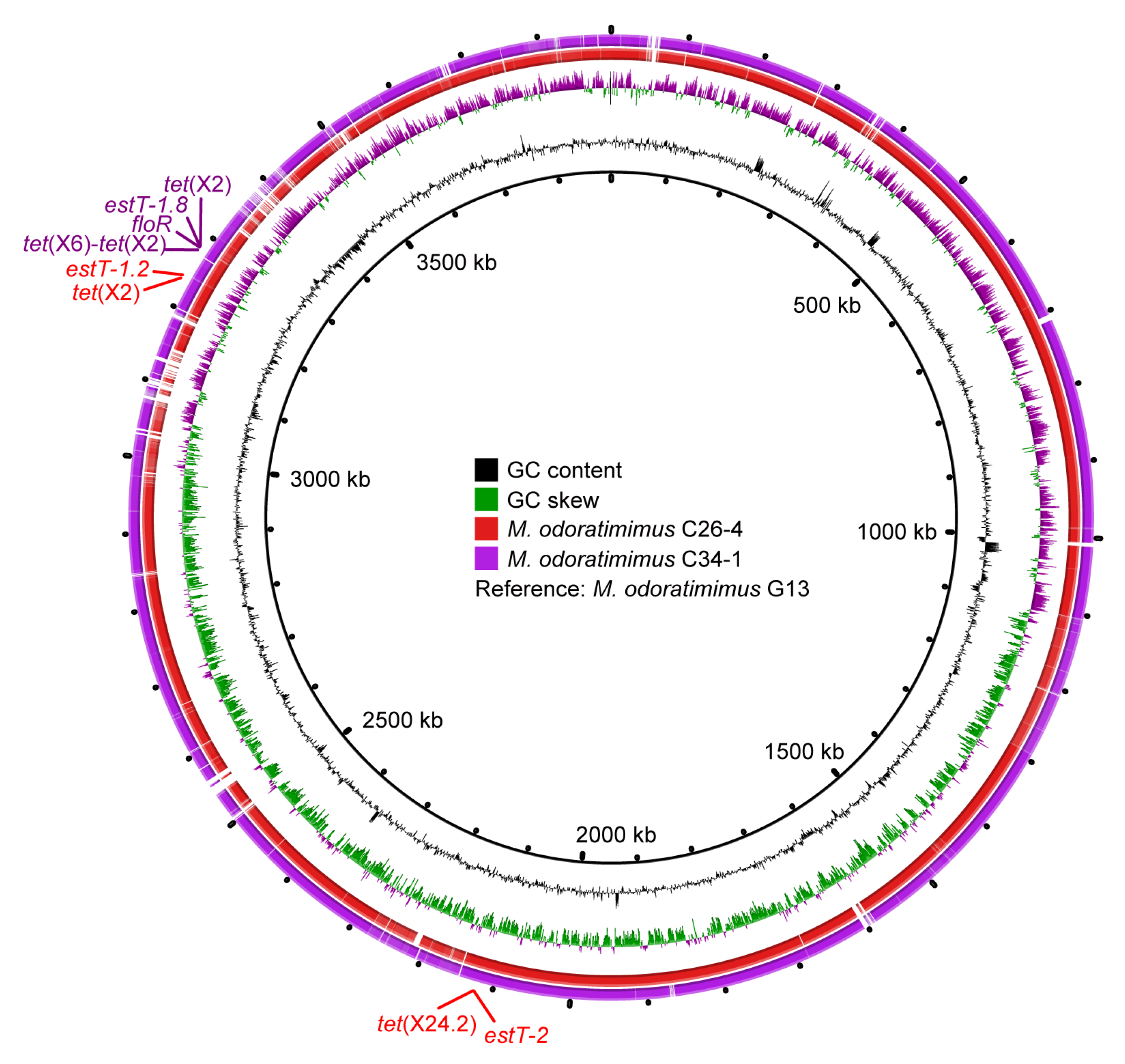
To analyze the molecular location of tet(X) and estT genes, NovaSeq and Nanopore sequencing of M. odoratimimus C26-4 and C34-1 were conducted. WGS results of M. odoratimimus C26-4 indicated the tet(X2) and tet(X24.2) genes were distantly located on a single chromosome (3,956,267 bp, CP182307), together with macrolide resistance genes estT-1.2 and estT-2, respectively (Figure 3). In M. odoratimimus C34-1, the tet(X2) and tet(X6)-tet(X2) genes were tandemly located on a single chromosome (3,983,038 bp, CP182237), together with estT-1.8 and phenicol resistance gene floR (Figure 3). A further comparative analysis confirmed they were highly homologous to the reference chromosome (3894807 bp, CP037427) of water-derived M. odoratimimus G13 (Figure 3), which also contained an untypable plasmid (43223 bp, CP037428).
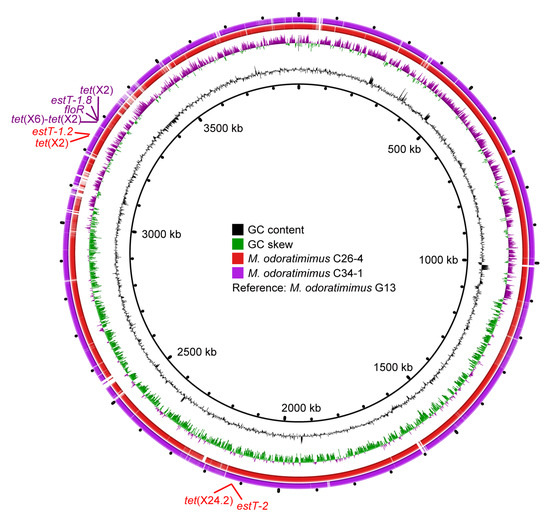
Figure 3.
Comparative analysis of the tet(X)-carrying Myroides chromosomes. GC content, GC skew, M. odoratimimus C26-4 (CP182307), and M. odoratimimus C34-1 (CP182237) are from inside out, with M. odoratimimus G13 (CP037427) as the reference strain. Antibiotic resistance gene clusters for M. odoratimimus C26-4 and M. odoratimimus C34-1 are also marked in red and purple, respectively.
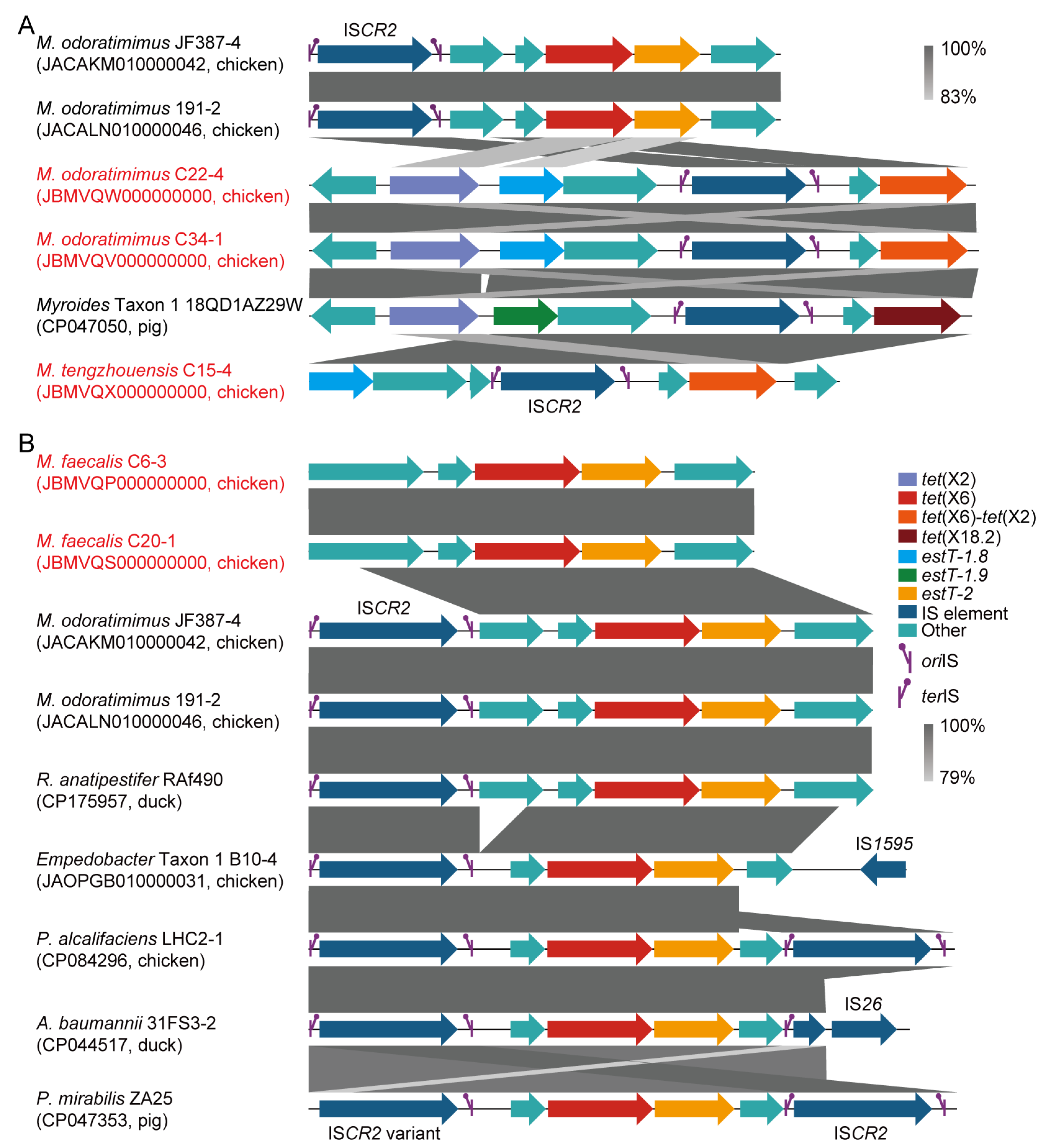
By analyzing the insertion sequence (IS), 6 out of 95 (6.3%) tet(X)-positive Myroides sp. strains were positive for ISCR2. In detail, the ISCR2-mediated transposon units of tet(X6)/estT-2, tet(X2)/tet(X6)-tet(X2)/estT-1.8, tet(X2)/tet(X18.2)/estT-1.9, and tet(X6)-tet(X2)/estT-1.8 were identified in M. odoratimimus (e.g., JACAKM010000042), M. tengzhouensis (e.g., JBMVQX000000000), and Myroides Taxon 1 (e.g., CP047050; Figure 4A). Additionally, an ISCR2-mediated transposon of tet(X6)/estT-2 was identified across M. odoratimimus (e.g., JACAKM010000042), Riemerella anatipestifer (e.g., CP175957), Empedobacter Taxon 1 (e.g., JAOPGB010000031), Providencia alcalifaciens (e.g., CP084296), Acinetobacter baumannii (e.g., CP044517), and Proteus mirabilis (e.g., CP047353) from food-producing animals in the NCBI database (Figure 4B). However, the tet(X) genes failed to be transferred into A. baylyi ADP1, E. coli C600, and M. odoratimimus ATCC BAA-634 by conjugation or natural transformation in this study. For the remaining 89 strains, all were negative for ISCR2, and their horizontal transmission risk also needed to be further confirmed.
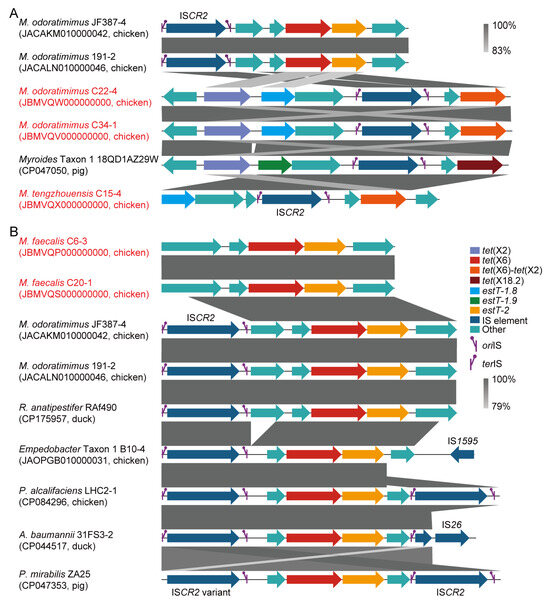
Figure 4.
Genetic characteristics of tet(X) and estT genes. (A) ISCR2-associated tet(X) and estT loci in Myroides species, with regions sharing >83% nucleotide identity highlighted in grey. (B) Interspecific occurrence of tet(X6) and estT-2, with conserved regions sharing >79% nucleotide identity colored in grey. The strains highlighted in red are Myroides spp. we isolated in this study. The tet(X), estT, IS element, ISCR2 replication initiation site (oriIS), ISCR2 replication termination site (terIS), and other genes are also indicated in different colors.
4. Discussion
Myroides spp., especially M. odoratimimus, appear as a group of sporadically reported clinical pathogens [2,6]. Before this study, the tet(X) genes had been reported in M. odoratimimus, M. odoratus, and M. phaeus isolates from homo sapiens, animal, and environmental samples [6,13,22,23,51]. It is noted that our study demonstrated a low prevalence of tet(X)-positive MDR M. odoratimimus, M. tengzhouensis, M. faecalis, and M. zaozhuangensis in chicken samples. Worrisomely, the tet(X)-positive M. odoratimimus, M. marinus, M. phaeus, M. pelagicus, M. injenensis, M. zaozhuangensis, and Myroides Taxon 1–4 genomes were detected in the public NCBI database. These tet(X)-positive Myroides species highlighted the genomic diversity, which is seriously underestimated. According to the isolation source, animals accounted for the majority (77.9%) of 95 tet(X)-positive Myroides sp. strains, and an SNP-based phylogenetic tree indicated that clonal transmission rarely occurs, underlying the zoonotic driver of horizontal transmission of tet(X) genes in Myroides species.
Since the first report in Bacteroides fragilis [52], a total of 54 non-duplicate tet(X) variants have been detected in a variety of Gram-negative bacteria, indicating the rapid evolutionary rate over the past decades (Figure S3). Structurally, a combination of six key amino acid substitutions of Tet(X) proteins has been reported, leading to enhanced degradation activity [49,50]. In this study, we identified eight tet(X) variants in Myroides spp., of which tet(X6), tet(X6)-tet(X2), and tet(X24.2) were confirmed to confer tigecycline resistance, and tet(X18.2) with six similar amino acid substitutions was also speculated for tigecycline resistance. With the continuous emergence of tet(X) variants, their differences in resistance phenotypes need to be evaluated and focused [53,54].
For estT genes (Figure S4), there have been 15 non-duplicate variants since the first report in Sphingobacterium faecium from water in 2023 [55]. Here, we reported eight estT variants in tet(X)-positive Myroides sp. strains, including four novel variants, and estT-1.2, estT-1.8, and estT-2 exhibited 2–4-fold increases of MICs against 16-atom-containing macrolides. In contrast, the estT genes have a similar three-dimensional structure and antibacterial activity to those of previously reported tet(X)-positive Empedobacter spp., but their hydrolyzation mechanisms need to be confirmed [18]. All the available data indicated that Myroides species act as the reservoir of tet(X) and estT gene clusters. There is no evidence for their co-expression or co-regulation; however, the co-existence of functional tet(X) and estT genes in this study signifies the co-dissemination risk.
ISCR2 is an atypical insertion sequence that can transpose adjacent ARGs [e.g., floR and sul2] through a rolling-circle transposition process [56]. Despite the low frequency of Myroides species, 99% and 100% of tet(X) genes were mediated by ISCR2 in Acinetobacter species and E. coli, respectively [45,57]. Our previous studies confirmed the ISCR2-mediated transposition ability of tet(X3), tet(X4), and tet(X5) genes in Acinetobacter species and Aeromonas caviae [12,45,58]. Although failing to be transferred by conjugation in this study, we identified a series of ISCR2-mediated transposon units of tet(X) and estT variants in M. odoratimimus, M. tengzhouensis, and Myroides Taxon 1. Additionally, a similar ISCR2-mediated transposition structure of tet(X6) and estT-2 was detected in public Myroides, Riemerella, Empedobacter, Providencia, Acinetobacter, and Proteus genomes, indicating a risk of interspecies dissemination [12,16,18]. With the widespread use and residue in animals, humans, and environments, tetracycline and macrolide antibiotics may facilitate the formation and mobilization of tet(X)/estT gene clusters, but further exploration is needed.
5. Conclusions
In summary, this study highlights Myroides spp. as critical reservoirs of diverse tet(X) and estT variants, driving resistance to tetracyclines and macrolides, especially in poultry settings. The identification of novel tet(X), estT, and Myroides species, alongside ISCR2-mediated transposons, underscores the genomic plasticity enabling resistance dissemination across species boundaries. Despite non-conjugative tet(X) genes, their integration with ISCR2 poses a significant risk for horizontal gene transfer. Our findings advocate stricter antimicrobial stewardship and genomic monitoring to mitigate the global spread of MDR Myroides sp. pathogens, reinforcing the One Health framework in addressing antibiotic resistance crises.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms13061180/s1, Figure S1: Three-dimensional structure of Tet(X) proteins; Figure S2: Three-dimensional structure of EstT proteins; Figure S3: Phylogenetic tree of the tet(X) variants; Figure S4: Phylogenetic tree of the estT variants; Table S1: Primers designed in this study; Table S2: Phenotypic characteristics of the novel Myroides species.
Author Contributions
Conceptualization, C.C.; methodology, C.C. and T.W.; formal analysis, C.C., T.W. and J.L.; investigation, T.W. and J.L.; writing—original draft preparation, T.W.; writing—review and editing, C.C.; visualization, T.W. and Y.L.; project administration, C.C.; funding acquisition, C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was jointly supported by the National Natural Science Foundation of China (grant number 32402890) and the China Postdoctoral Science Foundation (grant number 2023M732993).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Whole-genome sequences of nine tet(X)-positive Myroides sp. isolates are available in the NCBI BioProject repository (PRJNA1224301). Circular chromosome sequences of M. odoratimimus strains C26-4 (PRJNA1224307) and C34-1 (PRJNA1224310) are also submitted.
Acknowledgments
Thanks to Yanan Guo (Ningxia Academy of Agriculture and Forestry Sciences) for sample collection.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| MDR | Multidrug-resistant |
| MICs | Minimum inhibitory concentrations |
| LB | Luria–Bertani |
| MH | Mueller–Hinton |
| WGS | Whole genome sequencing |
| ANI | Average nucleotide identity |
| SNP | Single-nucleotide polymorphism |
| isDDH | in silico DNA-DNA hybridization |
| ARGs | Antibiotic resistance genes |
| ERIC-PCR | Enterobacterial repetitive intergenic consensus PCR |
| IS | Insertion sequence |
References
- LPSN. Available online: https://lpsn.dsmz.de/genus/myroides (accessed on 10 May 2025).
- Gunzer, F.; Rudolph, W.W.; Bunk, B.; Schober, I.; Peters, S.; Muller, T.; Oberheitmann, B.; Schrottner, P. Whole-genome sequencing of a large collection of Myroides odoratimimus and Myroides odoratus isolates and antimicrobial susceptibility studies. Emerg. Microbes Infect. 2018, 7, 61. [Google Scholar] [CrossRef]
- Aworh, M.K.; Colín-Castro, C.A.; Ortiz-Álvarez, J.M.; Hernández-Pérez, C.F.; Hernández-Durán, M.; García-Hernández, M.d.L.; Martínez-Zavaleta, M.G.; Becerra-Lobato, N.; Cervantes-Hernández, M.I.; Rosas-Alquicira, G.; et al. Myroides species, pathogenic spectrum and clinical microbiology sight in Mexican isolates. PLoS ONE 2024, 19, e0310262. [Google Scholar] [CrossRef]
- Kurt, A.F.; Mete, B.; Houssein, F.M.; Tok, Y.; Kuskucu, M.A.; Yucebag, E.; Urkmez, S.; Tabak, F.; Aygun, G. A pan-resistant Myroides odoratimimus catheter-related bacteremia in a COVID-19 patient and review of the literature. Acta Microbiol. Immunol. Hung. 2022, 69, 164–170. [Google Scholar] [CrossRef]
- Sahu, C.; Patel, S.S.; Chaudhary, R.; Bhartiya, C.; Bhatnagar, N. A Retrospective Study on UTI by Myroides Species: An Emerging Drug Resistant Nosocomial Pathogen. Indian J. Crit. Care M 2024, 28, 399–403. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, M.; Fu, J.; Zhong, C.; Zong, G.; Cao, G. Identification of a mobilizable, multidrug-resistant genomic island in Myroides odoratimimus isolated from Tibetan pasture. Sci. Total Environ. 2020, 723, 137970. [Google Scholar] [CrossRef]
- Aygar, I.S.; Aydogan, C.N.; Ozcan, H.; Unat, I.; Fatsa, T.; Tekin, K.; Yalci, A.; Hosbul, T.; Sahiner, F.; Gumral, R. Myroides odoratimimus: A New Threat with Persistent Infections, Multidrug Resistance, and the Potential for Hospital Outbreaks. Jpn. J. Infect. Dis. 2023, 76, 335–342. [Google Scholar] [CrossRef]
- Yartasi, E.; Durmaz, R.; Ari, O.; Mumcuoglu, I.; Dinc, B. Molecular characterization of the multi-drug resistant Myroides odoratimimus isolates: A whole genome sequence-based study to confirm carbapenem resistance. Int. Microbiol. 2023, 27, 1169–1180. [Google Scholar] [CrossRef]
- Seifert, H.; Blondeau, J.; Lucassen, K.; Utt, E.A. Global update on the in vitro activity of tigecycline and comparators against isolates of Acinetobacter baumannii and rates of resistant phenotypes (2016–2018). J. Glob. Antimicrob. Res. 2022, 31, 82–89. [Google Scholar] [CrossRef]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Donhofer, A.; Wilson, D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014, 395, 559–575. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Huband, M.D.; Streit, J.M.; Flamm, R.K.; Sader, H.S. Surveillance of tigecycline activity tested against clinical isolates from a global (North America, Europe, Latin America and Asia-Pacific) collection (2016). Int. J. Antimicrob. Agents 2018, 51, 848–853. [Google Scholar] [CrossRef]
- Chen, C.; Cui, C.Y.; Wu, X.T.; Fang, L.X.; He, Q.; He, B.; Long, T.F.; Liao, X.P.; Chen, L.; Liu, Y.H.; et al. Spread of tet(X5) and tet(X6) genes in multidrug-resistant Acinetobacter baumannii strains of animal origin. Vet. Microbiol. 2021, 253, 108954. [Google Scholar] [CrossRef]
- He, T.; Wang, R.; Liu, D.J.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.B.; Ji, Q.J.; Wei, R.C.; Liu, Z.H.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef]
- Sun, J.; Chen, C.; Cui, C.Y.; Zhang, Y.; Liu, X.; Cui, Z.H.; Ma, X.Y.; Feng, Y.; Fang, L.X.; Lian, X.L.; et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 2019, 4, 1457–1464. [Google Scholar] [CrossRef]
- Wang, L.; Liu, D.; Lv, Y.; Cui, L.; Li, Y.; Li, T.; Song, H.; Hao, Y.; Shen, J.; Wang, Y.; et al. Novel Plasmid-Mediated tet(X5) Gene Conferring Resistance to Tigecycline, Eravacycline, and Omadacycline in a Clinical Acinetobacter baumannii Isolate. Antimicrob. Agents Chemother. 2019, 64, e01326-19. [Google Scholar] [CrossRef]
- He, D.; Wang, L.; Zhao, S.; Liu, L.; Liu, J.; Hu, G.; Pan, Y. A novel tigecycline resistance gene, tet(X6), on an SXT/R391 integrative and conjugative element in a Proteus genomospecies 6 isolate of retail meat origin. J. Antimicrob. Chemother. 2020, 75, 1159–1164. [Google Scholar] [CrossRef]
- Li, R.; Peng, K.; Xiao, X.; Wang, Y.; Wang, Z. Characterization of novel ISAba1-bounded tet(X15)-bearing composite transposon Tn6866 in Acinetobacter variabilis. J. Antimicrob. Chemother. 2021, 76, 2481–2483. [Google Scholar] [CrossRef]
- Chen, C.; Lv, Y.; Wu, T.; Liu, J.; Guo, Y.; Huang, J. Concurrence of Inactivation Enzyme-Encoding Genes tet(X), blaEBR, and estT in Empedobacter Species from Chickens and Surrounding Environments. Foods 2024, 13, 3201. [Google Scholar] [CrossRef]
- Li, R.; Jiang, Y.; Peng, K.; Wang, Y.; Wang, M.; Liu, Y.; Wang, Z. Phenotypic and genomic analysis reveals Riemerella anatipestifer as the potential reservoir of tet(X) variants. J. Antimicrob. Chemother. 2022, 77, 374–380. [Google Scholar] [CrossRef]
- Jin, H.; Jia, Q.; Jin, X.; Zhu, X.; Wang, M.-G.; Sun, R.-Y.; Cui, C. Identification of novel Tet(X6)-Tet(X2) recombinant variant in Elizabethkingia meningoseptica from a bullfrog farm and downstream river in China. Front. Microbiol. 2024, 15, 1453801. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, L.; Peng, K.; Wang, Q.; Liu, R.; Wang, Z.; Li, R. Characterisation of a Novel Tigecycline Resistance Gene tet(X22) and its Coexistence with blaNDM-1 in a Pseudomonas caeni Isolate. Int. J. Antimicrob. Agents 2023, 62, 106961. [Google Scholar] [CrossRef]
- Liu, D.; Zhai, W.; Song, H.; Fu, Y.; Schwarz, S.; He, T.; Bai, L.; Wang, Y.; Walsh, T.R.; Shen, J. Identification of the novel tigecycline resistance gene tet(X6) and its variants in Myroides, Acinetobacter and Proteus of food animal origin. J. Antimicrob. Chemother. 2020, 75, 1428–1431. [Google Scholar] [CrossRef]
- Dong, N.; Zeng, Y.; Cai, C.; Sun, C.; Lu, J.; Liu, C.; Zhou, H.; Sun, Q.; Shu, L.; Wang, H.; et al. Prevalence, transmission, and molecular epidemiology of tet(X)-positive bacteria among humans, animals, and environmental niches in China: An epidemiological, and genomic-based study. Sci. Total Environ. 2022, 818, 151767. [Google Scholar] [CrossRef]
- Hsieh, Y.C.; Wu, J.W.; Chen, Y.Y.; Quyen, T.L.T.; Liao, W.C.; Li, S.W.; Chen, Y.C.; Pan, Y.J. An Outbreak of tet(X6)-Carrying Tigecycline-Resistant Acinetobacter baumannii Isolates with a New Capsular Type at a Hospital in Taiwan. Antibiotics 2021, 10, 1239. [Google Scholar] [CrossRef]
- Miyoshi, T.; Iwatsuki, T.; Naganuma, T. Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters. Appl. Environ. Microb. 2005, 71, 1084–1088. [Google Scholar] [CrossRef]
- CLSI. CLSI Performance Standards for Antimicrobial Susceptibility Testing Guideline M100-Ed32; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- Al Atrouni, A.; Joly-Guillou, M.L.; Hamze, M.; Kempf, M. Reservoirs of Non-baumannii Acinetobacter Species. Front. Microbiol. 2016, 7, 49. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- NCBI. Available online: https://www.ncbi.nlm.nih.gov/datasets/genome/ (accessed on 10 January 2025).
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, Y.; Fan, G.; Sun, D.; Zhang, X.; Yu, Z.; Wang, J.; Wu, L.; Shi, W.; Ma, J. IPGA: A handy integrated prokaryotes genome and pan-genome analysis web service. iMeta 2022, 1, e55. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antimicrobial Resistance or Virulence Genes. Available online: https://github.com/tseemann/abricate (accessed on 27 February 2025).
- Xu, S.; Dai, Z.; Guo, P.; Fu, X.; Liu, S.; Zhou, L.; Tang, W.; Feng, T.; Chen, M.; Zhan, L.; et al. ggtreeExtra: Compact Visualization of Richly Annotated Phylogenetic Data. Mol. Biol. Evol. 2021, 38, 4039–4042. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.M.; Schwarz, S. Resistance gene naming and numbering: Is it a new gene or not? J. Antimicrob. Chemother. 2016, 71, 569–571. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Ram, H.; Kumar, A.; Thomas, L.; Dastager, S.G.; Mawlankar, R.; Singh, V.P. Myroides indicus sp. nov., isolated from garden soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 4008–4012. [Google Scholar] [CrossRef]
- Chen, C.; Cui, C.Y.; Yu, J.J.; He, Q.; Wu, X.T.; He, Y.Z.; Cui, Z.H.; Li, C.; Jia, Q.L.; Shen, X.G.; et al. Genetic diversity and characteristics of high-level tigecycline resistance Tet(X) in Acinetobacter species. Genome Med. 2020, 12, 111. [Google Scholar] [CrossRef] [PubMed]
- Versalovic, J.; Koeuth, T.; Lupski, R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial genomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef] [PubMed]
- Grundmann, H.J.; Towner, K.J.; Dijkshoorn, L.; Gernersmidt, P.; Maher, M.; Seifert, H.; Vaneechoutte, M. Multicenter Study Using Standardized Protocols and Reagents for Evaluation of Reproducibility of PCR-Based Fingerprinting of Acinetobacter spp. J. Clin. Microbiol. 1997, 35, 3071–3077. [Google Scholar] [CrossRef]
- Cui, C.Y.; Chen, C.; Liu, B.T.; He, Q.; Wu, X.T.; Sun, R.Y.; Zhang, Y.; Cui, Z.H.; Guo, W.Y.; Jia, Q.L.; et al. Co-occurrence of Plasmid-Mediated Tigecycline and Carbapenem Resistance in Acinetobacter spp. from Waterfowls and Their Neighboring Environment. Antimicrob. Agents Chemother. 2020, 64, e02502-19. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.Y.; He, Q.; Jia, Q.L.; Li, C.; Chen, C.; Wu, X.T.; Zhang, X.J.; Lin, Z.Y.; Zheng, Z.J.; Liao, X.P.; et al. Evolutionary Trajectory of the Tet(X) Family: Critical Residue Changes towards High-Level Tigecycline Resistance. mSystems 2021, 6, e00050-21. [Google Scholar] [CrossRef]
- Cheng, Q.; Cheung, Y.; Liu, C.; Chan, E.W.C.; Wong, K.Y.; Zhang, R.; Chen, S. Functional and phylogenetic analysis of TetX variants to design a new classification system. Commun. Biol. 2022, 5, 522. [Google Scholar] [CrossRef]
- Ming, D.; Chen, Q.Q.; Chen, X.T. Analysis of resistance genes in pan-resistant Myroides odoratimimus clinical strain PR63039 using whole genome sequencing. Microb. Pathog. 2017, 112, 164–170. [Google Scholar] [CrossRef]
- Speer, B.S.; Bedzyk, L.; Salyers, A.A. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J. Bacteriol. 1991, 173, 176–183. [Google Scholar] [CrossRef]
- Linkevicius, M.; Sandegren, L.; Andersson, D.I. Potential of Tetracycline Resistance Proteins To Evolve Tigecycline Resistance. Antimicrob. Agents Chemother. 2016, 60, 789–796. [Google Scholar] [CrossRef]
- Blake, K.S.; Xue, Y.-P.; Gillespie, V.J.; Fishbein, S.R.S.; Tolia, N.H.; Wencewicz, T.A.; Dantas, G. The tetracycline resistome is shaped by selection for specific resistance mechanisms by each antibiotic generation. Nat. Commun. 2025, 16, 1452. [Google Scholar] [CrossRef]
- Dhindwal, P.; Thompson, C.; Kos, D.; Planedin, K.; Jain, R.; Jelinski, M.; Ruzzini, A. A neglected and emerging antimicrobial resistance gene encodes for a serine-dependent macrolide esterase. Proc. Natl. Acad. Sci. USA 2023, 120, e2219827120. [Google Scholar] [CrossRef] [PubMed]
- Toleman, M.A.; Bennett, P.M.; Walsh, T.R. ISCR elements: Novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. R 2006, 70, 296–316. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhan, Z.; Shi, C. International Spread of Tet(X4)-Producing Escherichia coli Isolates. Foods 2022, 11, 2010. [Google Scholar] [CrossRef]
- Chen, C.; Chen, L.; Zhang, Y.; Cui, C.Y.; Wu, X.T.; He, Q.; Liao, X.P.; Liu, Y.H.; Sun, J. Detection of chromosome-mediated tet(X4)-carrying Aeromonas caviae in a sewage sample from a chicken farm. J. Antimicrob. Chemother. 2019, 74, 3628–3630. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).