Faecalibacterium prausnitzii Is Associated with Disease Severity in MASLD but Its Supplementation Does Not Improve Diet-Induced Steatohepatitis in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Graphical Abstract
2.2. Study Subjects
2.3. Faecalibacterium Abundance by 16S rRNA Sequencing
2.4. Mice
2.5. Bacteria
2.6. Real-Time Quantitative PCR
2.7. Histological Staining Procedures
2.8. Biochemical Analysis
2.9. Random Amplified Polymorphic DNA
2.10. Whole Genome Sequencing and Sanger Sequencing
2.11. Statistical Analysis
3. Results
3.1. Demographics and Clinical Data
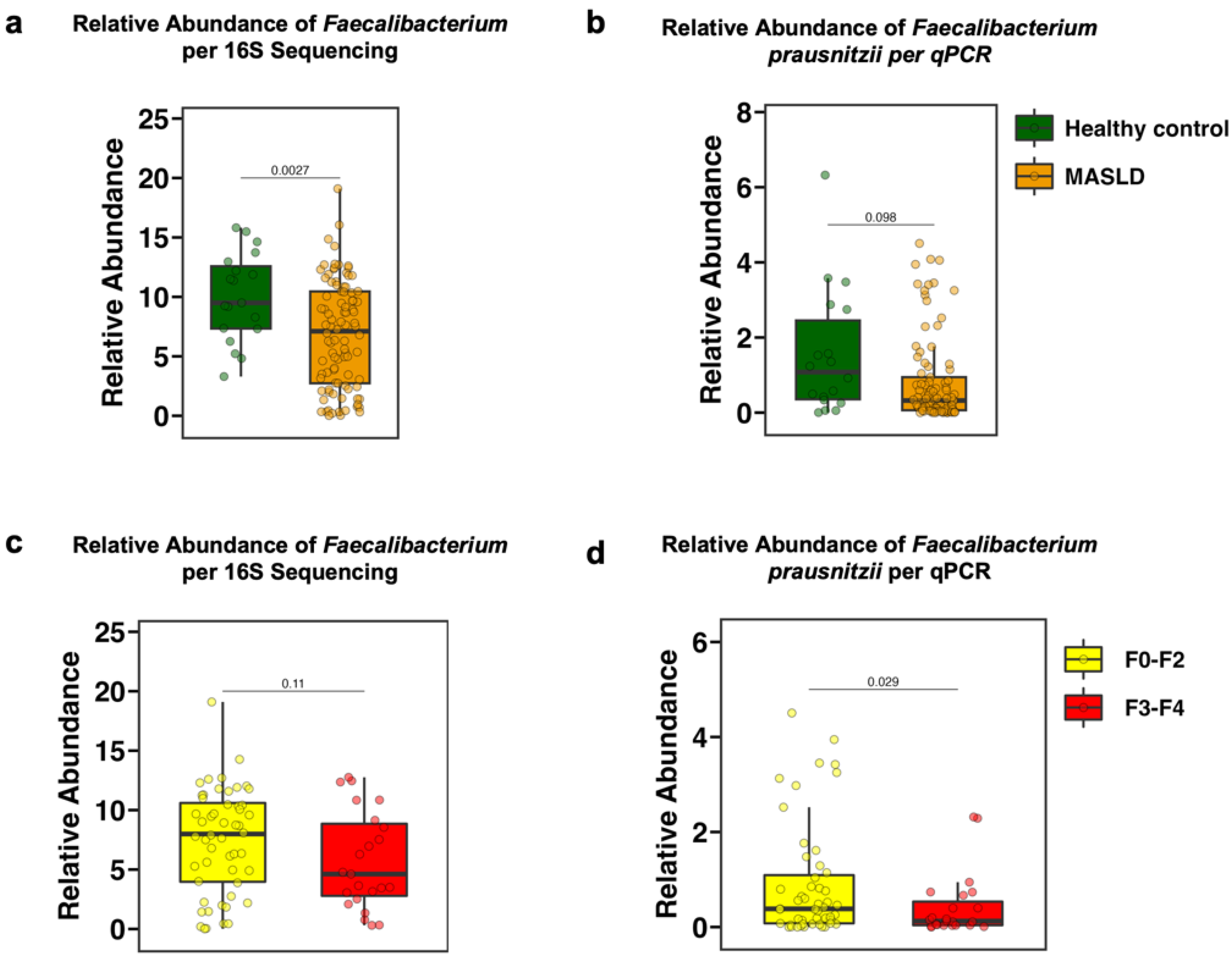
3.2. The Fecal Relative Abundance of Faecalibacterium Is Decreased in Patients with MASLD and in Particular with Advanced Fibrosis
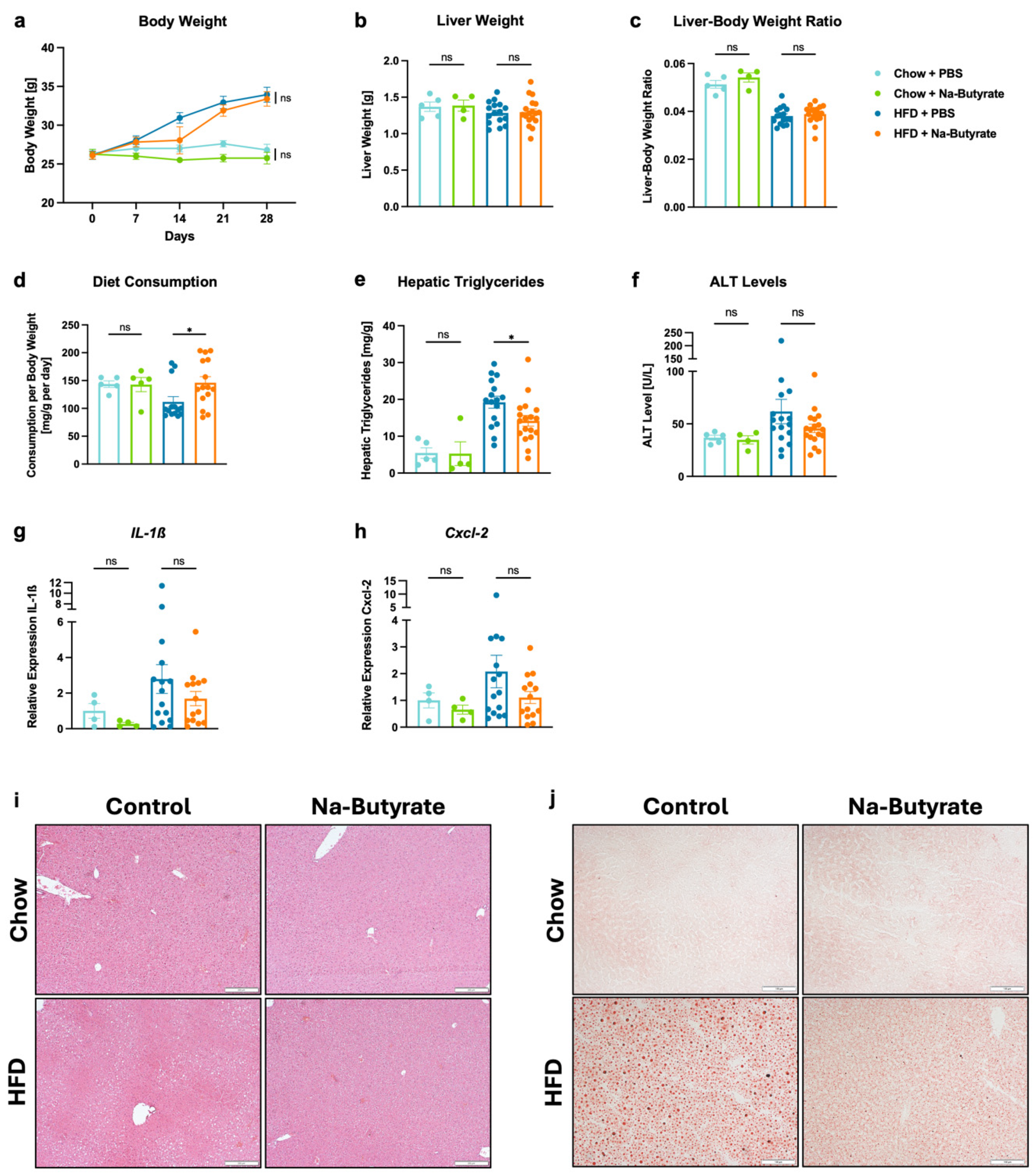
3.3. Sodium-Butyrate Reduces HFD-Induced Steatohepatitis
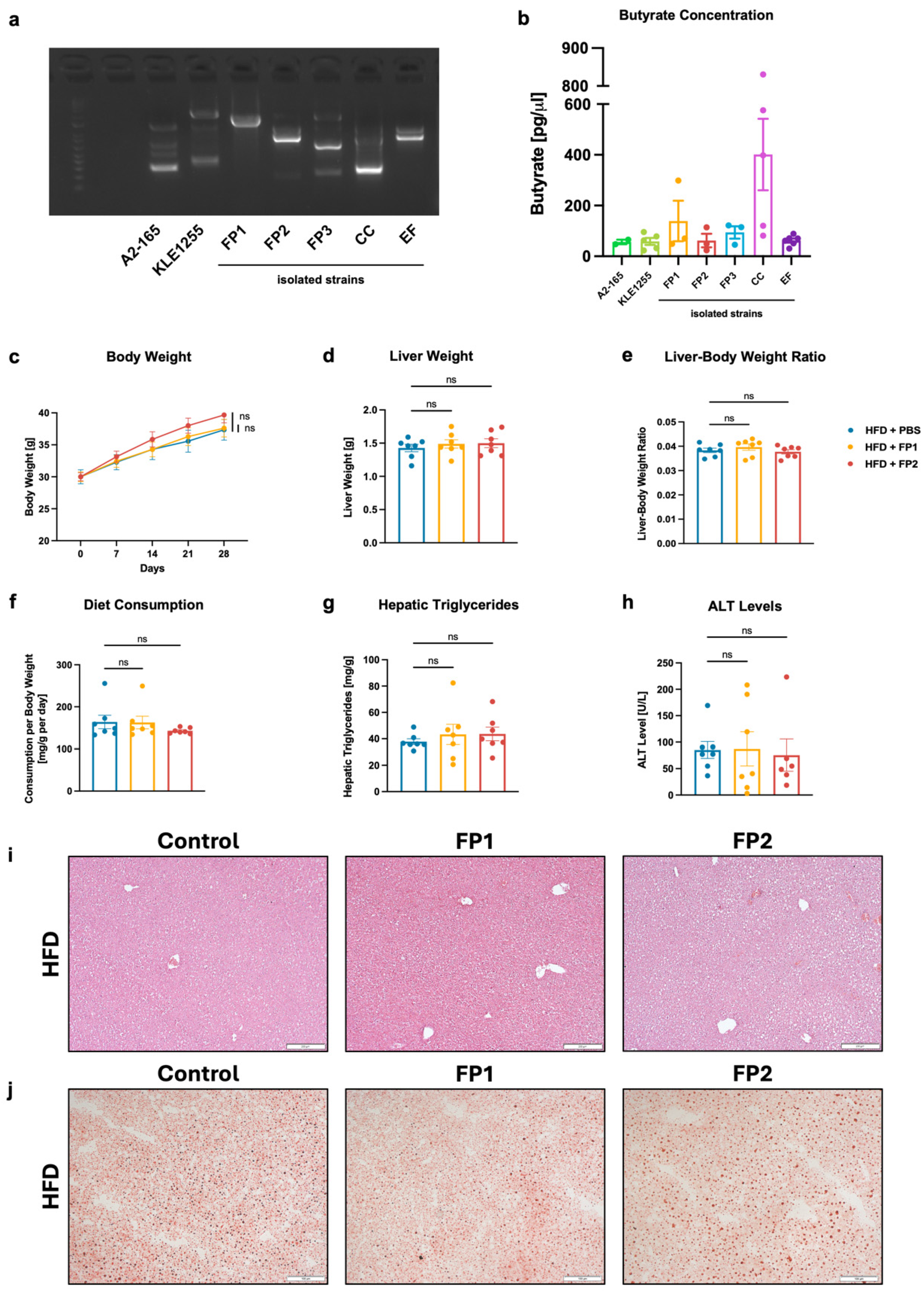
3.4. Limited Effects of Orogastric Gavage with Faecalibacterium prausnitzii on Liver Function and Metabolism in HFD-Fed Mice
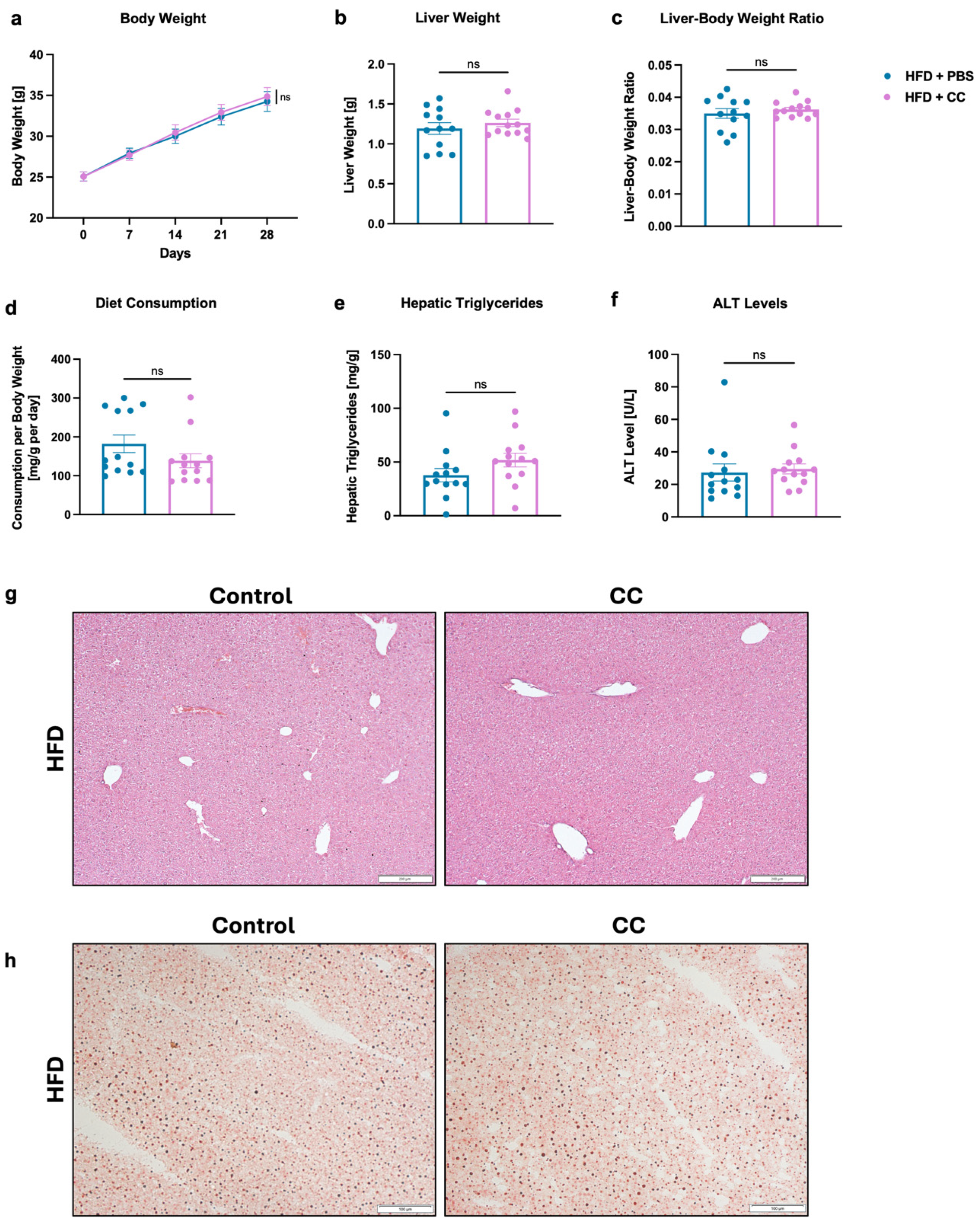
3.5. High-Dose Gavage of Faecalibacterium prausnitzii Shows No Significant Impact on HFD-Induced Liver Disease in Mice
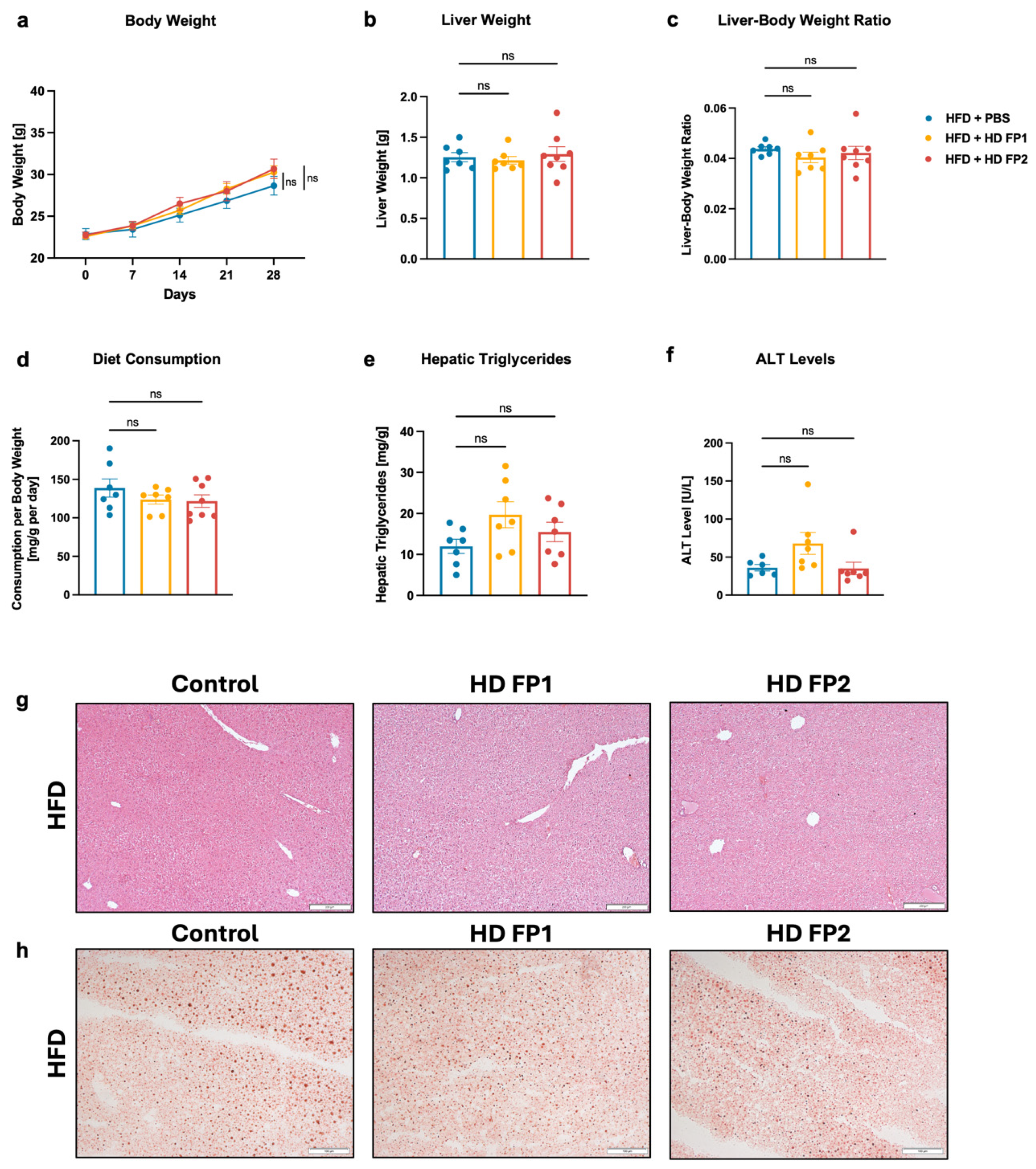
3.6. Gavage of High-Butyrate-Producing Coprococcus Comes Does Not Improve Liver Disease in HFD-Fed Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Younossi, Z.M.; Golabi, P.; Paik, J.M.; Henry, A.; Van Dongen, C.; Henry, L. The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): A systematic review. Hepatology 2023, 77, 1335–1347. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, P.; Zhang, X.; Loomba, R.; Schnabl, B. Global and national prevalence of nonalcoholic fatty liver disease in adolescents: An analysis of the global burden of disease study 2019. Hepatology 2023, 78, 1168–1181. [Google Scholar] [CrossRef]
- Sumida, Y.; Yoneda, M. Current and future pharmacological therapies for NAFLD/NASH. J. Gastroenterol. 2018, 53, 362–376. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Zong, Y.; Ma, Y.; Tian, Y.; Pang, Y.; Zhang, C.; Gao, J. Glucagon-like peptide-1 receptor: Mechanisms and advances in therapy. Signal Transduct. Target. Ther. 2024, 9, 234. [Google Scholar] [CrossRef] [PubMed]
- Vilar-Gomez, E.; Nephew, L.D.; Vuppalanchi, R.; Gawrieh, S.; Mladenovic, A.; Pike, F.; Samala, N.; Chalasani, N. High-quality diet, physical activity, and college education are associated with low risk of NAFLD among the US population. Hepatology 2022, 75, 1491–1506. [Google Scholar] [CrossRef]
- Hartmann, P.; Schnabl, B. New Developments in Microbiome in Alcohol-Associated and Nonalcoholic Fatty Liver Disease. Semin. Liver Dis. 2021, 41, 87–102. [Google Scholar] [CrossRef]
- Hartmann, P.; Schnabl, B. Risk factors for progression of and treatment options for NAFLD in children. Clin. Liver Dis. (Hoboken) 2018, 11, 11–15. [Google Scholar] [CrossRef]
- Harrison, S.A.; Bashir, M.R.; Guy, C.D.; Zhou, R.; Moylan, C.A.; Frias, J.P.; Bansal, M.B.; Baum, S.; Neuschwander-Tetri, B.A.; Taub, R.; et al. Resmetirom (MGL-3196) for the treatment of non-alcoholic steatohepatitis: A multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2019, 394, 2012–2024. [Google Scholar] [CrossRef]
- Harrison, S.A.; Taub, R.; Neff, G.W.; Lucas, K.J.; Labriola, D.; Moussa, S.E.; Alkhouri, N.; Bashir, M.R. Resmetirom for nonalcoholic fatty liver disease: A randomized, double-blind, placebo-controlled phase 3 trial. Nat. Med. 2023, 29, 2919–2928. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef]
- Layden, B.T.; Angueira, A.R.; Brodsky, M.; Durai, V.; Lowe, W.L. Short chain fatty acids and their receptors: New metabolic targets. Transl. Res. 2013, 161, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Xiong, R.G.; Zhou, D.D.; Wu, S.X.; Huang, S.Y.; Saimaiti, A.; Yang, Z.J.; Shang, A.; Zhao, C.N.; Gan, R.Y.; Li, H.B. Health Benefits and Side Effects of Short-Chain Fatty Acids. Foods 2022, 11, 2863. [Google Scholar] [CrossRef]
- Thomas, S.P.; Denu, J.M. Short-chain fatty acids activate acetyltransferase p300. Elife 2021, 10, e72171. [Google Scholar] [CrossRef]
- Mann, E.R.; Lam, Y.K.; Uhlig, H.H. Short-chain fatty acids: Linking diet, the microbiome and immunity. Nat. Rev. Immunol. 2024, 24, 577–595. [Google Scholar] [CrossRef]
- Kaufmann, B.; Seyfried, N.; Hartmann, D.; Hartmann, P. Probiotics, prebiotics, and synbiotics in nonalcoholic fatty liver disease and alcohol-associated liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2023, 325, G42–G61. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Ichimura, A.; Ohue-Kitano, R.; Igarashi, M. Free Fatty Acid Receptors in Health and Disease. Physiol. Rev. 2020, 100, 171–210. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Y.; de Zoete, M.R.; van Putten, J.P.; Strijbis, K. Redirection of Epithelial Immune Responses by Short-Chain Fatty Acids through Inhibition of Histone Deacetylases. Front. Immunol. 2015, 6, 554. [Google Scholar] [CrossRef]
- Kullberg, R.F.J.; Wikki, I.; Haak, B.W.; Kauko, A.; Galenkamp, H.; Peters-Sengers, H.; Butler, J.M.; Havulinna, A.S.; Palmu, J.; McDonald, D.; et al. Association between butyrate-producing gut bacteria and the risk of infectious disease hospitalisation: Results from two observational, population-based microbiome studies. Lancet Microbe 2024, 5, 100864. [Google Scholar] [CrossRef]
- Singhal, R.; Donde, H.; Ghare, S.; Stocke, K.; Zhang, J.; Vadhanam, M.; Reddy, S.; Gobejishvili, L.; Chilton, P.; Joshi-Barve, S.; et al. Decrease in acetyl-CoA pathway utilizing butyrate-producing bacteria is a key pathogenic feature of alcohol-induced functional gut microbial dysbiosis and development of liver disease in mice. Gut Microbes 2021, 13, 1946367. [Google Scholar] [CrossRef]
- Singh, V.; Lee, G.; Son, H.; Koh, H.; Kim, E.S.; Unno, T.; Shin, J.H. Butyrate producers, “The Sentinel of Gut”: Their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics. Front. Microbiol. 2022, 13, 1103836. [Google Scholar] [CrossRef] [PubMed]
- Detman, A.; Mielecki, D.; Chojnacka, A.; Salamon, A.; Błaszczyk, M.K.; Sikora, A. Cell factories converting lactate and acetate to butyrate: Clostridium butyricum and microbial communities from dark fermentation bioreactors. Microb. Cell Fact. 2019, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; de Los Reyes-Gavilán, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [PubMed]
- Demir, M.; Lang, S.; Martin, A.; Farowski, F.; Wisplinghoff, H.; Vehreschild, M.J.G.T.; Krawczyk, M.; Nowag, A.; Scholz, C.J.; Kretzschmar, A.; et al. Phenotyping non-alcoholic fatty liver disease by the gut microbiota: Ready for prime time? J. Gastroenterol. Hepatol. 2020, 35, 1969–1977. [Google Scholar] [CrossRef]
- Demir, M.; Lang, S.; Hartmann, P.; Duan, Y.; Martin, A.; Miyamoto, Y.; Bondareva, M.; Zhang, X.; Wang, Y.; Kasper, P.; et al. The fecal mycobiome in non-alcoholic fatty liver disease. J. Hepatol. 2022, 76, 788–799. [Google Scholar] [CrossRef]
- Viebahn, G.; Hartmann, P.; Lang, S.; Demir, M.; Zhang, X.; Fouts, D.E.; Stärkel, P.; Schnabl, B. Fungal signature differentiates alcohol-associated liver disease from nonalcoholic fatty liver disease. Gut Microbes 2024, 16, 2307586. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Hepatology 2023, 78, 1966–1986. [Google Scholar] [CrossRef]
- Alberti, K.G.; Zimmet, P.; Shaw, J.; Group, I.E.T.F.C. The metabolic syndrome—A new worldwide definition. Lancet 2005, 366, 1059–1062. [Google Scholar] [CrossRef]
- Miquel, S.; Martín, R.; Lashermes, A.; Gillet, M.; Meleine, M.; Gelot, A.; Eschalier, A.; Ardid, D.; Bermúdez-Humarán, L.G.; Sokol, H.; et al. Anti-nociceptive effect of Faecalibacterium prausnitzii in non-inflammatory IBS-like models. Sci. Rep. 2016, 6, 19399. [Google Scholar] [CrossRef]
- Miyazaki, K.; Martin, J.C.; Marinsek-Logar, R.; Flint, H.J. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B(1)4. Anaerobe 1997, 3, 373–381. [Google Scholar] [CrossRef]
- Wang, L.; Hartmann, P.; Haimerl, M.; Bathena, S.P.; Sjöwall, C.; Almer, S.; Alnouti, Y.; Hofmann, A.F.; Schnabl, B. Nod2 deficiency protects mice from cholestatic liver disease by increasing renal excretion of bile acids. J. Hepatol. 2014, 60, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, P.; Chen, P.; Wang, H.J.; Wang, L.; McCole, D.F.; Brandl, K.; Stärkel, P.; Belzer, C.; Hellerbrand, C.; Tsukamoto, H.; et al. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology 2013, 58, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, P.; Seebauer, C.T.; Mazagova, M.; Horvath, A.; Wang, L.; Llorente, C.; Varki, N.M.; Brandl, K.; Ho, S.B.; Schnabl, B. Deficiency of intestinal mucin-2 protects mice from diet-induced fatty liver disease and obesity. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G310–G322. [Google Scholar] [CrossRef]
- Hartmann, P.; Schnabl, B. Inexpensive, Accurate, and Stable Method to Quantitate Blood Alanine Aminotransferase (ALT) Levels. Methods Protoc. 2022, 5, 81. [Google Scholar] [CrossRef]
- Torriani, S.; Zapparoli, G.; Dellaglio, F. Use of PCR-based methods for rapid differentiation of Lactobacillus delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis. Appl. Environ. Microbiol. 1999, 65, 4351–4356. [Google Scholar] [CrossRef]
- Akopyanz, N.; Bukanov, N.O.; Westblom, T.U.; Kresovich, S.; Berg, D.E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992, 20, 5137–5142. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, P.; Duan, Y.; Miyamoto, Y.; Demir, M.; Lang, S.; Hasa, E.; Stern, P.; Yamashita, D.; Conrad, M.; Eckmann, L.; et al. Colesevelam ameliorates non-alcoholic steatohepatitis and obesity in mice. Hepatol. Int. 2022, 16, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef]
- Salipante, S.J.; SenGupta, D.J.; Cummings, L.A.; Land, T.A.; Hoogestraat, D.R.; Cookson, B.T. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J. Clin. Microbiol. 2015, 53, 1072–1079. [Google Scholar] [CrossRef]
- Aron-Wisnewsky, J.; Vigliotti, C.; Witjes, J.; Le, P.; Holleboom, A.G.; Verheij, J.; Nieuwdorp, M.; Clément, K. Gut microbiota and human NAFLD: Disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 279–297. [Google Scholar] [CrossRef]
- Yang, C.; Wu, J.; Yang, L.; Hu, Q.; Li, L.; Yang, Y.; Hu, J.; Pan, D.; Zhao, Q. Altered gut microbial profile accompanied by abnormal short chain fatty acid metabolism exacerbates nonalcoholic fatty liver disease progression. Sci. Rep. 2024, 14, 22385. [Google Scholar] [CrossRef] [PubMed]
- Duarte, S.M.B.; Stefano, J.T.; Miele, L.; Ponziani, F.R.; Souza-Basqueira, M.; Okada, L.S.R.R.; de Barros Costa, F.G.; Toda, K.; Mazo, D.F.C.; Sabino, E.C.; et al. Gut microbiome composition in lean patients with NASH is associated with liver damage independent of caloric intake: A prospective pilot study. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 369–384. [Google Scholar] [CrossRef]
- Da Silva, H.E.; Teterina, A.; Comelli, E.M.; Taibi, A.; Arendt, B.M.; Fischer, S.E.; Lou, W.; Allard, J.P. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci. Rep. 2018, 8, 1466. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef]
- Wong, V.W.; Tse, C.H.; Lam, T.T.; Wong, G.L.-H.; Chim, A.M.-L.; Chu, W.C.-W.; Yeung, D.K.-W.; Law, P.T.-W.; Kwan, H.-S.; Yu, J.; et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis--a longitudinal study. PLoS ONE 2013, 8, e62885. [Google Scholar] [CrossRef] [PubMed]
- Loomba, R.; Seguritan, V.; Li, W.; Long, T.; Klitgord, N.; Bhatt, A.; Dulai, P.S.; Caussy, C.; Bettencourt, R.; Highlander, S.K.; et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017, 25, 1054–1062.e5. [Google Scholar] [CrossRef]
- Caussy, C.; Tripathi, A.; Humphrey, G.; Bassirian, S.; Singh, S.; Faulkner, C.; Bettencourt, R.; Rizo, E.; Richards, L.; Xu, Z.Z.; et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat. Commun. 2019, 10, 1406. [Google Scholar] [CrossRef]
- Iebba, V.; Guerrieri, F.; Di Gregorio, V.; Levrero, M.; Gagliardi, A.; Santangelo, F.; Sobolev, A.P.; Circi, S.; Giannelli, V.; Mannina, L.; et al. Combining amplicon sequencing and metabolomics in cirrhotic patients highlights distinctive microbiota features involved in bacterial translocation, systemic inflammation and hepatic encephalopathy. Sci. Rep. 2018, 8, 8210. [Google Scholar] [CrossRef]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef]
- Simental-Mendía, L.E.; Simental-Mendía, E.; Rodríguez-Hernández, H.; Rodríguez-Morán, M.; Guerrero-Romero, F. The product of triglycerides and glucose as biomarker for screening simple steatosis and NASH in asymptomatic women. Ann. Hepatol. 2016, 15, 715–720. [Google Scholar] [CrossRef]
- Zhou, D.; Pan, Q.; Xin, F.Z.; Zhang, R.-N.; He, C.-X.; Chen, G.-Y.; Liu, C.; Chen, Y.-W.; Fan, J.-G. Sodium butyrate attenuates high-fat diet-induced steatohepatitis in mice by improving gut microbiota and gastrointestinal barrier. World J. Gastroenterol. 2017, 23, 60–75. [Google Scholar] [CrossRef]
- Sato, F.T.; Yap, Y.A.; Crisma, A.R.; Portovedo, M.; Murata, G.M.; Hirabara, S.M.; Ribeiro, W.R.; Ferreira, C.M.; Cruz, M.M.; Pereira, J.N.B.; et al. Tributyrin Attenuates Metabolic and Inflammatory Changes Associated with Obesity through a GPR109A-Dependent Mechanism. Cells 2020, 9, 2007. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Chen, Y.W.; Zhao, Z.H.; Yang, R.-X.; Xin, F.-Z.; Liu, X.-L.; Pan, Q.; Zhou, H.; Fan, J.-G. Sodium butyrate reduces high-fat diet-induced non-alcoholic steatohepatitis through upregulation of hepatic GLP-1R expression. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef]
- Lin, H.V.; Frassetto, A.; Kowalik, E.J.; Nawrocki, A.R.; Lu, M.M.; Kosinski, J.R.; Hubert, J.A.; Szeto, D.; Yao, X.; Forrest, G.; et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 2012, 7, e35240. [Google Scholar] [CrossRef] [PubMed]
- den Besten, G.; Bleeker, A.; Gerding, A.; van Eunen, K.; Havinga, R.; van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.-J.; et al. Short-Chain Fatty Acids Protect Against High-Fat Diet-Induced Obesity via a PPARγ-Dependent Switch From Lipogenesis to Fat Oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef]
- Chai, L.J.; Lu, Z.M.; Zhang, X.J.; Ma, J.; Xu, P.-X.; Qian, W.; Xiao, C.; Wang, S.-T.; Shen, C.-H.; Shi, J.-S.; et al. Zooming in on Butyrate-Producing Clostridial Consortia in the Fermented Grains of. Front. Microbiol. 2019, 10, 1397. [Google Scholar] [CrossRef]
- Geirnaert, A.; Calatayud, M.; Grootaert, C.; Laukens, D.; Devriese, S.; Smagghe, G.; De Vos, M.; Boon, N.; Van de Wiele, T. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017, 7, 11450. [Google Scholar] [CrossRef]
- Dil-Afroze Misra, A.; Sulaiman, I.M.; Sinha, S.; Sarkar, C.; Mahapatra, A.K.; Hasnain, S.E. Genetic alterations in brain tumors identified by RAPD analysis. Gene 1998, 206, 45–48. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Q.; Ye, F. Genetic instability in cancer tissues analyzed by random amplified polymorphic DNA PCR. Chin. Med. J. 2002, 115, 430–432. [Google Scholar]
- Jones, C.; Kortenkamp, A. RAPD library fingerprinting of bacterial and human DNA: Applications in mutation detection. Teratog. Carcinog. Mutagen. 2000, 20, 49–63. [Google Scholar] [CrossRef]
- Welsh, J.; McClelland, M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990, 18, 7213–7218. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Khan, T.M.; Duncan, S.H.; Harmsen, H.J.; Garcia-Gil, L.J.; Flint, H.J. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl. Environ. Microbiol. 2012, 78, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Lee, Y.; Song, E.J.; Lee, D.; Jang, S.-Y.; Byeon, H.R.; Hong, M.-G.; Lee, S.-N.; Kim, H.-J.; Seo, J.-G.; et al. Faecalibacterium prausnitzii prevents hepatic damage in a mouse model of NASH induced by a high-fructose high-fat diet. Front. Microbiol. 2023, 14, 1123547. [Google Scholar] [CrossRef] [PubMed]
- Munukka, E.; Rintala, A.; Toivonen, R.; Nylund, M.; Yang, B.; Takanen, A.; Hänninen, A.; Vuopio, J.; Huovinen, P.; Jalkanen, S.; et al. Faecalibacterium prausnitzii treatment improves hepatic health and reduces adipose tissue inflammation in high-fat fed mice. ISME J. 2017, 11, 1667–1679. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Gao, W.; Liu, Z.; Fang, Z.; Wang, H.; Zhao, J.; Zhang, H.; Lu, W.; Chen, W. Specific Strains of Faecalibacterium prausnitzii Ameliorate Nonalcoholic Fatty Liver Disease in Mice in Association with Gut Microbiota Regulation. Nutrients 2022, 14, 2945. [Google Scholar] [CrossRef]
- Hu, W.; Gao, W.; Liu, Z.; Fang, Z.; Zhao, J.; Zhang, H.; Lu, W.; Chen, W. Biodiversity and Physiological Characteristics of Novel. Microorganisms 2022, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Zhou, Y.; He, L.; Li, Y.; Shahzad, M.; Li, D. Coprococcus protects against high-fat diet-induced nonalcoholic fatty liver disease in mice. J. Appl. Microbiol. 2024, 135, lxae125. [Google Scholar] [CrossRef]
- Maeda, H.; Fujimoto, C.; Haruki, Y.; Maeda, T.; Kokeguchi, S.; Petelin, M.; Arai, H.; Tanimoto, I.; Nishimura, F.; Takashiba, S. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol. Med Microbiol. 2003, 39, 81–86. [Google Scholar] [CrossRef]
- Roh, Y.S.; Zhang, B.; Loomba, R.; Seki, E. TLR2 and TLR9 contribute to alcohol-mediated liver injury through induction of CXCL1 and neutrophil infiltration. Am. J. Physiol.-Gastrointest. Liver Physiol. 2015, 309, G30–G41. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Luo, Z.; Gong, S.; Win, S.; Kaplowitz, N.; Jiang, Y.; Chen, P. Intestinal Epithelial Chemokine (C-C Motif) Ligand 7 Overexpression Enhances Acetaminophen-Induced Hepatotoxicity in Mice. Am. J. Pathol. 2020, 190, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, C.B.; Shkoporov, A.N.; Sutton, T.D.S.; Chaplin, A.V.; Velayudhan, V.; Ross, R.P.; Hill, C. Comparative analysis of Faecalibacterium prausnitzii genomes shows a high level of genome plasticity and warrants separation into new species-level taxa. BMC Genom. 2018, 19, 931. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Münte, E.; Viebahn, G.; Khurana, A.; Fujiki, J.; Nakamura, T.; Lang, S.; Demir, M.; Schnabl, B.; Hartmann, P. Faecalibacterium prausnitzii Is Associated with Disease Severity in MASLD but Its Supplementation Does Not Improve Diet-Induced Steatohepatitis in Mice. Microorganisms 2025, 13, 675. https://doi.org/10.3390/microorganisms13030675
Münte E, Viebahn G, Khurana A, Fujiki J, Nakamura T, Lang S, Demir M, Schnabl B, Hartmann P. Faecalibacterium prausnitzii Is Associated with Disease Severity in MASLD but Its Supplementation Does Not Improve Diet-Induced Steatohepatitis in Mice. Microorganisms. 2025; 13(3):675. https://doi.org/10.3390/microorganisms13030675
Chicago/Turabian StyleMünte, Eliane, Greta Viebahn, Amit Khurana, Jumpei Fujiki, Tomohiro Nakamura, Sonja Lang, Münevver Demir, Bernd Schnabl, and Phillipp Hartmann. 2025. "Faecalibacterium prausnitzii Is Associated with Disease Severity in MASLD but Its Supplementation Does Not Improve Diet-Induced Steatohepatitis in Mice" Microorganisms 13, no. 3: 675. https://doi.org/10.3390/microorganisms13030675
APA StyleMünte, E., Viebahn, G., Khurana, A., Fujiki, J., Nakamura, T., Lang, S., Demir, M., Schnabl, B., & Hartmann, P. (2025). Faecalibacterium prausnitzii Is Associated with Disease Severity in MASLD but Its Supplementation Does Not Improve Diet-Induced Steatohepatitis in Mice. Microorganisms, 13(3), 675. https://doi.org/10.3390/microorganisms13030675





