Cross-Sectional Assessment on Carbapenem-Resistant Gram-Negative Bacteria Isolated from Patients in Moldova
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Sample Collection and Available Epidemiological Information
2.2. Identification and Phenotypic Resistance Testing
2.3. Next-Generation Sequencing
2.4. Bioinformatic Analysis
2.5. Ethical Clearance
3. Results
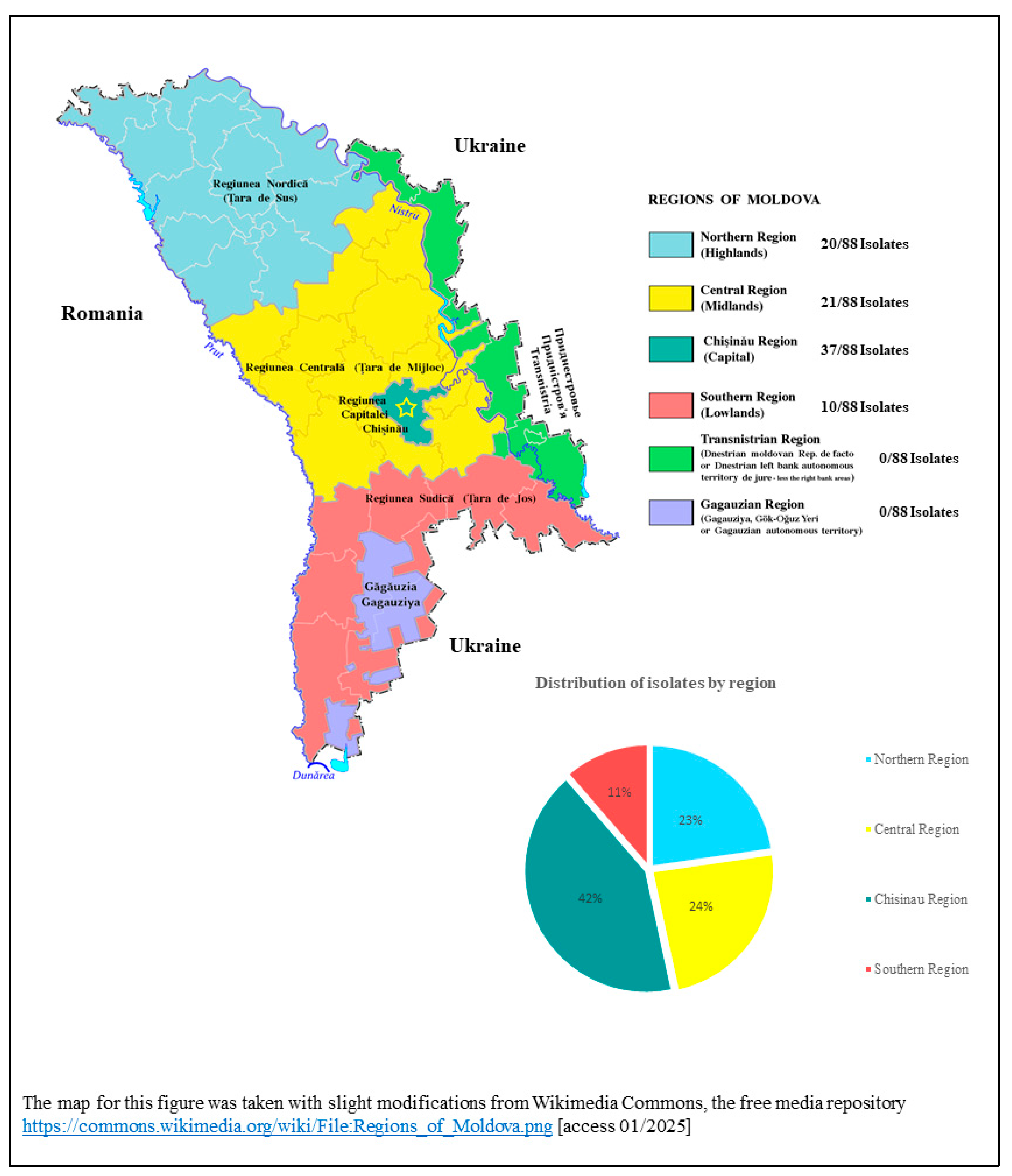
3.1. Sampling Sites, Epidemiological Information and Results of Phenotypic Resistance Testing
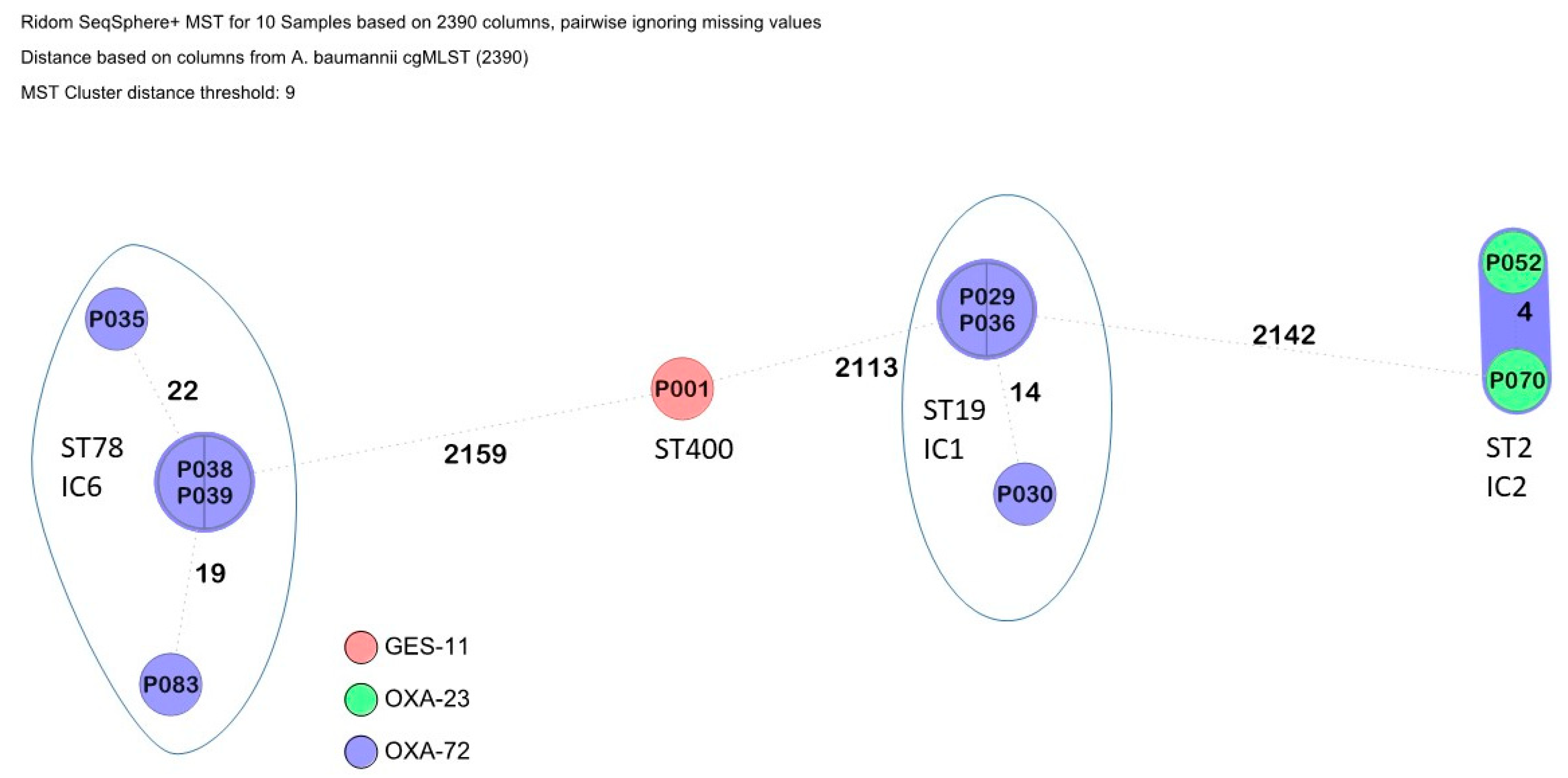
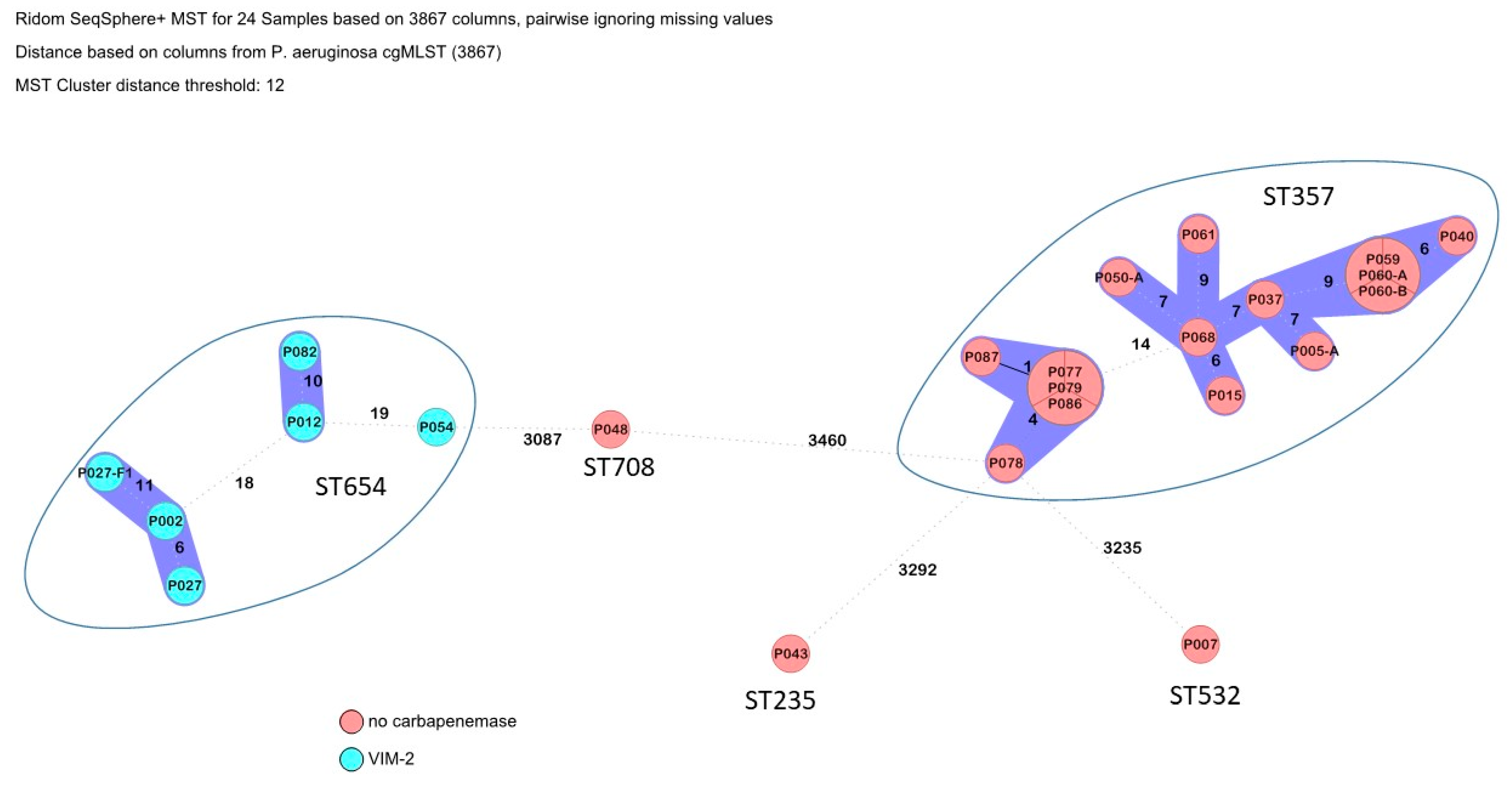
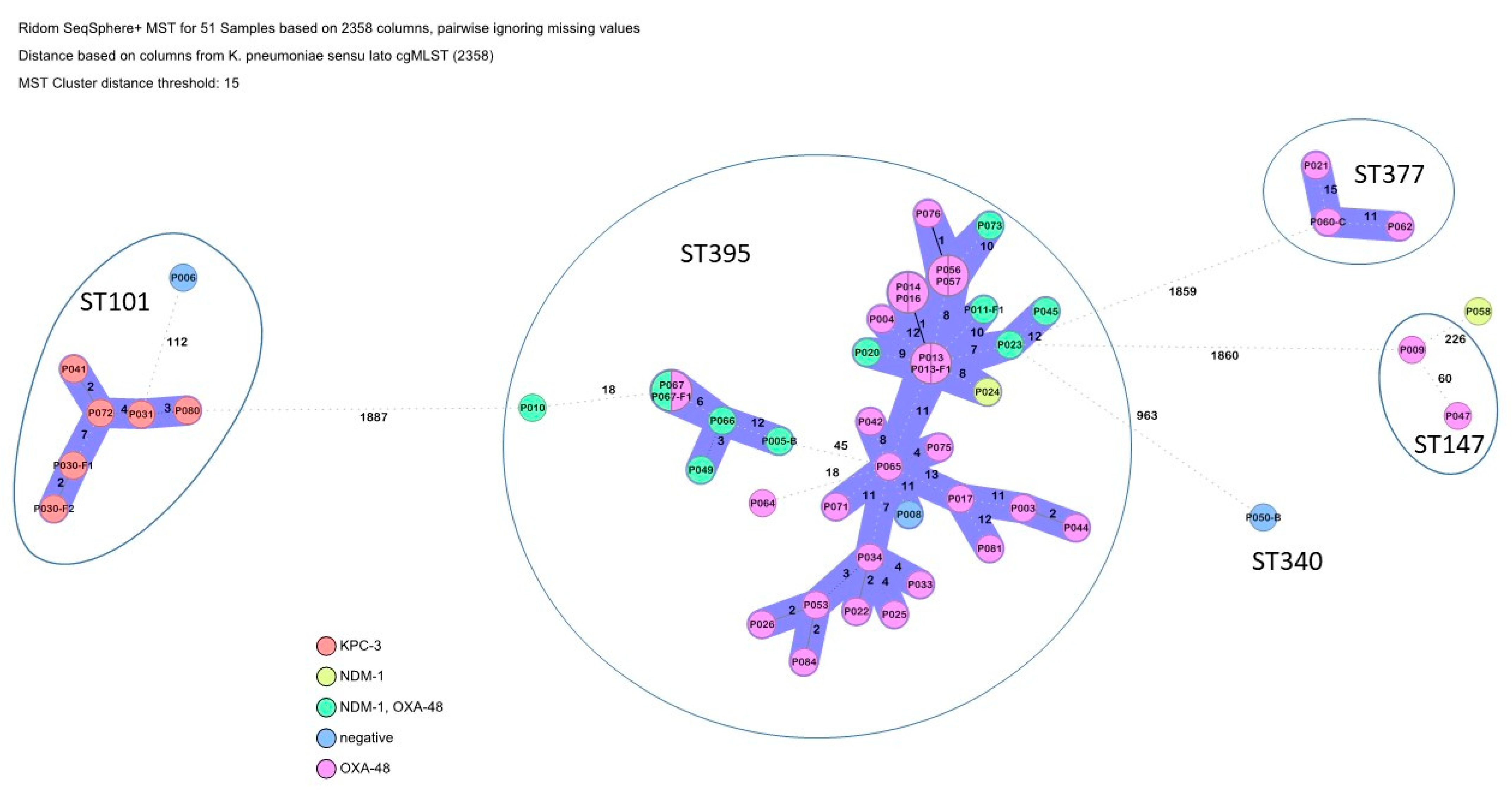
3.2. Phylogenetic Trees and Detected Multi-Locus Sequence Types
3.3. Molecular Resistance Determinants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sample Number | Collection Date (Month/Year) | Bacterial Species | Biomaterial | Patient Age | Patient Sex | In-Patient or Out-Patient | District |
|---|---|---|---|---|---|---|---|
| P001 | 01/2023 | Acinetobacter baumannii | surgical wound | 56 | f | In-patient | Northern Region (n = 20) Bălți (n = 9), Sîngerei (n = 3), Soroca, Floresti, Edinet, Fălești, Sîngerei, Drochia (n = 8) |
| P008 | 02/2023 | Klebsiella pneumoniae | urine | 74 | f | In-patient | |
| P038 | 06/2023 | Acinetobacter baumannii | surgical wound | 54 | f | In-patient | |
| P039 | 06/2023 | Acinetobacter baumannii | surgical wound | 55 | f | In-patient | |
| P040 | 06/2023 | Pseudomonas aeruginosa | urine | 73 | m | out-patient | |
| P041 | 06/2023 | Klebsiella pneumoniae | surgical wound | 57 | m | In-patient | |
| P045 | 06/2023 | Klebsiella pneumoniae | urine | 75 | m | out-patient | |
| P048 | 07/2023 | Pseudomonas aeruginosa | urine | 52 | m | out-patient | |
| P056 | 08/2023 | Klebsiella pneumoniae | surgical wound | 64 | m | In-patient | |
| P072 | 09/2023 | Klebsiella pneumoniae | urine | 70 | f | In-patient | |
| P076 | 10/2023 | Klebsiella pneumoniae | urine | 67 | m | In-patient | |
| P078 | 10/2023 | Pseudomonas aeruginosa | urine | 48 | m | In-patient | |
| P077 | 10/2023 | Pseudomonas aeruginosa | urine | 60 | f | In-patient | |
| P079 | 10/2023 | Pseudomonas aeruginosa | urine | 62 | f | In-patient | |
| P080 | 10/2023 | Klebsiella pneumoniae | surgical wound | 39 | m | In-patient | |
| P081 | 11/2023 | Klebsiella pneumoniae | urine | 81 | f | out-patient | |
| P085 | 11/2023 | Escherichia coli | urine | 66 | m | In-patient, ICU | |
| P086 | 11/2023 | Pseudomonas aeruginosa | urine | 39 | f | In-patient | |
| P087 | 11/2023 | Pseudomonas aeruginosa | urine | 36 | m | In-patient | |
| P083 | 11/2023 | Acinetobacter baumannii | surgical wound | 70 | f | In-patient | |
| P003 | 01/2023 | Klebsiella pneumoniae | urine | 75 | m | out-patient | Central Region (n = 21) Orhei (n = 10), Ialoveni, Hincesti, Nisporeni, Dubasari, Anenii Noi Ungheni (n = 11) |
| P004 | 01/2023 | Klebsiella pneumoniae | urine | 71 | m | out-patient | |
| P018 | 03/2023 | Providencia stuartii | urine | 54 | m | out-patient | |
| P021 | 04/2023 | Klebsiella pneumoniae | urine | 68 | m | out-patient | |
| P022 | 04/2023 | Klebsiella pneumoniae | feces | 0.5 | m | out-patient | |
| P011-F1 | 04/2023 | Klebsiella pneumoniae | urine | 63 | f | out-patient | |
| P024 | 04/2023 | Klebsiella pneumoniae | urine | 56 | m | out-patient | |
| P026 | 04/2023 | Klebsiella pneumoniae | feces | 62 | f | In-patient | |
| P035 | 05/2023 | Acinetobacter baumannii | sputum | 52 | m | In-patient | |
| P037 | 06/2023 | Pseudomonas aeruginosa | feces | 71 | m | out-patient | |
| P044 | 06/2023 | Klebsiella pneumoniae | feces | 56 | f | In-patient | |
| P047 | 06/2023 | Klebsiella pneumoniae | urine | 65 | m | out-patient | |
| P049 | 07/2023 | Klebsiella pneumoniae | surgical wound | 43 | f | In-patient | |
| P052 | 07/2023 | Acinetobacter baumannii | sputum | 53 | f | In-patient | |
| P053 | 07/2023 | Klebsiella pneumoniae | feces | 1 | f | In-patient | |
| P057 | 08/2023 | Klebsiella pneumoniae | feces | 64 | m | out-patient | |
| P061 | 08/2023 | Pseudomonas aeruginosa | wound | 63 | m | out-patient | |
| P062 | 08/2023 | Klebsiella pneumoniae | urine | 72 | m | out-patient | |
| P066 | 08/2023 | Klebsiella pneumoniae | sputum | 66 | f | In-patient, ICU | |
| P070 | 09/2023 | Acinetobacter baumannii | wound | 41 | m | out-patient | |
| P082 | 11/2023 | Pseudomonas aeruginosa | urine | 64 | m | out-patient | |
| P002 | 01/2023 | Pseudomonas aeruginosa | blood | 58 | m | In-patient, ICU | Chisinau Region (n = 37) |
| P005-A | 02/2023 | Pseudomonas aeruginosa | sputum | 61 | m | In-patient | |
| P005-B | 02/2023 | Klebsiella pneumoniae | sputum | 61 | m | In-patient | |
| P006 | 02/2023 | Klebsiella pneumoniae | urine | 84 | m | In-patient | |
| P007 | 02/2023 | Pseudomonas aeruginosa | urine | 33 | f | out-patient | |
| P009 | 02/2023 | Klebsiella pneumoniae | urine | 57 | f | out-patient | |
| P012 | 03/2023 | Pseudomonas aeruginosa | urine | 73 | m | out-patient | |
| P013 | 03/2023 | Klebsiella pneumoniae | body fluid | 73 | m | In-patient | |
| P014 | 03/2023 | Klebsiella pneumoniae | urine | 41 | m | out-patient | |
| P016 | 03/2023 | Klebsiella pneumoniae | urine | 79 | m | out-patient | |
| P020 | 03/2023 | Klebsiella pneumoniae | urine | 64 | f | out-patient | |
| P013-F1 | 03/2023 | Klebsiella pneumoniae | sputum | 73 | m | In-patient | |
| P023 | 04/2023 | Klebsiella pneumoniae | urine | 65 | m | out-patient | |
| P025 | 04/2023 | Klebsiella pneumoniae | feces | 1 | m | out-patient | |
| P027 | 04/2023 | Pseudomonas aeruginosa | sputum | 16 | m | out-patient | |
| P030 | 05/2023 | Acinetobacter baumannii | feces | 77 | m | out-patient | |
| P031 | 05/2023 | Klebsiella pneumoniae | urine | 63 | m | In-patient | |
| P033 | 05/2023 | Klebsiella pneumoniae | urine | 69 | m | out-patient | |
| P034 | 05/2023 | Klebsiella pneumoniae | urine | 71 | f | out-patient | |
| P042 | 06/2023 | Klebsiella pneumoniae | urine | 53 | f | out-patient | |
| P043 | 06/2023 | Pseudomonas aeruginosa | urine | 57 | f | out-patient | |
| P050-A | 07/2023 | Pseudomonas aeruginosa | wound | 57 | f | In-patient | |
| P050-B | 07/2023 | Klebsiella pneumoniae | urine | 57 | f | In-patient | |
| P030-F1 | 07/2023 | Klebsiella pneumoniae | urine | 77 | m | In-patient | |
| P027-F1 | 07/2023 | Pseudomonas aeruginosa | sputum | 17 | m | out-patient | |
| P054 | 07/2023 | Pseudomonas aeruginosa | urine | 84 | m | out-patient | |
| P030-F2 | 07/2023 | Klebsiella pneumoniae | urine | 77 | m | In-patient | |
| P058 | 08/2023 | Klebsiella pneumoniae | urine | 68 | m | out-patient | |
| P064 | 08/2023 | Klebsiella pneumoniae | urine | 80 | m | out-patient | |
| P065 | 08/2023 | Klebsiella pneumoniae | urine | 70 | m | out-patient | |
| P067 | 08/2023 | Klebsiella pneumoniae | urine | 65 | m | out-patient | |
| P068 | 09/2023 | Pseudomonas aeruginosa | urine | 76 | f | out-patient | |
| P071 | 09/2023 | Klebsiella pneumoniae | blood | 79 | m | In-patient, ICU | |
| P073 | 10/2023 | Klebsiella pneumoniae | urine | 44 | m | out-patient | |
| P075 | 10/2023 | Klebsiella pneumoniae | urine | 75 | m | out-patient | |
| P067-F1 | 10/2023 | Klebsiella pneumoniae | urine | 65 | m | out-patient | |
| P084 | 11/2023 | Klebsiella pneumoniae | urine | 17 | m | out-patient | |
| P010 | 02/2023 | Klebsiella pneumoniae | urine | 56 | m | out-patient | Southern Region (n = 10) Cimislia (n = 7), Basarabeasca, Leova, Cahul (n = 3) |
| P015 | 03/2023 | Pseudomonas aeruginosa | urine | 71 | m | out-patient | |
| P017 | 03/2023 | Klebsiella pneumoniae | urine | 50 | f | out-patient | |
| P029 | 05/2023 | Acinetobacter baumannii | sputum | 53 | m | In-patient, ICU | |
| P036 | 05/2023 | Acinetobacter baumannii | wound | 70 | f | In-patient, ICU | |
| P055 | 07/2023 | Pseudomonas monteilii | feces | 21 | f | out-patient | |
| P059 | 08/2023 | Pseudomonas aeruginosa | body fluid | 69 | m | In-patient, ICU | |
| P060 | 08/2023 | Pseudomonas aeruginosa | urine | 85 | m | In-patient, ICU | |
| P060-B | 08/2023 | Pseudomonas aeruginosa | feces | 85 | m | In-patient, ICU | |
| P060-C | 08/2023 | Klebsiella pneumoniae | feces | 85 | m | In-patient, ICU |
| Isolate Background (N = 90) | Sequence Type (ST) | Genes Mediating Resistance | ||||||
| Study ID | Species | Beta-Lactam (Carbapenemases in Bold) | Aminoglycoside | Chloramphenicol, Fosfomycin | Trimethoprime, Sulfonamide | Fluoroquinlone | Tetracycline | |
| P003 | Klebsiella pneumoniae (n = 51) | ST 395 (n = 36) | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5 | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) |
| P004 | blaOXA-48, blaTEM-1, blaSHV-11 | aadA1 | catA1 | dfrA1, sul1 | GGyrA_S83I, pParC_S80I | - | ||
| P005-B | blaOXA-48, blaNDM-1 blaCTX-M-15, blaTEM-1, blaOXA-1, blaSHV-11 | aac(6′)-Ib-cr5,rmtC, ant(2″)-Ia, aadA1, | catA1 | dfrA1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P008 | ompK36_D135DGD blaTEM-1, blaSHV-11, blaCTX-M-15, blaOXA-1 | aac(3)-IIe, aac(6′)-Ib-cr5, | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | |||
| P010 | blaOXA-48, blaNDM-1 blaTEM-1 | rmtC | catA1 | dfrA1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P013 | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5 | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P014 | blaOXA-48, ompK36_D135DGD, blaSHV-11 | - | catA1 | dfrA1 | GyrA_S83I, ParC_S80I | tet(A) | ||
| P016 | blaOXA-48, ompK36_D135DGD, blaSHV-11 | - | catA1 | dfrA1 | GyrA_S83I, ParC_S80I | tet(A) | ||
| P017 | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5 | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P020 | blaOXA-48, blaNDM-1 blaCTX-M-15, blaTEM-1, blaOXA-1, blaSHV-11 | aac(6′)-Ib-cr5, rmtC, ant(3)-IIe, | catA1 | dfrA1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P013-F1 | …Klebsiella pneumoniae (n = 51) | …ST 395 (n = 36) | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5 | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) |
| P022 | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, aac(3)-IIe | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P011-F1 | blaOXA-48, blaNDM-1 blaCTX-M-15, blaTEM-1, blaSHV-11 | rmtC, | catA1 | dfrA1 | GyrA_S83I, ParC_S80I, | tet(A) | ||
| P023 | blaOXA-48, blaNDM-1 blaCTX-M-15, blaTEM-1, blaOXA-1, blaSHV-11 | aac(6′)-Ib-cr5, rmtC, | catA1 | dfrA1 | GyrA_S83I, ParC_S80I, | tet(A) | ||
| P024 | blaNDM-1 blaSHV-11 | rmtC, | catA1 | sul1 | GyrA_S83I, ParC_S80I, | - | ||
| P025 | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, aac(3)-IIe | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P026 | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, aac(3)-IIe | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P033 | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, aac(3)-IIe | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P034 | blaOXA-48, ompK36_D135DGD, blaSHV-11, blaTEM-1 | - | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P042 | blaOXA-48, blaTEM-1, blaSHV-11 | aadA1 | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | - | ||
| P044 | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | |||
| P045 | blaOXA-48, blaNDM-1 blaCTX-M-15, blaTEM-1, blaOXA-1, blaSHV-11 | aac(6′)-Ib-cr5, rmtC, aac(3)-IIe | catA1 | dfrA1 | GyrA_S83I, GyrA_D87G, ParC_S80I, | tet(A) | ||
| P049 | blaOXA-48, blaNDM-1 blaCTX-M-15, blaTEM-1, blaOXA-1, blaSHV-11 | aac(6′)-Ib-cr5, rmtC, | dfrA1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | |||
| P053 | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, aac(3)-IIe | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P056 | …Klebsiella
pneumoniae (n = 51) | ST 395 (n = 36) | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) |
| P057 | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P065 | blaOXA-48, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, aac(3)-IIe | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P066 | blaOXA-48, blaNDM-1 blaCTX-M-15, blaTEM-1, blaOXA-1, blaSHV-11 | aac(6′)-Ib-cr5, rmtC, | dfrA1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | |||
| P067 | blaOXA-48, blaCTX-M-15 blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, aadA1 aac(2″)-Ia, rmtC | catA1 | dfrA1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P071 | blaOXA-48, blaCTX-M-15 blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, aac(3)-IIe | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P073 | blaOXA-48, blaNDM-1 blaCTX-M-15, blaTEM-1, blaOXA-1, blaSHV-11 | aac(6′)-Ib-cr5, rmtC, | catA1 | dfrA1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P075 | blaOXA-48, blaCTX-M-15 blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, aac(3)-IIe | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P076 | blaOXA-48, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5, | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | |||
| P067-F1 | blaOXA-48, blaNDM-1 blaCTX-M-15, blaTEM-1, blaOXA-1, blaSHV-11 | aac(6′)-Ib-cr5,rmtC, ant(2″)-Ia, aadA1, | catA1 | dfrA1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | ||
| P081 | blaOXA-48, blaTEM-1, blaSHV-11 | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | |||
| P084 | blaOXA-48, blaCTX-M-15 blaOXA-1, blaSHV-11 | aac(6′)-Ib-cr5, aac(3)-IIe | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, | tet(A) | ||
| P006 | ST 101 (n = 7) | ompK36_D135DGD, blaCTX-M-15 blaTEM-1, blaSHV-1, blaOXA-9 | aac(6′)-Ib, rmtB1, aadA1 | - | GyrA_D87G, GyrA_S83Y, ParC_S80I, qepA1 | - | ||
| P030-F1 | blaKPC-3, blaSHV-1, | armA, | - | GyrA_D87G, GyrA_S83Y, ParC_S80I | - | |||
| P030-F2 | …Klebsiella pneumoniae (n = 51) | …ST 101 (n = 7) | blaKPC-3, blaSHV-1, | armA, | - | GyrA_D87G, GyrA_S83Y, ParC_S80I | - | |
| P031 | blaKPC-3, blaSHV-1, | armA, | - | GyrA_D87G, GyrA_S83Y, ParC_S80I | - | |||
| P041 | blaKPC-3, blaSHV-1, | armA, | - | GyrA_D87G, GyrA_S83Y, ParC_S80I | - | |||
| P072 | blaKPC-3, blaSHV-1, | armA, | - | GyrA_D87G, GyrA_S83Y, ParC_S80I | - | |||
| P080 | blaKPC-3, blaSHV-1, | armA, | - | GyrA_D87G, GyrA_S83Y, ParC_S80I | - | |||
| P021 | ST 377 (n = 3) | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-110 | aac(6′)-Ib-cr5, armA aph(3′)-VIa, aac(3)-IIe | catA1 | dfrA1 | GyrA_D87G, GyrA_S83Y, ParC_S80I | - | |
| P60-C | blaOXA-48, blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-110 | aac(6′)-Ib-cr5, armA aph(3′)-VIa, aac(3)-IIe | catA1 | dfrA1 | GyrA_D87G, GyrA_S83Y, ParC_S80I | - | ||
| P62 | blaOXA-48, blaCTX-M-15, blaSHV-110 | aac(6′)-Ib, armA aac(3)-IIe | catA1 | dfrA1 | GyrA_D87G, GyrA_S83Y, ParC_S80I | ramR_T162I | ||
| P009 | ST 147 (n = 2) | blaOXA-48, blaTEM-1, blaSHV-11 | rmtC | - | sul1 | GyrA_S83I, ParC_S80I | - | |
| P047 | blaOXA-48, blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5 | - | - | GyrA_S83I, ParC_S80I | - | ||
| P050-B | ST 340 (n = 1) | blaCTX-M-15 blaTEM-1, blaSHV-11, blaOXA-1 | aac(6′)-Ib-cr5, aac(3)-IIe | catA1 | dfrA1, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | tet(A) | |
| P058 | Singleton (n = 1) | blaNDM-1, blaCTX-M-15 blaSHV-11, blaOXA-9, | aac(6′)-Ib, aph(3′)-VI, aadA1 | - | dfrA5, sul1 | GyrA_S83I, ParC_S80I, qnrS1 | - | |
| P064 | Singleton (n = 1) | blaOXA-48, blaCTX-M-15 blaOXA-1, blaTEM-1, blaSHV-11 | aac(6′)-Ib-cr5 | catA1 | dfrA1 | GyrA_S83I, ParC_S80I | tet(A) | |
| P002 | P. aeruginosa (n = 24) | ST 654 (n = 6) | blaVIM-2, blaPDC-3 | aph(3′)-VI, aph(3′)-IIb | catB7, floR2 | GyrA_T83I, ParC_S87L | tet(A), tet(G) | |
| P012 | blaVIM-2, blaPDC-3 | aph(3′)-VI, aph(3′)-IIb | catB7, floR2 | GyrA_T83I, ParC_S87L, crpP | tet(A), tet(G) | |||
| P027 | blaVIM-2, blaPDC-3 | aph(3′)-VI, aph(3′)-IIb | catB7, floR2 | GyrA_T83I, ParC_S87L | tet(A), tet(G) | |||
| P027-F1 | blaVIM-2, blaPDC-3 | aph(3′)-VI, aph(3′)-IIb | catB7, floR2 | GyrA_T83I, ParC_S87L | tet(A), tet(G) | |||
| P054 | blaVIM-2, blaPDC-3 | aph(3′)-IIb | catB7, floR2 | GyrA_T83I, ParC_S87L | tet(A), tet(G) | |||
| P082 | blaVIM-2, blaPDC-3 | aph(3′)-VI, aph(3′)-IIb | catB7, floR2 | GyrA_T83I, ParC_S87L, crpP | tet(A), tet(G) | |||
| P007 | ST 532 | blaOXA906, blaTEM-1, blaPDC-59 | aac(3)-IIe | catB7 | sul1 | GyrA_T83I, ParC_S87L, crpP | - | |
| P043 | ST 235 | blaOXA-488, blaOXA-14, blaPDC-35 | aac(6′)-Ib4, aadA1 | catB7 | sul1 | GyrA_T83I, ParC_S87L, crpP | - | |
| P048 | ST 708 | blaOXA-50, blaTEM-1, blaCTX-M-15, blaPDC-5 | aac(3)-Iie, armA | catB7 | sul1 | GyrA_T83I, ParC_S87L, crpP | - | |
| P005-A | ST 357 (n = 15) | blaOXA-677, blaOXA-846, blaVEB-9, blaPDC-11 | ant(2″)-Ia, aph(3′)-Ib, aadA1 | catB7 | dfrB2, sul1 | GyrA_T83I, ParC_S87L, crpP | - | |
| P015 | blaOXA-677, blaOXA-846, blaVEB-9, blaPDC-11 | aadA1, aph(3′)-IIb | catB7 | dfrB2, sul1 | GyrA_T83I,ParC_S87L, crpP, GyrA, ParC | - | ||
| P037 | blaOXA-677, blaOXA-846, blaVEB-9, blaPDC-11 | aac(6′)-II, aph(3′)-Iib | catB7 | dfrB2, sul1 | GyrA_T83I, ParC_S87L, crpP | - | ||
| P040 | blaOXA-677, blaOXA-846, blaVEB-9, blaPDC-11 | aadA1 | catB7 | dfrB2, sul1 | GyrA_T83I, ParC_S87L, crpP | - | ||
| P050-A | blaOXA-677, blaOXA-846, blaVEB-9, blaPDC-11 | aadA1 | catB7 | dfrB2, sul1 | GyrA_T83I, ParC_S87L, crpP | - | ||
| P059 | blaOXA-677, blaOXA-846, blaVEB-9, blaPDC-11 | aadA1 | catB7 | dfrB2, sul1 | GyrA_T83I, ParC_S87L, crpP | - | ||
| P068 | blaOXA-677, blaOXA-846, blaVEB-9, blaPDC-11 | aadA1 | catB7 | dfrB2, sul1 | GyrA_T83I, ParC_S87L, crpP | - | ||
| P060-A | blaOXA-677, blaOXA-846, blaVEB-9, blaPDC-11 | aadA1 | catB7 | dfrB2, sul1 | GyrA_T83I, ParC_S87L, crpP | - | ||
| P060-B | blaOXA-677, blaOXA-846, blaVEB-9, blaPDC-11 | aadA1 | catB7 | dfrB2, sul1 | GyrA_T83I, ParC_S87L, crpP | - | ||
| P061 | blaOXA-677, blaOXA-846, blaVEB-9, blaPDC-11 | ant(2″)-Ia, aph(3′)-Iib | catB7 | dfrB2, sul1 | GyrA_T83I, ParC_S87L, crpP | - | ||
| P077 | blaOXA-846, blaVEB-9, blaPDC-11 | aac(6′)-II, aph(3′)-Iib | catB7 | sul1 | GyrA_T83I, ParC_S87L, crpP | - | ||
| P078 | blaOXA-846, blaVEB-9, blaPDC-11 | aac(6′)-II, aph(3′)-Iib | catB7 | sul1 | GyrA_T83I, ParC_S87L, crpP | - | ||
| P079 | …P. aeruginosa | …ST 357 (n = 15) | blaOXA-846, blaVEB-9, blaPDC-11 | aac(6′)-II, aph(3′)-Iib | catB7 | sul1 | GyrA_T83I, ParC_S87L, crpP | - |
| P086 | blaOXA-846, blaVEB-9, blaPDC-11 | aac(6′)-II, aph(3′)-Iib | catB7 | sul1 | GyrA_T83I, ParC_S87L, crpP | - | ||
| P087 | (n = 24) | blaOXA-846, blaVEB-9, blaPDC-11 | aac(6′)-II, aph(3′)-IIb | catB7 | sul1 | GyrA_T83I, ParC_S87L, crpP | - | |
| P035 | Acinetobacter baumannii (n = 10) | ST 78 (n = 4) | blaOXA-72, blaCTX-M-115, blaADC-152 | aac(6′)-Ian, aadA1, armA, ant(3″)-IIa | catA1, floR | sul1 | GyrA_T81L, ParC_S84L | - |
| P038 | blaOXA-72, blaADC-152 | aac(6′)-Ian, aadA1, armA, ant(3″)-IIa | catA1, floR | sul1 | GyrA_T81L, ParC_S84L | - | ||
| P039 | blaOXA-72, blaADC-152 | aac(6′)-Ian, aadA1, armA, ant(3″)-IIa | catA1, floR | sul1 | GyrA_T81L, ParC_S84L | - | ||
| P083 | blaOXA-72, blaCTX-M-115, blaADC-152 | aac(6′)-Ian, aadA1, armA, ant(3″)-IIa | catA1 | sul1 | GyrA_T81L, ParC_S84L | - | ||
| P029 | ST 19 (n = 3) | blaOXA-72, blaADC-185 | ant(3″)-IIa | catA1 | sul2 | GyrA_T81L, ParC_S84L | tet(B) | |
| P030 | blaOXA-72, blaADC-185 | ant(3″)-IIa, aph(3′)-VIa | catA1 | sul2 | GyrA_T81L, ParC_S84L | tet(B) | ||
| P036 | blaOXA-72, blaADC-185 | ant(3″)-IIa | catA1 | sul2 | GyrA_T81L, ParC_S84L | tet(B) | ||
| P052 | ST 2 (n = 2) | blaOXA-23, blaADC-30 | aac(6′)-Ib′, aadA1, armA, ant(3″)-Iia, aph(3″)-Ib | catB8, cmlA5 | sul2 | GyrA_T81L, ParC_S84L | - | |
| P070 | blaOXA-23, blaADC-30 | aac(6′)-Ib′, aadA1, armA, ant(3″)-Iia, aph(3″)-Ib | catB8, cmlA5 | sul2 | GyrA_T81L, ParC_S84L | - | ||
| P001 | ST 400 (n = 1) | blaGES-11, blaADC | aac(6′)-Ib3, aadA2, ant(2′)-Ia, aph(3″)-VIa | cmlA1 | Sul1, dfrA7 | GyrA_T81L, ParC_S84L | - | |
| P085 | E. coli (n = 1) | ST 648 | blaNDM-5, blaCTX-M-15 blaOXA-1, blaTEM-1, | aac(3)-IIe, aadA5,aac(6′)-Ib-cr5 | catB3, catA1 | sul1, dfrA17 | GyrA_D87N | tet(B) |
| P018 | Providencia stuartii (n = 1) | blaOXA-48, blaNDM-1 blaCTX-M-15, blaCMY-4, | aph(3′)-VI, aac(2′)-Ia, armA | catA1, catA3 | dfrA1, dfrA14, sul1 | - | tet(A), tet(B) | |
| P055 | Pseudomonas monteilii (n = 1) | blaIMP-1 | ant(2′)-Ia, aadA1, aadA2 | - | - | - | - | |
References
- Lange, C.; Dheda, K.; Chesov, D.; Mandalakas, A.M.; Udwadia, Z.; Horsburgh, C.R., Jr. Management of drug-resistant tuberculosis. Lancet 2019, 394, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Prisacari, V.; Berdeu, I. Antimicrobial resistance in septic-purulent infections. Rev. Med. Chir. Soc. Med. Nat. Iasi. 2013, 117, 457–464. [Google Scholar] [PubMed]
- ECDC; WHO. Antimicrobial Resistance Surveillance in Europe. 2023. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Antimicrobial%20resistance%20surveillance%20in%20Europe%202023%20-%202021%20data.pdf (accessed on 1 October 2024).
- Frickmann, H.; Podbielski, A.; Kreikemeyer, B. Resistant Gram-Negative Bacteria and Diagnostic Point-of-Care Options for the Field Setting during Military Operations. Biomed. Res. Int. 2018, 2018, 9395420. [Google Scholar] [CrossRef]
- Pallett, S.J.C.; Boyd, S.E.; O’Shea, M.K.; Martin, J.; Jenkins, D.R.; Hutley, E.J. The contribution of human conflict to the development of antimicrobial resistance. Commun. Med. 2023, 3, 153. [Google Scholar] [CrossRef]
- Sandfort, M.; Hans, J.B.; Fischer, M.A.; Reichert, F.; Cremanns, M.; Eisfeld, J.; Pfeifer, Y.; Heck, A.; Eckmanns, T.; Werner, G.; et al. Increase in NDM-1 and NDM-1/OXA-48-producing Klebsiella pneumoniae in Germany associated with the war in Ukraine, 2022. Eurosurveillance 2022, 27, 2200926. [Google Scholar] [CrossRef]
- Wang, J.L.; Lai, C.C.; Tsai, Y.W.; Hsueh, C.C.; Ko, W.C.; Hsueh, P.R. Global trends in nonsusceptibility rates of Escherichia coli isolates to meropenem and ceftazidime/avibactam: Data from the Antimicrobial Testing Leadership and Surveillance (ATLAS) programme, 2014–2021. Int. J. Antimicrob. Agents 2024, 63, 107103. [Google Scholar] [CrossRef]
- Jones, R.N.; Flonta, M.; Gurler, N.; Cepparulo, M.; Mendes, R.E.; Castanheira, M. Resistance surveillance program report for selected European nations (2011). Diagn. Microbiol. Infect. Dis. 2014, 78, 429–436. [Google Scholar] [CrossRef]
- Salmanov, A.G.; Voronenko, Y.V.; Vozianov, S.O.; Shunko, Y.Y.; Mamenko, M.Y.; Verner, O.M.; Mykhalchuk, V.M.; Vydyborets, S.V.; Shkorbotun, V.O.; Beketova, H.V.; et al. Bloodstream infections and antimicrobial resistance of responsible pathogens in Ukraine: Results of a multicenter study (2013–2015). Wiad. Lek. 2019, 72, 2069–2075. [Google Scholar] [CrossRef]
- Higgins, P.G.; Hagen, R.M.; Podbielski, A.; Frickmann, H.; Warnke, P. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii Isolated from War-Injured Patients from the Eastern Ukraine. Antibiotics 2020, 9, 579. [Google Scholar] [CrossRef]
- Salmanov, A.; Shchehlov, D.; Svyrydiuk, O.; Bortnik, I.; Mamonova, M.; Korniyenko, S.; Rud, V.; Artyomenko, V.; Gudym, M.; Maliarchuk, R.; et al. Epidemiology of healthcare-associated infections and mechanisms of antimicrobial resistance of responsible pathogens in Ukraine: A multicentre study. J. Hosp. Infect. 2023, 131, 129–138. [Google Scholar] [CrossRef]
- Salmanov, A.; Shchehlov, D.; Artyomenko, V.; Svyrydiuk, O.; Maliarchuk, R.; Bortnik, I.; Mamonova, M.; Korniyenko, S.; Rud, V.; Gudym, M.; et al. Nosocomial transmission of multi-drug-resistant organisms in Ukrainian hospitals: Results of a multi-centre study (2019–2021). J. Hosp. Infect. 2023, 132, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Kondratiuk, V.; Jones, B.T.; Kovalchuk, V.; Kovalenko, I.; Ganiuk, V.; Kondratiuk, O.; Frantsishko, A. Phenotypic and genotypic characterization of antibiotic resistance in military hospital-associated bacteria from war injuries in the Eastern Ukraine conflict between 2014 and 2020. J. Hosp. Infect. 2021, 112, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Kovalchuk, V.; Kondratiuk, V.; McGann, P.; Jones, B.T.; Fomina, N.; Nazarchuk, O.; Fomin, O.; Kovalenko, I. Temporal evolution of bacterial species and their antimicrobial resistance characteristics in wound infections of war-related injuries in Ukraine from 2014 to 2023. J. Hosp. Infect. 2024, 152, 99–104. [Google Scholar] [CrossRef]
- Martin, M.J.; Luo, T.L.; Kovalchuk, V.; Kondratiuk, V.; Dao, H.D.; Kovalenko, I.; Plaza, B.J.; Kettlewell, J.M.; Anderson, C.P.; Smedberg, J.R.; et al. Detection of cefiderocol and aztreonam/avibactam resistance in epidemic Escherichia coli ST-361 carrying blaNDM-5 and blaKPC-3 from foreign fighters evacuated from Ukraine. Antimicrob. Agents Chemother. 2024, 20, e0109024. [Google Scholar] [CrossRef]
- Stein, C.; Zechel, M.; Spott, R.; Pletz, M.W.; Kipp, F. Multidrug-resistant isolates from Ukrainian patients in a German health facility: A genomic surveillance study focusing on antimicrobial resistance and bacterial relatedness. Infection 2023, 51, 1731–1738. [Google Scholar] [CrossRef]
- Schultze, T.; Hogardt, M.; Velázquez, E.S.; Hack, D.; Besier, S.; Wichelhaus, T.A.; Rochwalsky, U.; Kempf, V.A.; Reinheimer, C. Molecular surveillance of multidrug-resistant Gram-negative bacteria in Ukrainian patients, Germany, March to June 2022. Euro. Surveill. 2023, 28, 2200850. [Google Scholar] [CrossRef]
- López-Hernández, I.; García Barrionuevo, A.; Díaz de Alba, P.; Clavijo, E.; Pascual, A. Characterization of NDM-1- and CMH-3-producing Enterobacter cloacae complex ST932 in a patient transferred from Ukraine to Spain. Enferm. Infecc. Microbiol. Clin. 2020, 38, 327–330. [Google Scholar] [CrossRef]
- Verkaik, N.J.; Wielders, C.C.H.; den Boer, H.; Langerak, D.; Vogel, M.; Witteveen, S.; de Haan, A.; Bos, J.; van Westreenen, M.; Notermans, D.W.; et al. Antimicrobial susceptibility to last-resort antibiotics in carbapenemase-producing bacteria from Ukrainian patients. Microbiol. Spectr. 2024, 24, e0114224. [Google Scholar] [CrossRef]
- Fuchs, F.; Xanthopoulou, K.; Burgwinkel, T.; Arazo Del Pino, R.; Wohlfarth, E.; Pavlu, F.; Hagen, R.M.; Higgins, P.G. Coexistence of seven different carbapenemase producers in a single hospital admission screening confirmed by whole-genome sequencing. J. Glob. Antimicrob. Resist. 2024, 39, 184–188. [Google Scholar] [CrossRef]
- Luo, T.L.; Martin, M.J.; Kovalchuk, V.; Kondratiuk, V.; Trapaidze, N.; Metreveli, M.; Hulseberg, C.E.; Dao, H.D.; Kwak, Y.I.; Maybank, R.; et al. Detection of carbapenemase producing Acinetobacter baumannii ST19 from Georgia and Ukraine carrying blaOXA-23, blaOXA-72, and/or blaNDM-5, December 2019 to June 2023. Euro. Surveill. 2024, 29, 2400259. [Google Scholar] [CrossRef]
- Hernández-García, M.; Cabello, M.; Ponce-Alonso, M.; Herrador-Gómez, P.M.; Gioia, F.; Cobo, J.; Cantón, R.; Ruiz-Garbajosa, P. First detection in Spain of NDM-1-producing Pseudomonas aeruginosa in two patients transferred from Ukraine to a university hospital. J. Glob. Antimicrob. Resist. 2024, 36, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Hernández-García, M.; González de Aledo, M.; Ponce-Alonso, M.; González-Blanco, B.; Viedma, E.; Villa, J.; Tomás, M.; Hendrickx, A.P.A.; Ruiz-Garbajosa, P.; Cantón, R. Simultaneous clonal spread of NDM-1-producing Pseudomonas aeruginosa ST773 from Ukrainian patients in the Netherlands and Spain. IJID Reg. 2024, 12, 100415. [Google Scholar] [CrossRef] [PubMed]
- Berger, F.K.; Schmartz, G.P.; Fritz, T.; Veith, N.; Alhussein, F.; Roth, S.; Schneitler, S.; Gilcher, T.; Gärtner, B.C.; Pirpilashvili, V.; et al. Occurrence, resistance patterns, and management of carbapenemase-producing bacteria in war-wounded refugees from Ukraine. Int. J. Infect. Dis. 2023, 132, 89–92. [Google Scholar] [CrossRef]
- Higgins, P.G.; Hagen, R.M.; Kreikemeyer, B.; Warnke, P.; Podbielski, A.; Frickmann, H.; Loderstädt, U. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii Isolates from Northern Africa and the Middle East. Antibiotics 2021, 10, 291. [Google Scholar] [CrossRef]
- Van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef]
- Kimbrough, J.H.; Maher, J.M.; Sader, H.S.; Castanheira, M.; Mendes, R.E. In vitro activity assessment of cefiderocol against Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter spp., including β-lactam nonsusceptible molecularly characterized isolates, collected from 2020 to 2021 in the United States and European hospitals. Microbiol. Spectr. 2024, 12, e0147424. [Google Scholar]
- Witteveen, S.; Hans, J.B.; Izdebski, R.; Hasman, H.; Samuelsen, Ø.; Dortet, L.; Pfeifer, Y.; Delappe, N.; Oteo-Iglesias, J.; Żabicka, D.; et al. Dissemination of extensively drug-resistant NDM-producing Providencia stuartii in Europe linked to patients transferred from Ukraine, March 2022 to March 2023. Euro Surveill. 2024, 29, 2300616. [Google Scholar] [CrossRef]
- Del Barrio-Tofiño, E.; López-Causapé, C.; Oliver, A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob. Agents 2020, 56, 106196. [Google Scholar] [CrossRef]
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
| Species | Number (n) | Sampling Sites (%) | Percentage of Cefiderocol Resistance | Percentage of Gentamicin Resistance | Percentage of Amikacin Resistance | Percentage of Ciprofloxacin Resistance | Percentage of Trimethoprim/Sulfamethoxazole Resistance | Percentage of Colistin Resistance | Percentage of Fosfomycin Resistance |
|---|---|---|---|---|---|---|---|---|---|
| Klebsiella pneumoniae * | 51 | 68.6% urine, 13.7% feces, 7.8% wound, 5.9% respiratory secretion, 2.0% blood, 2.0% bioptically taken body fluid | 35.3% | 68.0% | 54.0% | 100% | 80.4% | 5.7% | n.a. |
| Pseudomonas aeruginosa | 24 | 64.0% urine; 12.0% respiratory secretion, 8.0% wound, 8.0% feces, 4.0% blood, 4.0% bioptically taken body fluid | 45.8% | n.a. | 72.0% | 100% | n.a. | 0% | n.a. |
| Acinetobacter baumannii * | 10 | 60% wound, 30% respiratory secretion, 10% feces | n.a. | 30.0% | 20.0% | 100% | 100% | 0% | n.a. |
| Escherichia coli | 1 | 100% urine | 100% | 0% | 0% | 100% | 100% | n.m. | 0% |
| Providencia stuartii | 1 | 100% urine | 0% | 100% | 100% | 100% | 100% | n.a. | n.a. |
| Pseudomonas monteilii ° | 1 | 100% feces | 0% | n.a. | n.m. | 0% | n.a. | 0% | n.a. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nirca, V.; Fuchs, F.; Burgwinkel, T.; del Pino, R.A.; Zaharcenco, E.; Hagen, R.M.; Poppert, S.; Frickmann, H.; Higgins, P.G. Cross-Sectional Assessment on Carbapenem-Resistant Gram-Negative Bacteria Isolated from Patients in Moldova. Microorganisms 2025, 13, 421. https://doi.org/10.3390/microorganisms13020421
Nirca V, Fuchs F, Burgwinkel T, del Pino RA, Zaharcenco E, Hagen RM, Poppert S, Frickmann H, Higgins PG. Cross-Sectional Assessment on Carbapenem-Resistant Gram-Negative Bacteria Isolated from Patients in Moldova. Microorganisms. 2025; 13(2):421. https://doi.org/10.3390/microorganisms13020421
Chicago/Turabian StyleNirca, Vadim, Frieder Fuchs, Tessa Burgwinkel, Rocío Arazo del Pino, Ecaterina Zaharcenco, Ralf Matthias Hagen, Sven Poppert, Hagen Frickmann, and Paul G. Higgins. 2025. "Cross-Sectional Assessment on Carbapenem-Resistant Gram-Negative Bacteria Isolated from Patients in Moldova" Microorganisms 13, no. 2: 421. https://doi.org/10.3390/microorganisms13020421
APA StyleNirca, V., Fuchs, F., Burgwinkel, T., del Pino, R. A., Zaharcenco, E., Hagen, R. M., Poppert, S., Frickmann, H., & Higgins, P. G. (2025). Cross-Sectional Assessment on Carbapenem-Resistant Gram-Negative Bacteria Isolated from Patients in Moldova. Microorganisms, 13(2), 421. https://doi.org/10.3390/microorganisms13020421





