Identification, Pathogenicity, and Antimicrobial Resistance Analysis of Bacterial Pathogenesis Aeromonas hydrophila from Hybrid Sturgeon (Huso dauricus ♀ × A. schrenckii ♂) in Zhejiang, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation
2.2. Challenge Test
2.3. Physiological and Biochemical Characterization of Bacteria Isolates
2.4. Molecular Identification
2.5. Virulence Genes Analysis for ST-1902
2.6. Histopathology Observation
2.7. Antimicrobial Sensitivity Tests
2.8. Antimicrobial Drug Resistance Gene Analysis
3. Results
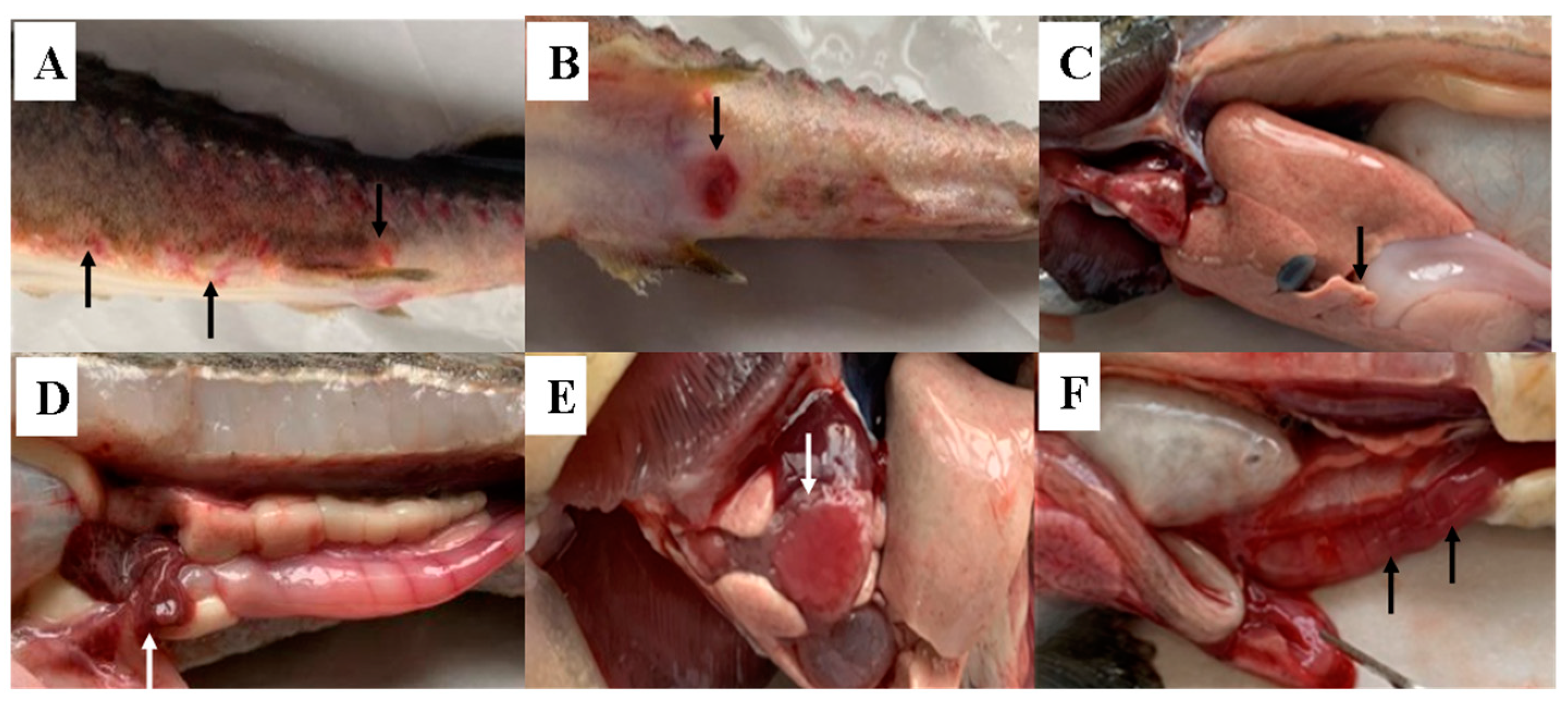
3.1. Symptoms of Diseased Sturgeon
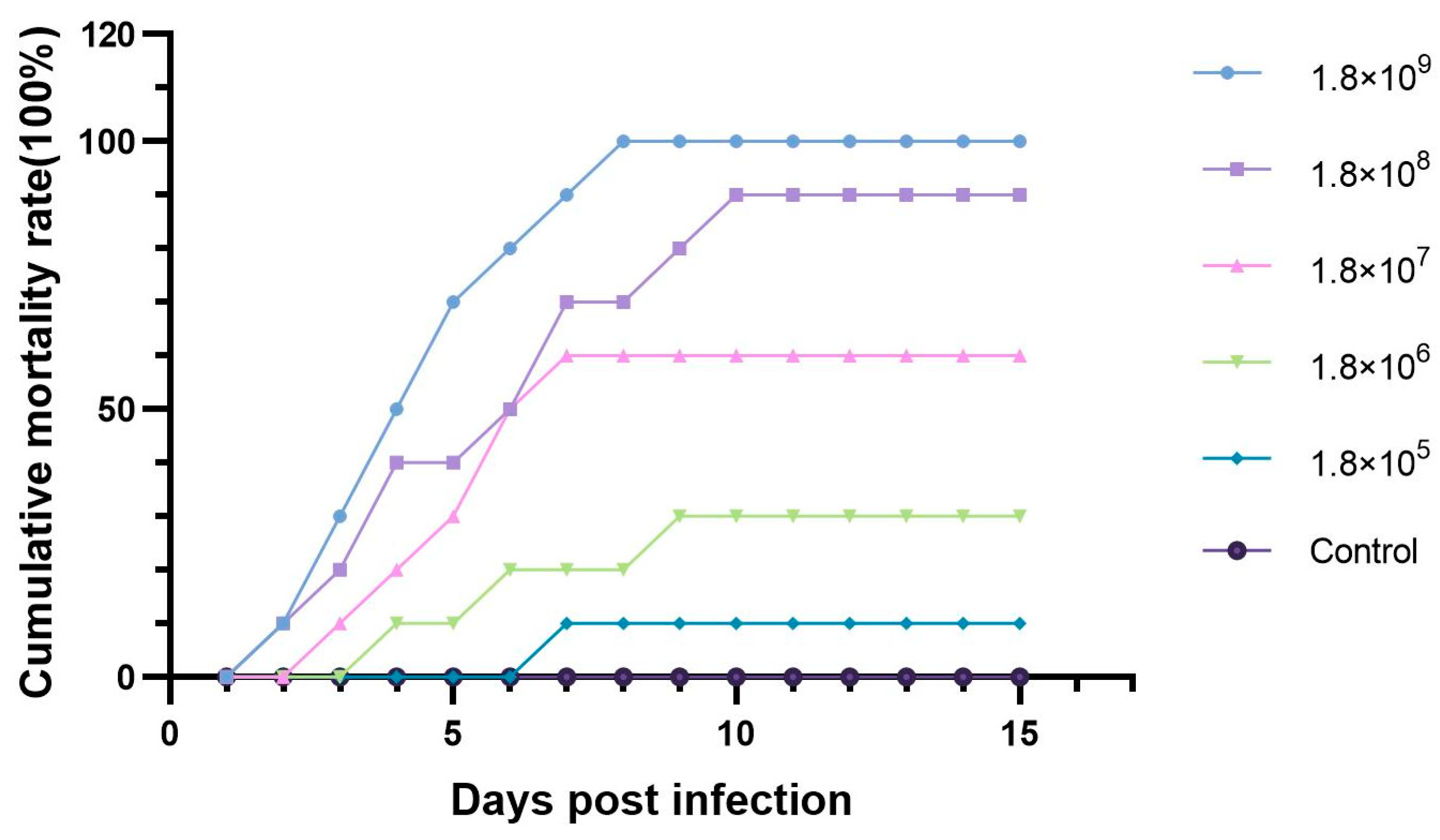
3.2. The Pathogenicity of ST-1902 to Hybrid Sturgeon
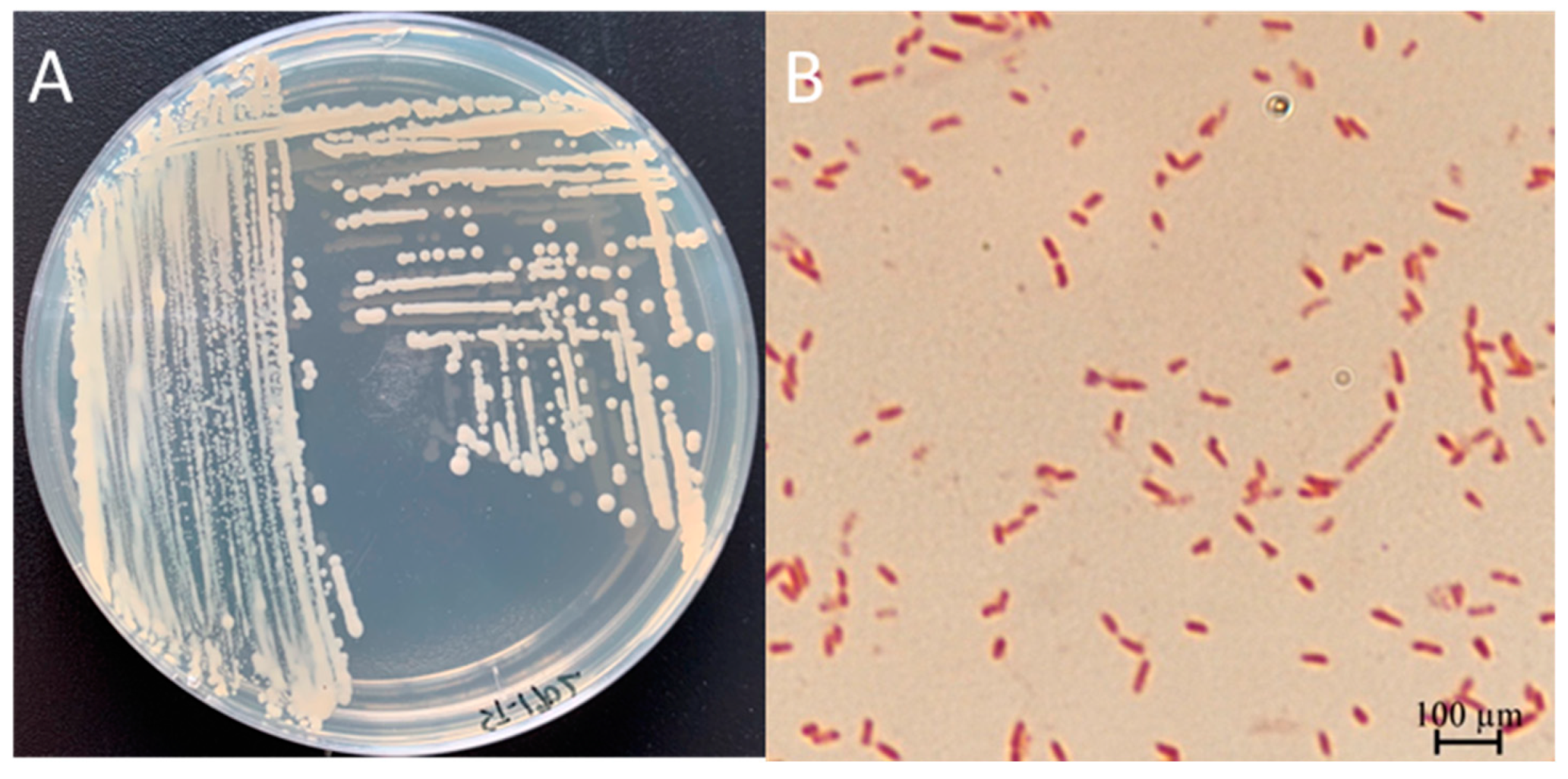
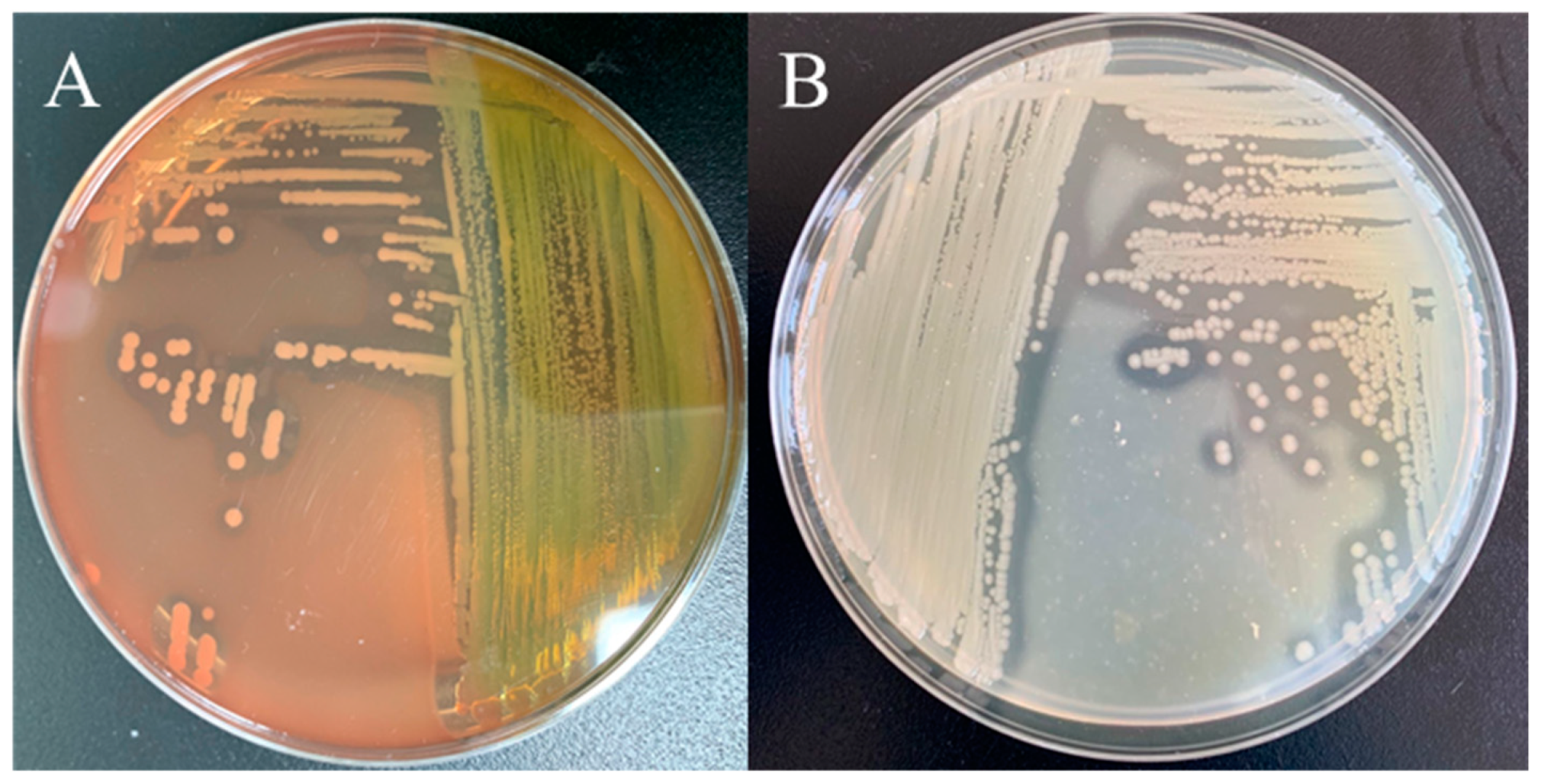
3.3. Physiological and Biochemical Characterization of ST-1902
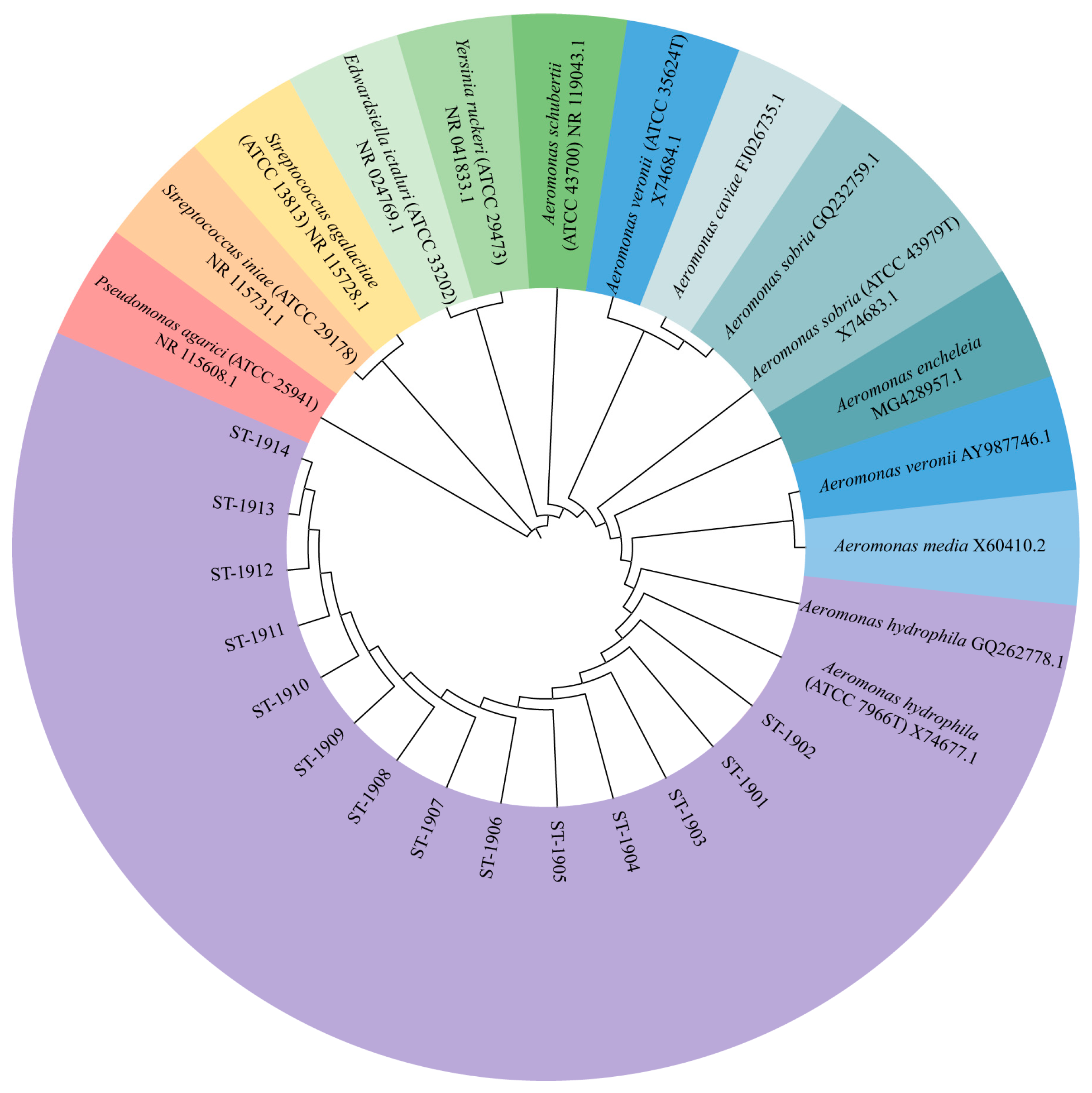
3.4. Molecular Identification and Phylogenetic Analysis
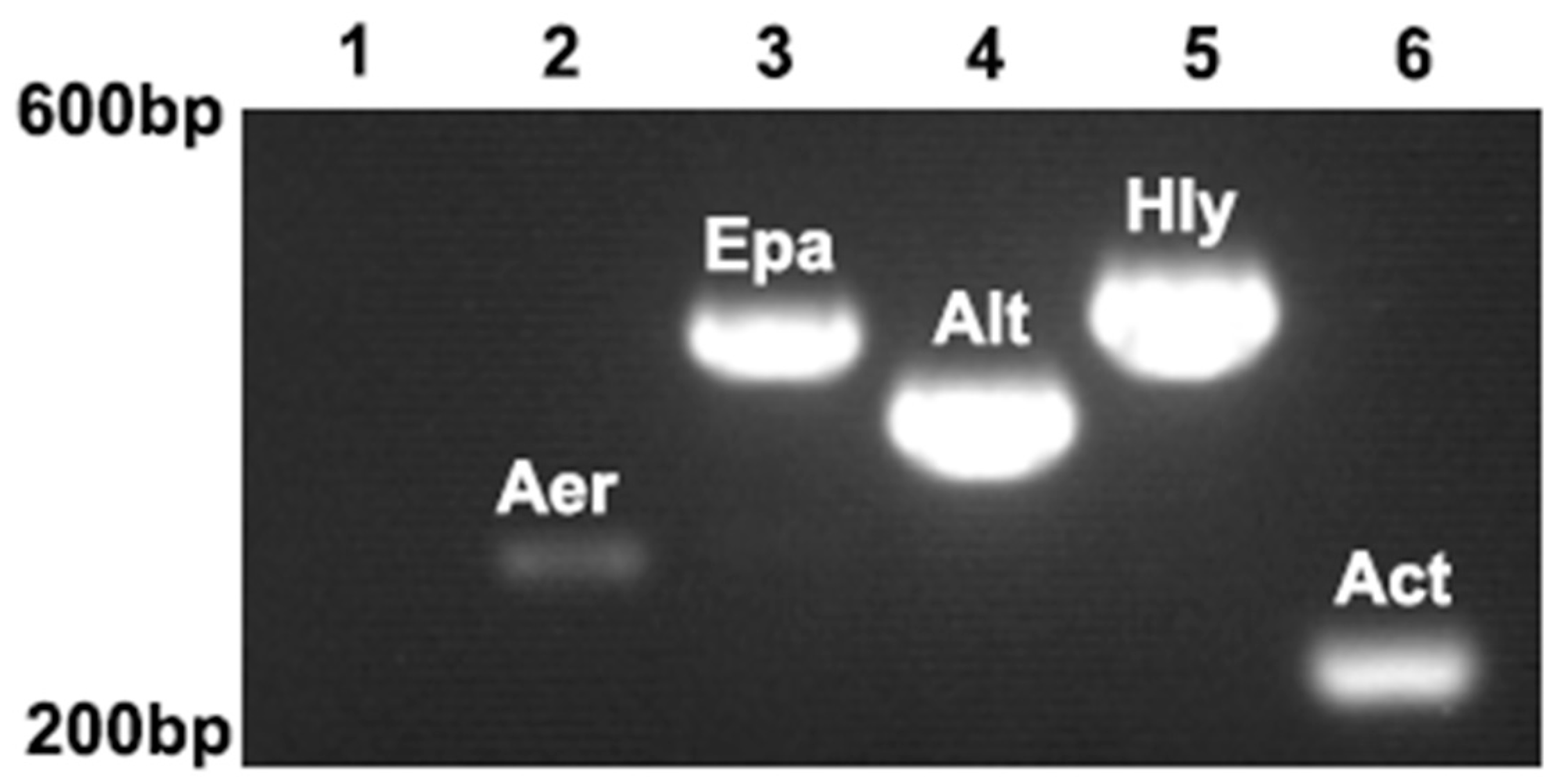
3.5. Main Virulence Genes for ST-1902
3.6. Histopathological Analysis
3.7. Antimicrobial Sensitivity and Resistance Gene Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
| Tissue | ||||||
|---|---|---|---|---|---|---|
| Date | Liver | Spleen | Kidney | Ascites | Total | Number of the Strains |
| 1 July 2019 | 1 | 1 | 1 | 4 | 7 | ST-1901~ST-1907 |
| 5 July 2019 | 1 | 1 | 1 | 2 | 5 | ST-1908~ST-1912 |
| 10 July 2019 | 1 | 0 | 0 | 1 | 2 | ST-1913~ST-1914 |
References
- Ludwig, A. Identification of Acipenseriformes species in trade. J. Appl. Ichthyol. 2008, 24, 2–19. [Google Scholar] [CrossRef]
- Bronzi, P.; Rosenthal, H.; Gessner, J. Global sturgeon aquaculture production: An overview. J. Appl. Ichthyol. 2011, 27, 169–175. [Google Scholar] [CrossRef]
- Williot, P.; Sabeau, L.; Gessner, J.; Arlati, G.; Bronzi, P.; Gulyas, T.; Berni, P. Sturgeon farming in Western Europe: Recent developments and perspectives. Aquat. Living Resour. 2001, 14, 367–374. [Google Scholar] [CrossRef]
- Fishery Administration Bureau of Ministry of Agriculture and Rural Affairs, National Fisheries Technology Extension Center and China Society of Fisheries. The China Fishery Statistical Yearbook; China Agricultural Press: Beijing, China, 2024; pp. 1–159. [Google Scholar]
- Chen, X.; Li, C.; Yang, C.; Zhang, S.; Wu, J. Status and prospects of techniques in the sturgeon aquaculture industry in China. Freshw. Fishries. 2017, 47, 108–112. [Google Scholar]
- Luo, L.; Ai, L.; Li, T.; Xue, M.; Wang, J.; Li, W.; Liang, X. The impact of dietary DHA/EPA ratio on spawning performance, egg and offspring quality in Siberian sturgeon (Acipenser baeri). Aquaculture 2015, 437, 140–145. [Google Scholar] [CrossRef]
- Tian, T.; Yuan-Jin, Y.; Jing-Shu, W.; Xu, Z. Research Progress in Sturgeon Diseases. Hubei Agric. Sci. 2012, 51, 559–563. [Google Scholar]
- Jiang, N.; Fan, Y.; Zhou, Y.; Ma, J.; Liu, W.; Zeng, L. Overview of sturgeon pathogenic disease research. J. Hydroecology 2016, 37, 1–9. [Google Scholar]
- Chu, W.H.; Yu, Y. The detection of Aeromonas hydrophila of sturgeon. Freshw. Fishries 2003, 33, 16–17. [Google Scholar]
- Li, Y.-Y.; Cao, H.-P.; He, S.; Yang, X.-L. Isolation and identification of Aeromonas hydrophila strain X1 from Acipenser baerii and its antibiotic sensitivity. Microbiol. China 2008, 35, 1186–1191. [Google Scholar]
- Yang, Z.G. Isolation and identification of the Aeromonas hydrophila pathogen the sturgeon. Freshw. Fishries 2001, 31, 41. [Google Scholar]
- Cao, H.; Yang, X.; Wang, Y.; Li, Y. Isolation and growth characteristics of pathogenic Aeromonas punctata caviae from Sturgeons. Chin. J. Zool. 2007, 42, 1–6. [Google Scholar]
- Modarres, M.B.S.; Akhlaghi, M.; Sharifiyazdi, H. Phenotypic and genetic diversity of motile aeromonads isolated from diseased fish and fish farms. Iran. J. Vet. Res. 2014, 15, 238–243. [Google Scholar]
- Liu, Y.; Yang, R.; Chen, Y.; Song, M.; Yan, Q.; Gong, Q.; Li, Q.; Yang, H.; Lai, J. Isolation identification of Aeromonas veronii in Acipenser dabryanus and its histological observations. J. South. Agric. 2018, 49, 1235–1241. [Google Scholar]
- Yang, Y.; Yang, Q.; Liu, Y.; Xu, N.; Yang, X.; Ai, X. Isolation, identification and drug sensitivity of Streptococcus dysgalactiae subsp. dysgalactiae from Acipenser gueldenstaedti. Chin. J. Prev. Vet. Med. 2017, 39, 717–721. [Google Scholar]
- Yang, Y.; Xia, Y.; Zhao, L.; Zheng, W.; Qiu, J.; Cao, H.; Hu, K.; Yang, X. Isolation and identification of Citrobacter freundii from Amur. Sturgeon (Acipenser schrenckii Brandt) and its antibiotic sensitivity. Acta Hydrobiol. Sin. 2013, 37, 766–771. [Google Scholar]
- Xu, J.; Zeng, X.; Jiang, N.; Zhou, Y.; Zeng, L. Pseudomonas alcaligenes infection and mortality in cultured Chinese sturgeon, Acipenser sinensis. Aquaculture 2015, 446, 37–41. [Google Scholar] [CrossRef]
- Kunming, Y.; Wenrun, Z.; JiangXia, M.; ChengRen, D.; Aimin, G.; Zhisheng, X.; Cheng, Y. Isolation, identification and antibiotic sensitivity of pathogenic bacterium Yersinia ruckeri from Siberian sturgeon Acipenser baerii. Fish. Sci. 2019, 1, 48–54. [Google Scholar]
- Zhang, D.-F.; Li, A.-H.; Gong, X.-N. Research on the mycobacteriosis and its pathogen in sturgeons. Acta Hydrobiol. Sin. 2014, 38, 495–504. [Google Scholar]
- Igbinosa, I.H.; Beshiru, A.; Odjadjare, E.E.; Ateba, C.N.; Igbinosa, E.O. Pathogenic potentials of Aeromonas species isolated from aquaculture and abattoir environments. Microb. Pathog. 2017, 107, 185–192. [Google Scholar] [CrossRef]
- Zmys Owska, I.; Korzekwa, K.; Szarek, J. Aeromonas hydrophila in Fish Aquaculture. J. Comp. Pathol. 2009, 141, 313. [Google Scholar] [CrossRef]
- Hu, A.; Zhang, M.; Zhang, P.; Yang, X.; Cheng, Z.; Jiang, H.; Wen, M. Isolation, identification, drug resistance and virulence genes detection of Aeromonas hydrophila from carp. China Anim. Husb. Vet. Med. 2019, 46, 2167–2175. [Google Scholar]
- Li, S.; Wu, L.S.; Wang, D.; Cao, Y.; Sheng, C.Y.; Liu, H.; Bai, L.H.; Lu, T.; Yan, L.T. Identification of the causative pathogen causing skin-ulcer in Silurus soldatovi and its pathogenicity. J. Fish. China 2018, 42, 1446–1453. [Google Scholar]
- Yang, W.; Li, A. Isolation and characterization of Streptococcus dysgalactiae from diseased Acipenser schrenckii. Aquaculture 2009, 294, 14–17. [Google Scholar] [CrossRef]
- Xuan, H.; Cai, J.; Xia, L. Research progress of cfr gene mediated bacteria resistance. J. Agric. Sci. Technol. 2019, 21, 43–50. [Google Scholar]
- Wang, J.; Zhong, W.J.; Zhong, J.; Ping, Z.J.; Jin, F.; Xiang, J.F.; Wang, H.; Yun, W.H.; Xu, W.; Fang, X.W.; et al. Virulence of resistant Aeromonas hydrophila and six kinds of antibiotic resistance components genes. Chin. J. Zoonoses 2018, 34, 912–919. [Google Scholar]
- Huang, W.; Xiao, S.; Li, S. Detection of antibiotics resistance and resistance genes in Vibrio parahaemolyticus from Litopenaeus vannamei of Guangxi. Southwest China J. Agric. Sci. 2018, 31, 1979–1988. [Google Scholar]
- Hamilton, M.; Russo, R.; Thurston, R. Trimmed Spearman-Karber method for estimating median lethal concentrations in bioassays. Environ. Sci. Technol. 1978, 12, 417. [Google Scholar] [CrossRef]
- Pelkola, K.; Vennerström, P.; Viljamaa-Dirks, S.; Kuronen, H. Bacterial infections of farmed sturgeon in Finland. Fin. Food Saf. Auth. Evira. 2012, 12, 122–127. [Google Scholar]
- TİMUR, G.; Akayli, T.; Korun, J.; Yardimcir, R.E. A study on bacterial haemorhagic septicemia in farmed young Russian sturgeon (Acipencer gueldenstaedtii) in Turkey. J. Aquat. Sci. 2010, 25, 19–26. [Google Scholar]
- Beaz-Hidalgo, R.; Figueras, M. Aeromonas spp. whole genomes and virulence factors implicated in fish disease. J. Fish. Dis. 2013, 36, 371–388. [Google Scholar]
- Long, S.; Han, S.; Niu, Z.; Liang, J.; Hu, D.; Huang, J.; Li, H.; Liu, Q.; Su, J. Isolation and identification of pathogenic Aeromonas in Clarias fuscus and analysis of the correlation between its pathogenicity and virulence genotypes. J. Fish. China 2016, 3, 308–317. [Google Scholar]
- Tomás, J. The main Aeromonas pathogenic factors. Isrn Microbiol. 2012, 2012, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Sreedharan, K.; Philip, R.; Singh, I.S.B. Characterization and virulence potential of phenotypically diverse Aeromonas veronii isolates recovered from moribund freshwater ornamental fishes of Kerala, India. Int. J. Gen. Mol. Microbiol. 2013, 103, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Yang, H.; Li, T.; Luo, L.; Gao, J. Isolation and identification of pathogenic Aeromonas veronii isolated from infected Siberian sturgeon (Acipenser baerii). Acta Microbiol. Sinica 2009, 49, 1289–1294. [Google Scholar]
- Pan, L.-D.; Li, S.-Y.; Xia, Y.-T. Preliminary studies on histopathogy epicardial septic granuloma of sturgeon. Acta Hydrobiol. Sin. 2000, 24, 563–566. [Google Scholar] [CrossRef]
- Chang, O.; Liu, X.; Pan, H.; Shi, C.; Sun, H.; Wu, S. Histopathological and pathogeny preliminary research on mulberry heart of Siberian sturgeon (Acipenser baeri). J. Fish. China 2014, 6, 904–911. [Google Scholar]
- Guarda, F.; Bertoja, G.; Zoccarato, I.; Tartari, E.; Biolatti, B. Spontaneous steatitis of epicardial fat in farmed white sturgeon (Acipenser transmontanus). Aquaculture 1997, 158, 167–177. [Google Scholar] [CrossRef]
- Ye, C.; Wang, K.; Liu, T.; He, Y.; Wang, E.; Yang, Q.; He, Q.; Xie, H.; He, S. Isolation and identification of pathogenic Aeromonas hydrophila isolated from Acipenser baerii. Chin. J. Prev. Vet. Med. 2018, 40, 195–199. [Google Scholar]
- Wang, Y.T.; Chen, C.F.; Lu, Y.H. Practical role of drug resistance detection data of pathogenic bacteria aquaculture animals. China Fish 2013, 10, 57–61. [Google Scholar] [CrossRef]
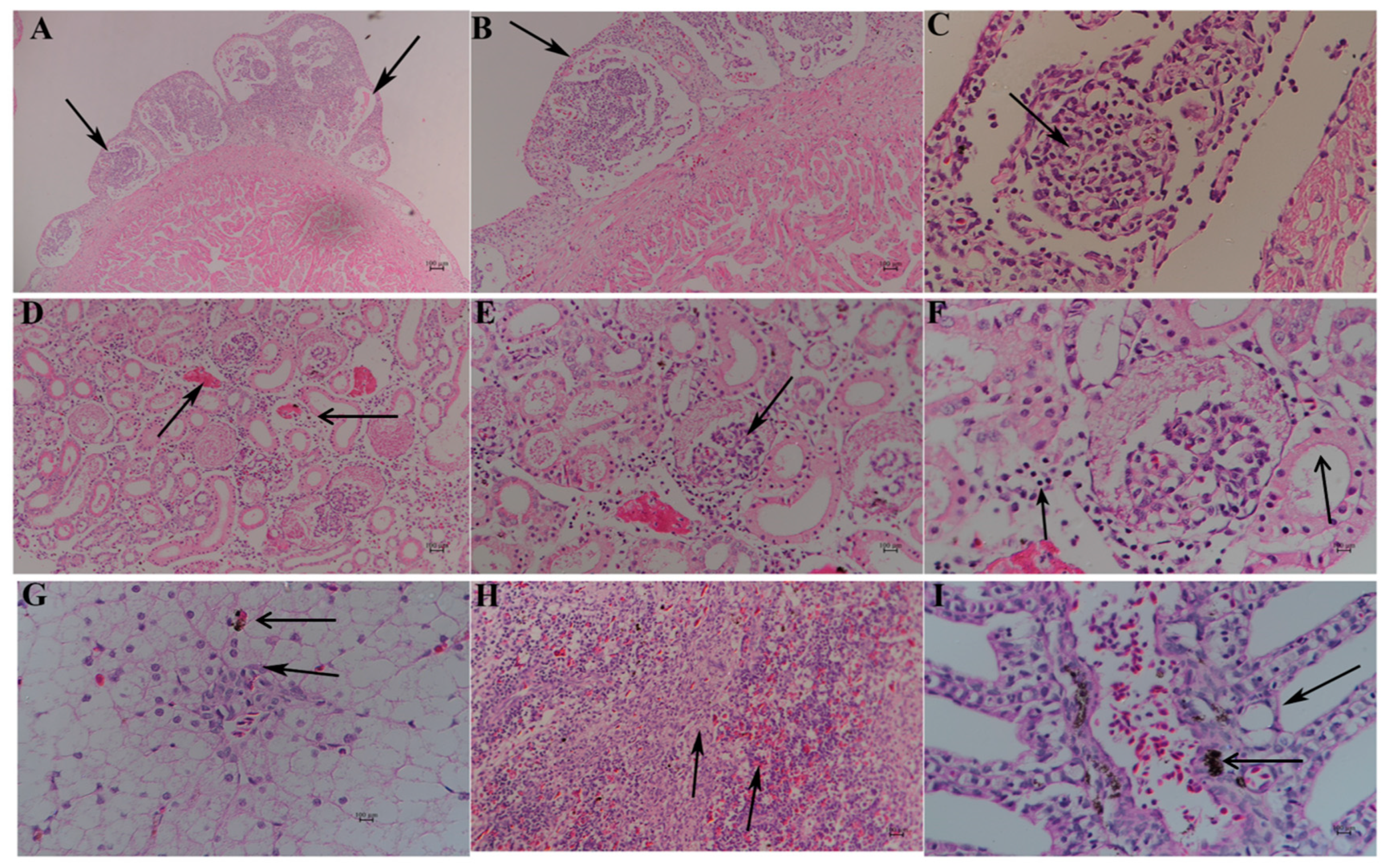
 ) (4×). (B): Undifferentiated spherical, spindle cells, and vascular tissue (
) (4×). (B): Undifferentiated spherical, spindle cells, and vascular tissue ( ) (20×). (C): Mononuclear cell (
) (20×). (C): Mononuclear cell ( ) (40×). (D): Enlarged renal interstitial space (
) (40×). (D): Enlarged renal interstitial space ( ) with hyperemia (
) with hyperemia ( ) (10×). (E): Atrophic glomeruli with inflammatory cell infiltration (
) (10×). (E): Atrophic glomeruli with inflammatory cell infiltration ( ) (20×). (F): Leucocytes, neutrophils around renal interstitial space (
) (20×). (F): Leucocytes, neutrophils around renal interstitial space ( ), irregular and disassembled nuclei in renal tubule (
), irregular and disassembled nuclei in renal tubule ( ) (20×). (G): Swollen, vacuolar, and necrotic hepatocytes (
) (20×). (G): Swollen, vacuolar, and necrotic hepatocytes ( ), with inflammatory cells around the central vein and hepatic sinus (
), with inflammatory cells around the central vein and hepatic sinus ( ) (10×). (H): The blurred boundary between the red pulp and white pulp (
) (10×). (H): The blurred boundary between the red pulp and white pulp ( ) (10×). (I): Cavitated gill filaments (
) (10×). (I): Cavitated gill filaments ( ) and hemosiderosis on epithelial cells (
) and hemosiderosis on epithelial cells ( ).
).
 ) (4×). (B): Undifferentiated spherical, spindle cells, and vascular tissue (
) (4×). (B): Undifferentiated spherical, spindle cells, and vascular tissue ( ) (20×). (C): Mononuclear cell (
) (20×). (C): Mononuclear cell ( ) (40×). (D): Enlarged renal interstitial space (
) (40×). (D): Enlarged renal interstitial space ( ) with hyperemia (
) with hyperemia ( ) (10×). (E): Atrophic glomeruli with inflammatory cell infiltration (
) (10×). (E): Atrophic glomeruli with inflammatory cell infiltration (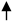 ) (20×). (F): Leucocytes, neutrophils around renal interstitial space (
) (20×). (F): Leucocytes, neutrophils around renal interstitial space (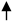 ), irregular and disassembled nuclei in renal tubule (
), irregular and disassembled nuclei in renal tubule (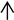 ) (20×). (G): Swollen, vacuolar, and necrotic hepatocytes (
) (20×). (G): Swollen, vacuolar, and necrotic hepatocytes (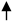 ), with inflammatory cells around the central vein and hepatic sinus (
), with inflammatory cells around the central vein and hepatic sinus ( ) (10×). (H): The blurred boundary between the red pulp and white pulp (
) (10×). (H): The blurred boundary between the red pulp and white pulp ( ) (10×). (I): Cavitated gill filaments (
) (10×). (I): Cavitated gill filaments ( ) and hemosiderosis on epithelial cells (
) and hemosiderosis on epithelial cells ( ).
).
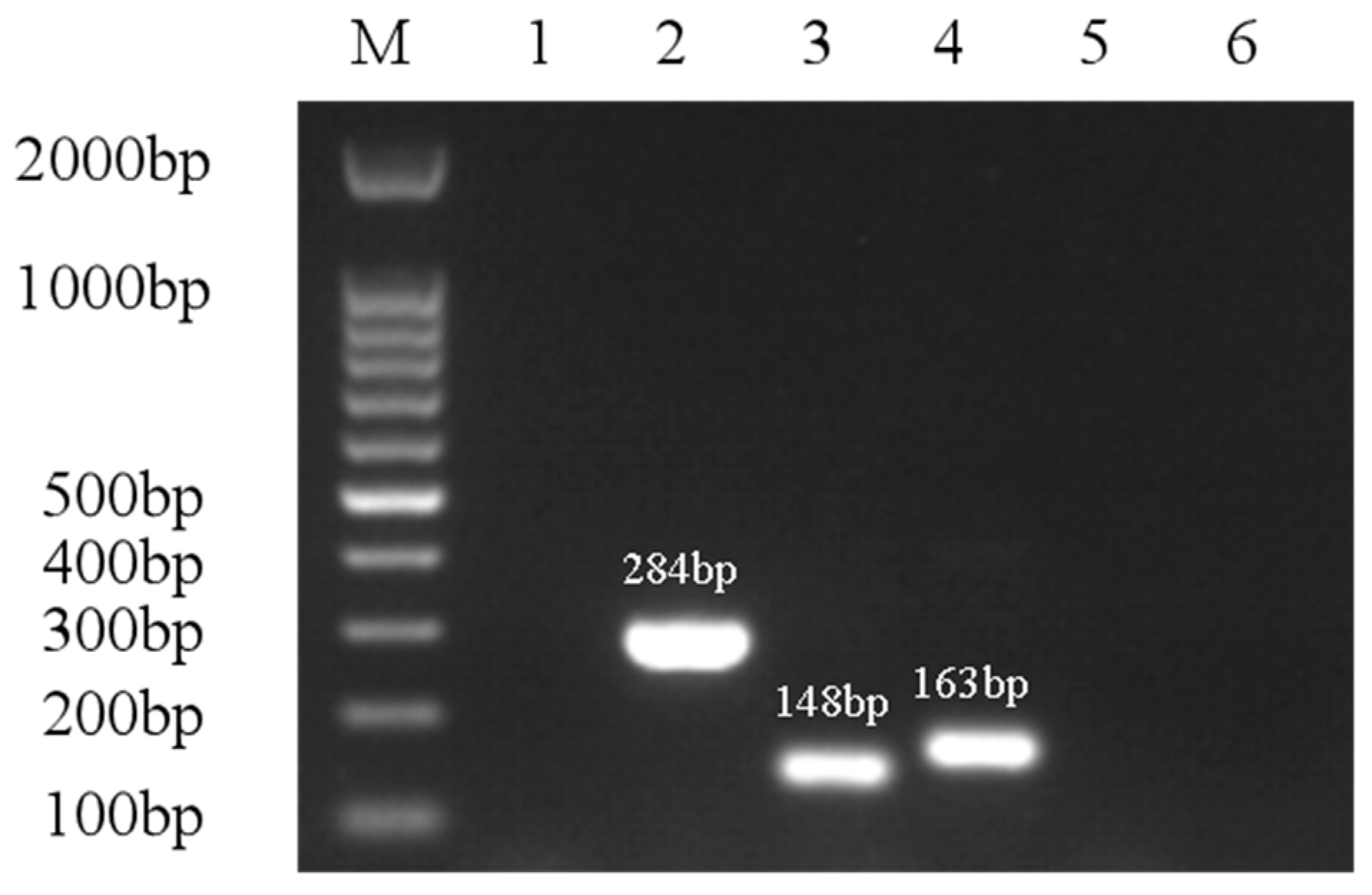
| Genes | Primer Sequences (5′→3′) | Amplicon Size/bp | Annealing Temperature/°C | GenBank Accession No. |
|---|---|---|---|---|
| Aer | F: CACAGCCAATATGTCGGTGAAG | 301 | 52 | MH607886.1 |
| R: GTCACCTTCTCGCTCCAGGC | ||||
| Hly | F: GGCCGGTGGCCCGAAGATAGGG | 584 | 64 | KP942382.1 |
| R: GGCGGCGCCGGACGAGACGGG | ||||
| Alt | F: ATGACCCAGTCCTGGCACGG | 482 | 58 | KC695750.1 |
| R: GCCGCTCAGGGCGAAGCCGC | ||||
| Act | F: TACCACCACCTCCCTGTCGC | 249 | 60 | KC687134.1 |
| R: ATGCTGCTCGCCTTGTGGTT | ||||
| Epa | F: TGGTTGTCGGCGTTGTTGAG | 543 | 61 | DQ137138.1 |
| R: TGTGGGTGGACGGAGTGAGT | ||||
| Sul1 | F: CGCACCGGAAACATCGCTGCAC | 163 | 56.3 | JN088221 |
| R: TGAAGTTCCGCCGCAAGGCTCG | ||||
| Intl1 | F: GGCTTCGTGATGCCTGCTT | 146 | 63 | MF769724.1 |
| R: CATTCCTGGCCGTGGTTCT | ||||
| Cfr | F: TGTGCTACAGGCAACATTGGAT | 148 | 55 | AJ249217 |
| R: CAAATACTTGACGGTTGGCTAGAG | ||||
| Ant (3″)-I | F: TGATTTGCTGGTTACGGTGAC | 284 | 53 | FJ644661.1 |
| R: CGCTATGTTCTCTTGCTTTTG | ||||
| FexA | F: ATTCTCCCGCAAATAACG | 156 | 52 | AJ549214 |
| R: TCGGCTCAGTAGCATCACG |
| Biochemical Items | ST-1902 | ATCC 7966 | Biochemical Items | ST-1902 | ATCC 7966 |
|---|---|---|---|---|---|
| Gram stain | − | − | Methyl red | + | + |
| Oxidase | + | + | Glucose | + | + |
| Catalase | + | + | Mannitol | + | + |
| Motility | + | + | Inositol | − | − |
| Glucose fermented | + | + | Sorbitol | + | − |
| Galactoside | − | − | Rhamnol | + | D |
| Arginine | + | + | Sucrase | + | + |
| Lysine | + | + | Melibiose | − | − |
| Ornithine | − | − | Amygdalin | − | − |
| Phenylalanine TDA | − | D | Arabinose | + | + |
| Citrate | + | D | Nitrate reduction | + | + |
| Hydrothion | + | + | Nitrite reduction | + | + |
| Urea | − | − | H2S | + | + |
| Indole | + | + | KCN | − | − |
| Pyruvate | + | D | SS Agar | + | + |
| Gelatin | + | + | MacConkey Agar | + | + |
| Drugs | IZD/mm | Resistance | Drugs | IZD/mm | Resistance |
|---|---|---|---|---|---|
| Sulfamethoxazole | 8 | R | Enrofloxacin | 24 | S |
| Amoxicillin | 8 | R | Levofloxacin | 22 | S |
| Ceftriaxone | 32 | S | Chloramphenicol | 22 | S |
| Vancomycin | 0 | R | Florfenicol | 24 | S |
| Neomycin | 6 | R | Azithromycin | 14 | I |
| Gentamicin | 10 | R | Acetylspiramycin | 4 | R |
| Kanamycin | 8 | R | Rifampicin | 18 | I |
| Tobramycin | 10 | R | Polymyxin B | 6 | R |
| Amikacin | 9 | R | Doxycycline | 18 | I |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, H.; Weng, X.; Pang, G.; Li, X.; Xia, J.; Gao, X.; He, J.; Li, J.; Qian, D. Identification, Pathogenicity, and Antimicrobial Resistance Analysis of Bacterial Pathogenesis Aeromonas hydrophila from Hybrid Sturgeon (Huso dauricus ♀ × A. schrenckii ♂) in Zhejiang, China. Microorganisms 2025, 13, 278. https://doi.org/10.3390/microorganisms13020278
Hu H, Weng X, Pang G, Li X, Xia J, Gao X, He J, Li J, Qian D. Identification, Pathogenicity, and Antimicrobial Resistance Analysis of Bacterial Pathogenesis Aeromonas hydrophila from Hybrid Sturgeon (Huso dauricus ♀ × A. schrenckii ♂) in Zhejiang, China. Microorganisms. 2025; 13(2):278. https://doi.org/10.3390/microorganisms13020278
Chicago/Turabian StyleHu, Haojie, Xinzhi Weng, Gang Pang, Xiaobing Li, Jing Xia, Xiu Gao, Jie He, Ji Li, and Dong Qian. 2025. "Identification, Pathogenicity, and Antimicrobial Resistance Analysis of Bacterial Pathogenesis Aeromonas hydrophila from Hybrid Sturgeon (Huso dauricus ♀ × A. schrenckii ♂) in Zhejiang, China" Microorganisms 13, no. 2: 278. https://doi.org/10.3390/microorganisms13020278
APA StyleHu, H., Weng, X., Pang, G., Li, X., Xia, J., Gao, X., He, J., Li, J., & Qian, D. (2025). Identification, Pathogenicity, and Antimicrobial Resistance Analysis of Bacterial Pathogenesis Aeromonas hydrophila from Hybrid Sturgeon (Huso dauricus ♀ × A. schrenckii ♂) in Zhejiang, China. Microorganisms, 13(2), 278. https://doi.org/10.3390/microorganisms13020278





