Comparative Analysis of Microbial Communities in Diseased and Healthy Sweet Cherry Trees (Prunus avium L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Material
2.2. DNA Extraction, PCR Analysis and MiSeq Illumina Sequencing
2.3. Sequencing Data Processing
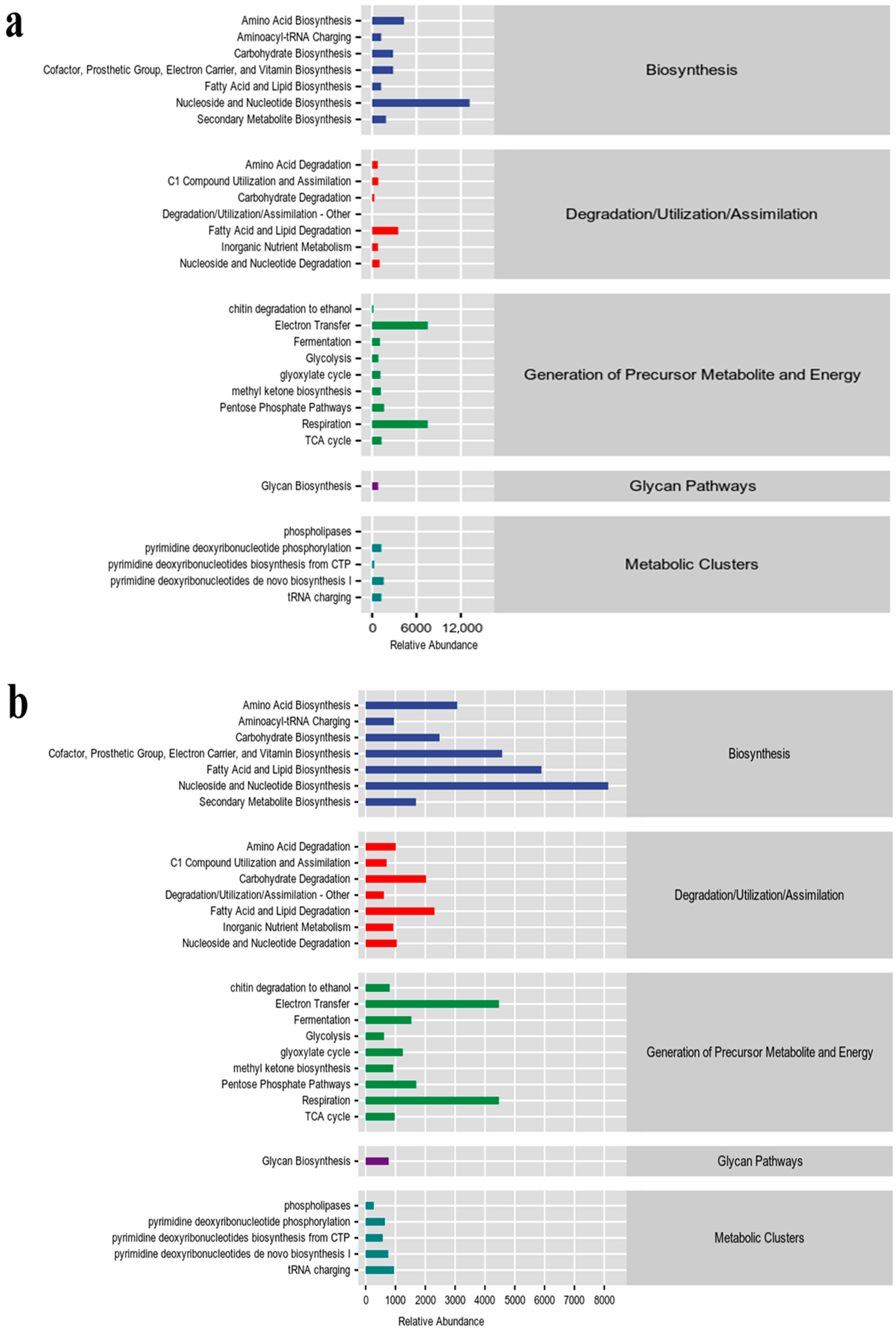
2.4. Functional Gene Prediction
2.5. Statistical Analysis
3. Results
3.1. Amplicon Sequencing and Community Diversity Overview
3.2. Changes in Bacterial Community Structure at Phylum and Genus Level
3.3. Changes in Bacterial Community Structure at the Phylum and Genus Level
3.4. Changes in Fungal Community Structure at the Phylum and Genus Level
3.5. Enrichment of Bacterial Colonies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, H.; Peng, X.; Jiang, P.; Xing, L.; Sun, X. Prediction of potential suitable distribution for sweet cherry (Prunus avium) based on the MaxEnt model. PLoS ONE 2024, 19, e0294098. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.W.; Li, M.; Tan, Y.; Zhang, X.M.; Wang, B.G.; Yan, G.H.; Wang, J.; Pan, F.R.; Liu, Q.Z.; Zhang, K.C. Fruit scientific research in New China in the past 70 years: Cherry. J. Fruit Sci. 2019, 36, 1339–1351. [Google Scholar]
- Butler, O. A study on gummosis of Prunus and Citrus, with observations on squamosis and exanthema of the Citrus. Ann. Bot. 1911, 25, 107–153. [Google Scholar] [CrossRef]
- Ezra, D.; Hershcovich, M.; Shtienberg, D. Insights Into the Etiology of Gummosis Syndrome of Deciduous Fruit Trees in Israel and its Impact on Tree Productivity. Plant Dis. 2017, 101, 1354–1361. [Google Scholar] [CrossRef]
- Chen, P.; Abeywickrama, P.D.; Ji, S.; Zhou, Y.; Li, X.; Zhang, W.; Yan, J. Molecular Identification and Pathogenicity of Diaporthe eres and D. hongkongensis (Diaporthales, Ascomycota) Associated with Cherry Trunk Diseases in China. Microorganisms 2023, 11, 2400. [Google Scholar] [CrossRef]
- Zhang, K.F.; Liu, P.W.; Xin, P.M. Effect of animal boneorganic fertilizer to control cherry gummosis. Shandong Agric. Sci. 2009, 12, 93–95. (In Chinese) [Google Scholar]
- Zhang, Y.C.; Ma, H.Y.; Chen, H.Y. Control and Prevention to cherry gummosis in Aba Tibetan Autonomous Prefecture. Sichuan Agric. Sci. Technol. 2011, 8, 40–41. (In Chinese) [Google Scholar]
- Wang, Y.; Song, X.; Xie, S.; Geng, Y.; Xu, C.; Yin, X.; Zang, R.; Guo, L.; Zhang, M.; Guo, Y. Diversity of Lasiodiplodia species associated with canker and dieback in fruit trees in the Henan and Shandong Provinces of China. Plant Dis. 2024, 108, 563–575. [Google Scholar] [CrossRef]
- Zhang, L.Z.; Zhang, Q.; Yang, P. First Report of Gummosis Disease of Sweet Cherry Caused by Botryosphaeria dothidea in China. Plant Dis. 2019, 103, 32–83. [Google Scholar] [CrossRef]
- Ding, L.; Xu, L.; Chu, X.; Yang, L.; Zhu, H.; Huang, J. Dissimilarity analysis of microbial communities in the rhizosphere and tissues of diseased and healthy cherry trees (Cerasus pseudocerasus). Can. J. Plant Pathol. 2021, 4, 43. [Google Scholar] [CrossRef]
- Li, H.Y.; Cao, R.B.; Mu, Y.T. In vitro inhibition of Botryosphaeria dothidea and Lasiodiplodia theobromae, and chemical control of gummosis disease of Japanese apricot and peach trees in Zhejiang Province, China. Crop Prot. 1995, 14, 187–191. [Google Scholar] [CrossRef]
- Beckman, T.G.; Pusey, P.L.; Bertrand, P.F. Impact of fungal gummosis on peach trees. HortScience 2003, 38, 1141–1143. [Google Scholar] [CrossRef]
- Lima, R.P.M.; Máximo, H.J.; Merfa, M.V.; Dalio, R.J.D.; Cristofani-Yaly, M.; Machado, M.A. Genetic tools and strategies for citrus breeding aiming at resistant rootstocks to gummosis disease. Trop. Plant Pathol. 2018, 43, 279–288. [Google Scholar] [CrossRef]
- Bouaziz, F.; Koubaa, M.; Ghorbel, R.E.; Chaabouni, S.E. Recent advances in Rosaceae gum exudates: From synthesis to food and non-food applications. Int. J. Biol. Macromol. 2016, 86, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.; Jung, D.R.; Park, T.H.; Lee, D.; Park, M.K.; Lim, K.; Shin, J.H. Changes in microbial community structure in response to gummosis in peach tree bark. Plants 2022, 11, 2834. [Google Scholar] [CrossRef]
- Liu, H.X.; Tan, W.P.; Sun, G.W. First report of gummosis disease of apricot (Prunus armeniaca) caused by Botryosphaeria obtusa in China. Plant Dis. 2015, 99, 888. [Google Scholar] [CrossRef]
- Baltazar, E.; Rodrigues, S.; Ares, A.; Camelo, A.; Brandão, I.; Espirito Santo, C.; Trovão, J.; Garcia, E.; Costa, J. Morphological, molecular and genomic identification and Characterisation of Monilinia fructicola in Prunus persica from Portugal. Agronomy 2023, 13, 1493. [Google Scholar] [CrossRef]
- Ogawa, J.M.; Zehr, E.I.; Bird, G.W.; Ritchie, D.F.; Uriu, K.I.; Uyemoto, J.K. Compendium of Stone Fruit Diseases; American Phytopathological Society: St. Paul, CA, USA, 1995; Volume 11, pp. 245–247. [Google Scholar]
- Al-Sadi, A.M.; Al-Ghaithi, A.G.; Al-Fahdi, N.; Al-Yahyai, R. Characterization and pathogenicity of fungal pathogens associated with root diseases of citrus in oman. Int. J. Agric. Biol. 2014, 16, 371–376. [Google Scholar]
- Ko, Y.; Liu, C.W.; Chen, S.S.; Chiu, K.Y.; Sun, Y.W.; Maruthasalam, S. First report of gummosis disease of japanese apricot caused by Botryosphaeria dothidea in Taiwan. Plant Diver. 2011, 95, 77. [Google Scholar] [CrossRef]
- William, C.O.; Martin, J.B. Ethephon-Induced Gummosis in Sour Cherry (Prunus cerasus L.) I. Effect on Xylem Function and Shoot Water Status. Plant Physiol. 1982, 70, 547–555. [Google Scholar]
- Elizabeth, A. Botryosphaeria canker and dieback of trees and shrubs in the landscape. Va. Coop. Ext. 2015, 5, 450–726. [Google Scholar]
- de Vries, D.P. Field resistance to bacterial canker in some cherry seedling populations. Euphytica 1965, 14, 78–82. [Google Scholar] [CrossRef]
- Xu, L.; Wang, J.W.; Chen, X.; Wei, H.R.; Zong, X.J.; Liu, Q.Z. Identification and pathogenicity detection of the cherry gummosis pathogen. Acta Phytopathol. Sin. 2015, 45, 350–355. [Google Scholar]
- Akbaba, M.; Ozaktan, H. Evaluation of bacteriophages in the biocontrol of Pseudomonas syringae pv. syringae isolated from cankers on sweet cherry (Prunus avium L.) in Turkey. Egypt. J. Biol. Pest Control 2021, 31, 35. [Google Scholar] [CrossRef]
- Marroni, M.V.; Casonato, S.; Pitman, A.R. Review of Pseudomonas species causing bacterial canker of Prunus species with emphasis on sweet cherry (Prunus avium) in New Zealand. Eur. J. Plant Pathol. 2023, 168, 297–314. [Google Scholar] [CrossRef]
- Griffin, F.L. A bacterial gummosis of Cherry. Science 1911, 34, 615–616. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Saniewski, M.; Ueda, J. Gummosis and leaf abscission in Yoshino cherry (Prunus yedoensis): Relevance to hormonal regulation and chemical composition of gums. ISHS Acta Hortic. 2019, 65, 467–474. [Google Scholar] [CrossRef]
- Germinara, G.S.; Pistillo, M.; Griffo, R.; Garonna, A.P.; Di Palma, A. Electroantennographic responses of Aromia bungii (Faldermann, 1835)(Coleoptera, Cerambycidae) to a range of volatile compounds. Insects 2019, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Tekpinar, A.D.; Kalmer, A. Utility of various molecular markers in fungal identification and phylogeny. Nova Hedwig. 2019, 109, 187–224. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. Dada2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Quensen, J.F.; Fish, J.A.; Lee, T.K.; Sun, Y.N.; Tiedje, J.M.; Cole, J.R. Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. mBio 2013, 4, e00592-13. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Köljalg, U.; Nilsson, R.H.; Abarenkov, K. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Hall, M.; Beiko, R.G. 16S rRNA Gene Analysis with QIIME2. In Microbiome Analysis: Methods and Protocols; Humana Press: New York, NY, USA, 2018; pp. 113–129. [Google Scholar]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- McArdle, B.H.; Anderson, M.J. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 2001, 82, 290–297. [Google Scholar] [CrossRef]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Warton, D.I.; Wright, S.T.; Wang, Y. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol. Evol. 2012, 3, 89–101. [Google Scholar] [CrossRef]
- Anderson, M.J. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 2006, 62, 245–253. [Google Scholar] [CrossRef]
- Huson, D.H.; Mitra, S.; Ruscheweyh, H.J. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef] [PubMed]
- Asnicar, F.; Weingart, G.; Tickle, T.L.; Huttenhower, C.; Segata, N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 2015, 3, 1029. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhu, W.; Zhu, K.; Ge, Y.; Li, W.; Liao, N. Similarities and differences in the microbial structure of surface soils of different vegetation types. PeerJ 2023, 11, e16260. [Google Scholar] [CrossRef]
- Deangelis, K.M.; Brodie, E.L.; Desantis, T.Z.; Andersen, G.L.; Lindow, S.E.; Firestone, M.K. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009, 3, 168–178. [Google Scholar] [CrossRef]
- Shang, Q.; Yang, G.; Wang, Y.; Wu, X.K.; Zhao, X.; Hao, H.T.; Li, Y.Y.; Xie, Z.K.; Zhang, Y.B.; Wang, R.Y. Illumina-based analysis of the rhizosphere microbial communities associated with healthy and wilted Lanzhou lily (Lilium davidii var. unicolor) plants grown in the field. World J. Microbiol. Biotechnol. 2016, 32, 95. [Google Scholar] [CrossRef] [PubMed]
- Wiedenbeck, J.; Cohan, F.M. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 2011, 35, 957–976. [Google Scholar] [CrossRef]
- Wang, G.; Huang, X.; Ng, T.B.; Lin, J.; Ye, X.Y. High phylogenetic diversity of glycosyl hydrolase family 10 and 11 xylanases in the sediment of Lake Dabusu in China. PLoS ONE 2014, 9, e112798. [Google Scholar] [CrossRef]
- O’Brien, H.E.; Parrent, J.L.; Jackson, J.A.; Moncalvo, J.M.; Vilgalys, R. Fungal community analysis by large-scale sequencing of environmental samples. Appl. Environ. Microbiol. 2005, 71, 5544–5550. [Google Scholar] [CrossRef]
- Okon, Y.; Labandera-Gonzalez, C.A. Agronomic applications of Azospirillum: An evaluation of 20 years worldwide field inoculation. Soil. Biol. Biochem. 1994, 26, 1591–1601. [Google Scholar] [CrossRef]
- Achouak, W.; Conrod, S.; Cohen, V.; Heulin, T. Phenotypic variation of Pseudomonas brassicacearum as a plant root-colonization strategy. Mol. Plant-Microbe Interact. 2004, 17, 872–879. [Google Scholar] [CrossRef]
- Bashan, Y.; de Bashan, L.E. How the plant growth-promoting bacterium Azospirillum promotes plant growth—A critical assessment. Adv. Agron. 2010, 108, 77–136. [Google Scholar]
- Fedorova, O.A.; Bezler, N.V. Influence of bacteria of the genus Azospirillum on sugar beet productivity. Future Biotechnol. 2020, 11, 101–123. [Google Scholar]
- Alvarez-Hubert, I.; Durán, R.E.; Vega-Celedón, P.; Vásconez, I.N.; Macaya, C.C.; Seeger, M. Draft genome sequences of Halotolerant Halomonas spp. SpR1 and SpR8, potential plant growth-promoting bacteria associated with Salicornia rhizosphere in a hydrothermal lagoon ecosystem of the Altiplano, Northern Chile. Microbiol. Resour. Ann. 2023, 13, 822–823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qin, X.J.; Guo, Y.P.; Zhang, Q.Q.; Ma, Q.G.; Ji, C.; Zhao, L.H. Enzymatic degradation of deoxynivalenol by a novel bacterium, Pelagibacterium halotolerans ANSP101. Food Chem. Toxicol. 2020, 140, 111–276. [Google Scholar] [CrossRef]
- Koul, M.; Singh, S. Penicillium spp.: Prolific producer for harnessing cytotoxic secondary metabolites. Anti-Cancer Drugs 2017, 28, 11–30. [Google Scholar] [CrossRef] [PubMed]
- López, S.N.; Sangorrín, M.P.; Pildain, M.B. Fruit rot of sweet cherries and raspberries caused by Penicillium crustosum and Mucor piriformis in South Patagonia, Argentina. Can. J. Plant Pathol. 2016, 3, 511–516. [Google Scholar] [CrossRef]
- Zhu, H.F.; Yang, B. Preliminary comparison of herbicidal activity of mycotoxins produced by Lecanosticta acicola and four species in genus Mycosphaerella. J. Southwest For. Univ. 2010, 30, 33–36. [Google Scholar]
- Onaebi, N.C.; Ugwuja, N.F.; Okoro, C.A.; Amujiri, N.A.; Ivoke, U.M. Mycoflora associated with post-harvest rot of onion (Allium cepa) and garlic (Allium sativum) bulbs. Res. Crops 2020, 21, 380–389. [Google Scholar]
- Sang, M.K.; Han, G.D.; Oh, J.Y.; Chun, S.C.; Kim, K.D. Penicillium brasilianum as a novel pathogen of onion (Allium cepa L.) and other fungi predominant on market onion in Korea. Crop Prot. 2014, 65, 138–142. [Google Scholar] [CrossRef]
- Yu, S.; Deng, J.; Paul, J. Reevaluation of Alternaria panax associated with leaf spot and blight of araliaceous plants. Phytopathology 2012, 102, 142. [Google Scholar]
- Zhang, J.H.; Yun, L.I.; Yu, H.; Yang, M.X.; Song, S.; Zhong, Q.Y.; Fan, L. Study on pathogen identification and biological characteristics of rice brown panicle disease in Heilongjiang Province. J. Northeast Agric. Univ. 2018, 49, 27–48. [Google Scholar]
- Branda, E.; Turchetti, B.; Diolaiuti, G.; Pecci, M.; Smiraglia, C.; Buzzini, P. Yeast and yeast-like diversity in the southernmost glacier of Europe (Calderone Glacier, Apennines, Italy). FEMS Microbiol. Ecol. 2010, 72, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.X.; Hu, Y.Y.; Wu, J.S.; Chen, J.H.; Yrjälä, K.; Yu, W.W. Change in microbial communities, soil enzyme and metabolic activity in a Torreya grandis plantation in response to root rot disease. For. Ecol. Manag. 2018, 432, 932–941. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; Qasim, M.; Haider, I.; Lou, Y. Plant-insect-microbe interaction: A love triangle between enemies in ecosystem. Sci. Total Environ. 2020, 699, 134181. [Google Scholar] [CrossRef]
- Katan, J. Diseases caused by soilborne pathogens: Biology, management and challenges. J. Plant Pathol. 2017, 99, 305–315. [Google Scholar]
- Hui, Y.; Cheng, X.; Qiang, L.; Zhang, Y.G. Present Situation and Prospect of Grub Control in Sweet Potato Field. Plant Dis. Pests 2015, 6, 1–4. [Google Scholar]
- Mostafa, S.; Wang, Y.; Zeng, W.; Jin, B. Plant responses to herbivory, wounding, and infection. Int. J. Mol. Sci. 2022, 23, 7031. [Google Scholar] [CrossRef]
- Sunamura, E.; Tamura, S.; Taki, H.; Buczkowski, G.; Shoda-Kagaya, E. Effects of neonicotinoid insecticide trunk injections on non-target arboreal ants, potential biological control agents for invasive longhorn beetle Aromia bungii on cherry trees. Appl. Entomol. Zool. 2023, 58, 401–407. [Google Scholar] [CrossRef]
- He, D.C.; Zhan, J.Y.; Xie, L.H. Problems, challenges and future of plant disease management: From an ecological point of view. J. Integr. Agric. 2016, 15, 705–715. [Google Scholar] [CrossRef]

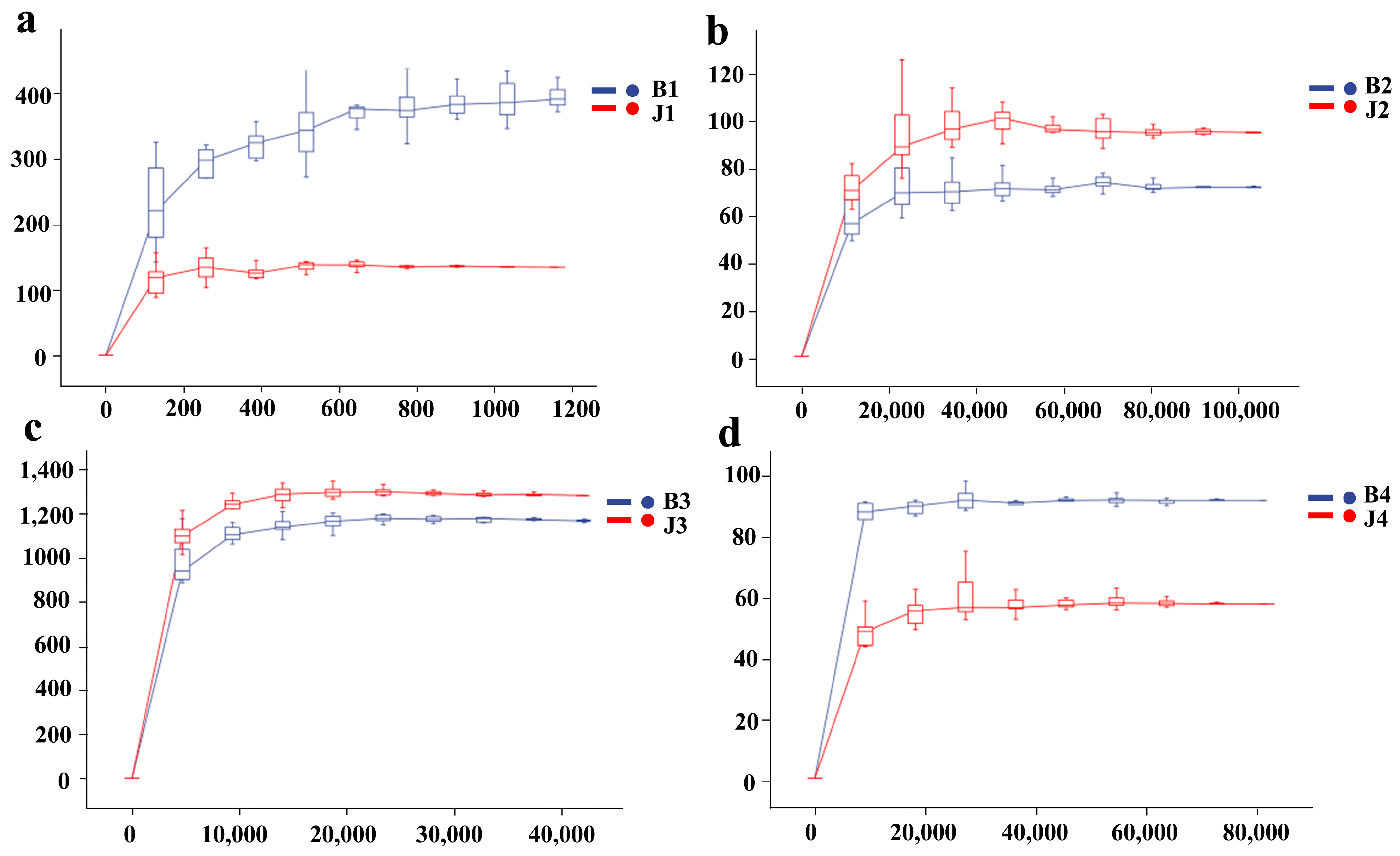
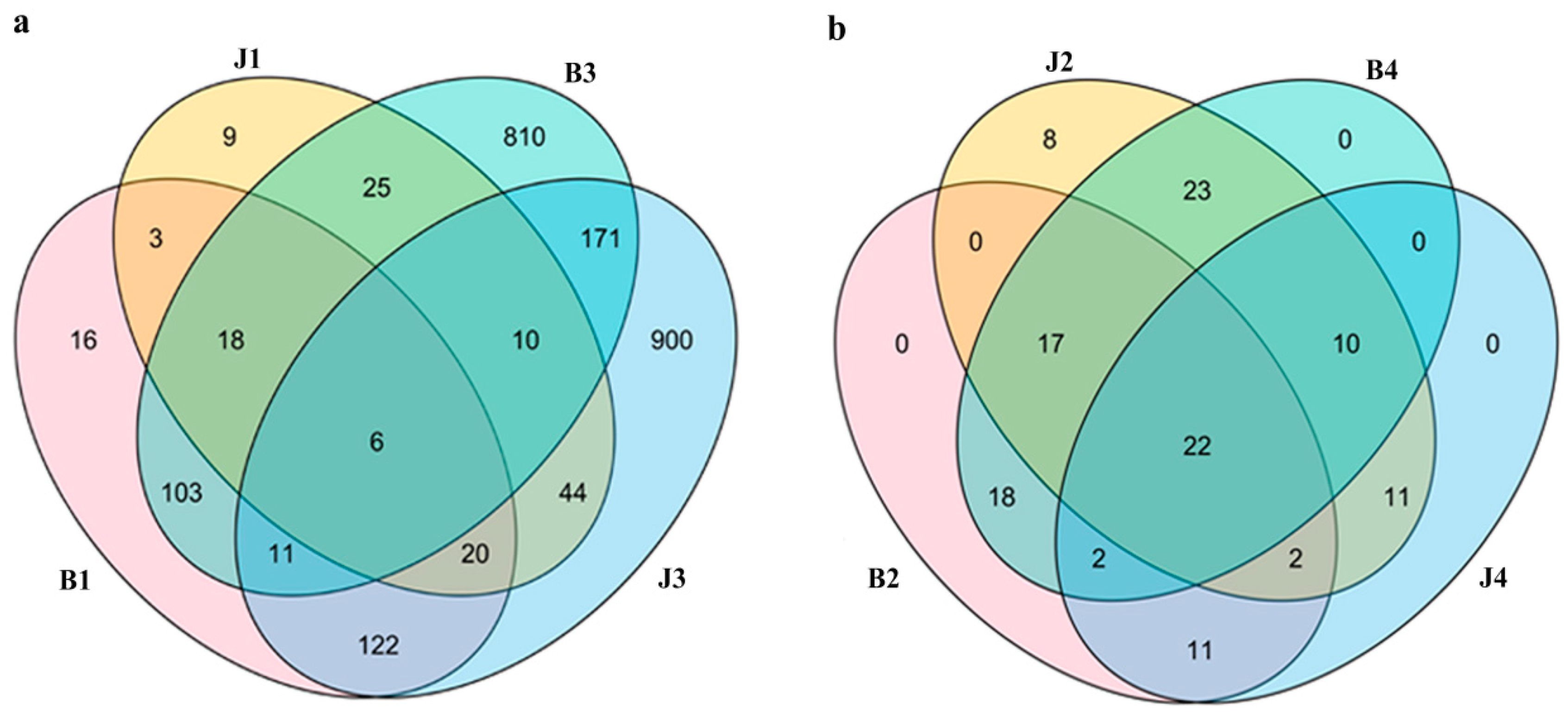

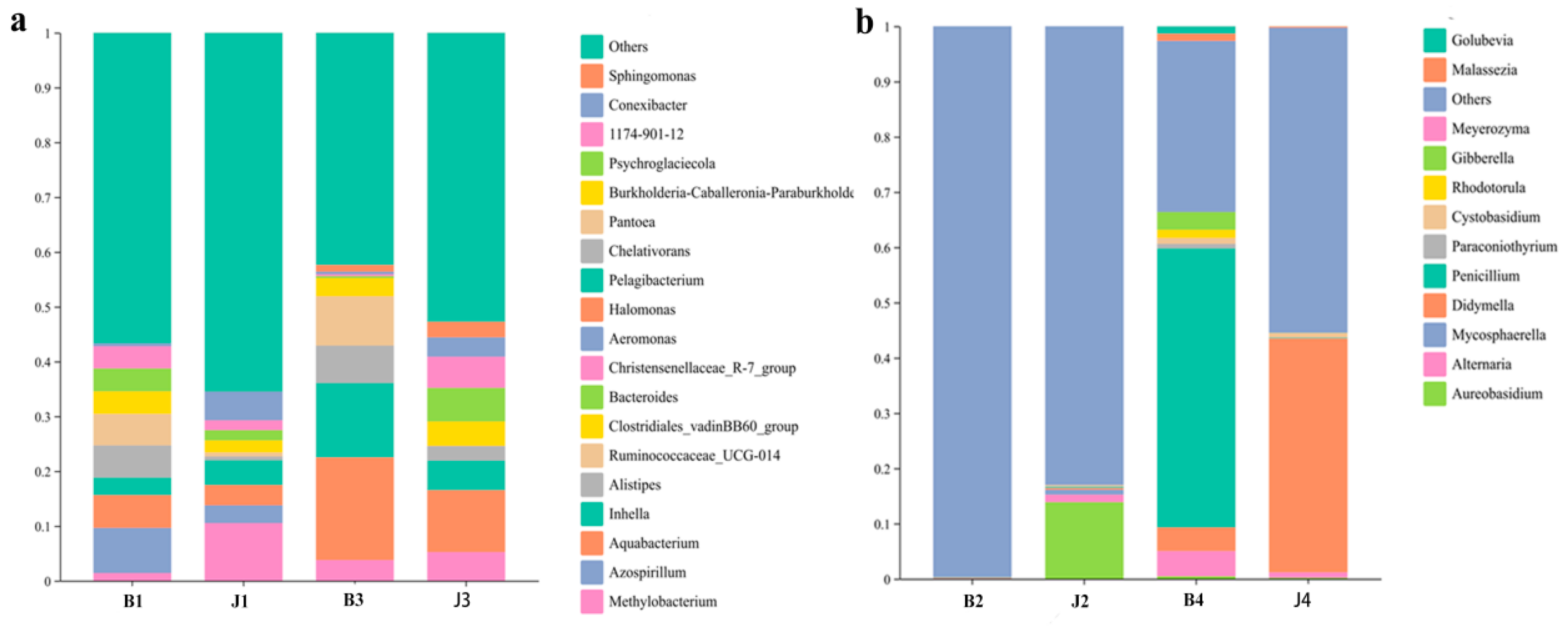
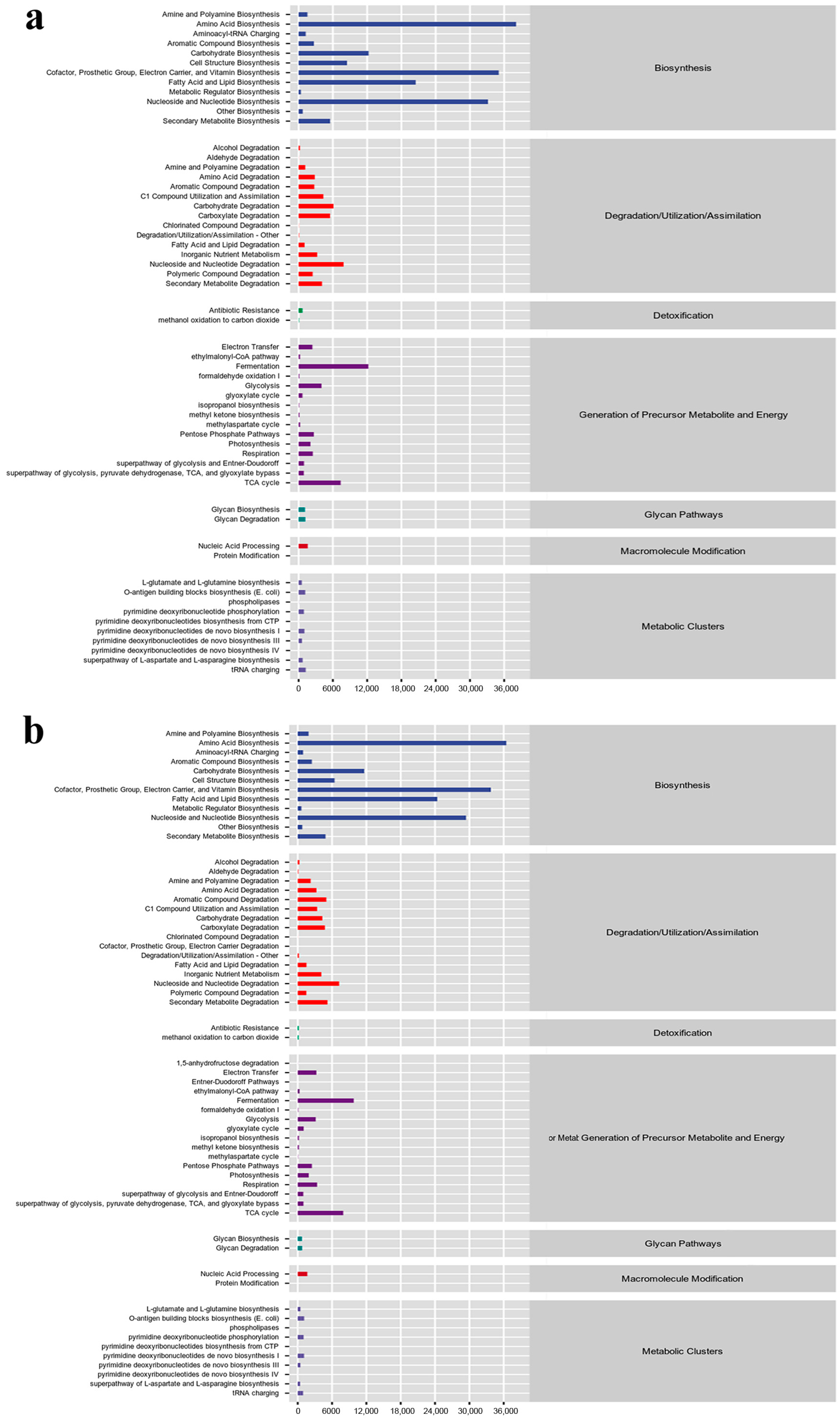
| Sample | Simpson | Chao1 | Shannon | Goods_Coverage | Observed_Species | Pielou_e | Faith_pd |
|---|---|---|---|---|---|---|---|
| J1 | 0.979 | 135.2 | 6.27 | 0.996735 | 135 | 0.886 | 14.53 |
| B1 | 0.982 | 394.8 | 7.09 | 0.900172 | 295 | 0.864 | 21.99 |
| J2 | 0.881 | 95.3 | 3.48 | 0.999970 | 95 | 0.530 | - |
| B2 | 0.859 | 72.1 | 3.15 | 0.999983 | 72 | 0.510 | - |
| J3 | 0.971 | 1284.9 | 6.98 | 0.999516 | 1284 | 0.676 | 91.05 |
| B3 | 0.931 | 1171.8 | 6.17 | 0.998383 | 1155 | 0.607 | 83.75 |
| J4 | 0.710 | 58.0 | 2.22 | 0.999995 | 58 | 0.379 | - |
| B4 | 0.723 | 92.1 | 3.10 | 0.999996 | 92 | 0.476 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, T.; Huang, X.; Zhu, D.; Tang, Y.; Xu, H.; Ran, F.; Ullah, H.; Tan, J. Comparative Analysis of Microbial Communities in Diseased and Healthy Sweet Cherry Trees (Prunus avium L.). Microorganisms 2024, 12, 1837. https://doi.org/10.3390/microorganisms12091837
Zhou T, Huang X, Zhu D, Tang Y, Xu H, Ran F, Ullah H, Tan J. Comparative Analysis of Microbial Communities in Diseased and Healthy Sweet Cherry Trees (Prunus avium L.). Microorganisms. 2024; 12(9):1837. https://doi.org/10.3390/microorganisms12091837
Chicago/Turabian StyleZhou, Tong, Xiaojuan Huang, Danyang Zhu, Yan Tang, Hongli Xu, Fanrong Ran, Hasin Ullah, and Jiangli Tan. 2024. "Comparative Analysis of Microbial Communities in Diseased and Healthy Sweet Cherry Trees (Prunus avium L.)" Microorganisms 12, no. 9: 1837. https://doi.org/10.3390/microorganisms12091837
APA StyleZhou, T., Huang, X., Zhu, D., Tang, Y., Xu, H., Ran, F., Ullah, H., & Tan, J. (2024). Comparative Analysis of Microbial Communities in Diseased and Healthy Sweet Cherry Trees (Prunus avium L.). Microorganisms, 12(9), 1837. https://doi.org/10.3390/microorganisms12091837







