Epibiotic Bacteria Isolated from the Non-Indigenous Species Codium fragile ssp. fragile: Identification, Characterization, and Biotechnological Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site
2.2. Taxonomic Identification of Seaweed Sample
2.3. Isolation of Seaweed Associated Bacteria
2.4. Morphological Characterization
2.5. Genotypic Characterization and Identification of Isolated Strains
2.6. Antibiotic Resistance Profile Analysis
Determination of MAR Index
2.7. Screening of Qualitative Enzymatic Production of CFF-Associated Bacteria
2.7.1. Lipase
2.7.2. DNase
2.7.3. Lecithinase
2.7.4. Amylase
2.7.5. Hemolysis
2.7.6. Gelatinase
2.7.7. Chitinase
2.7.8. Cellulase
2.7.9. Agarase
2.8. Antimicrobial Tests of CFF Associated Bacteria
3. Results
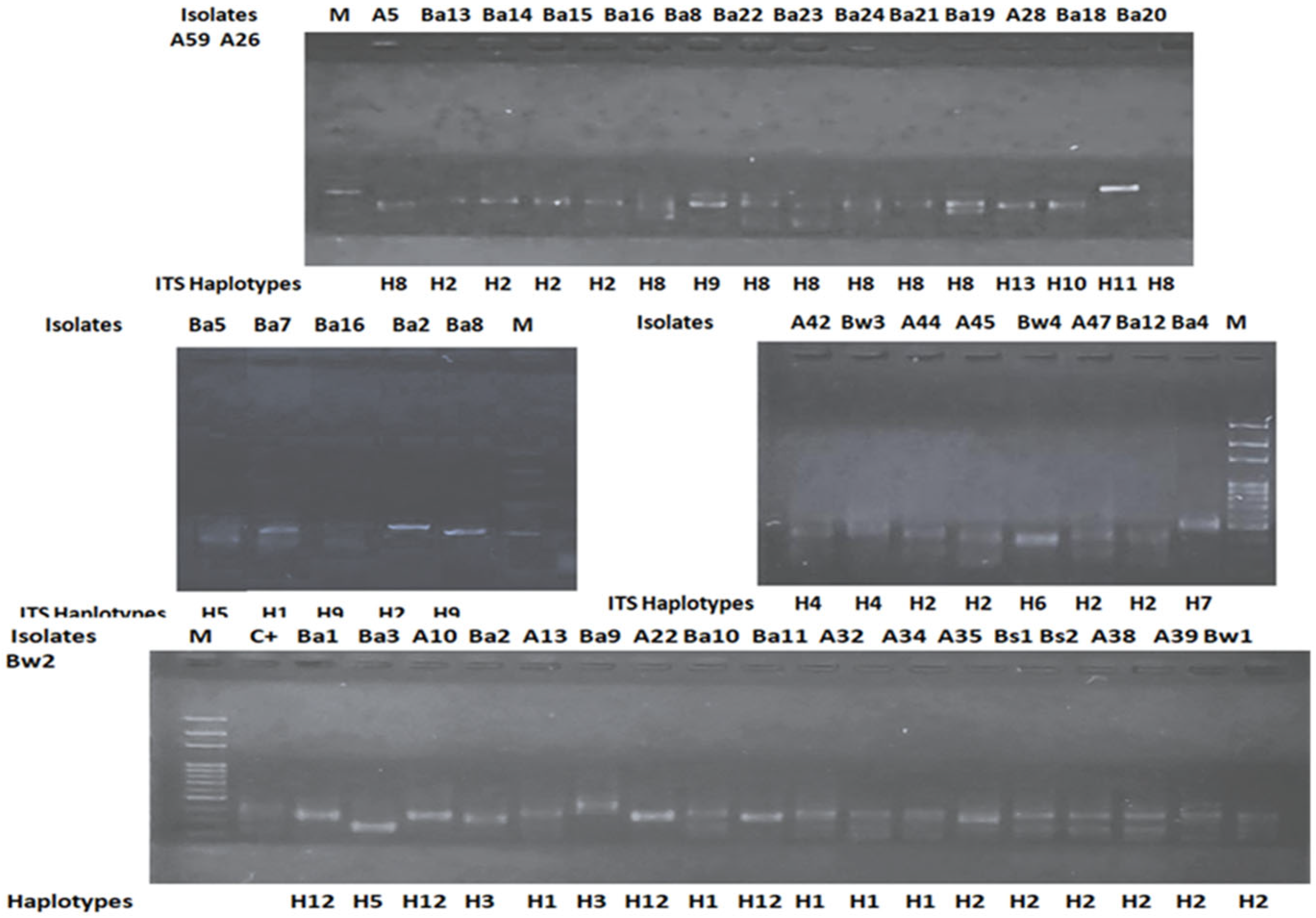
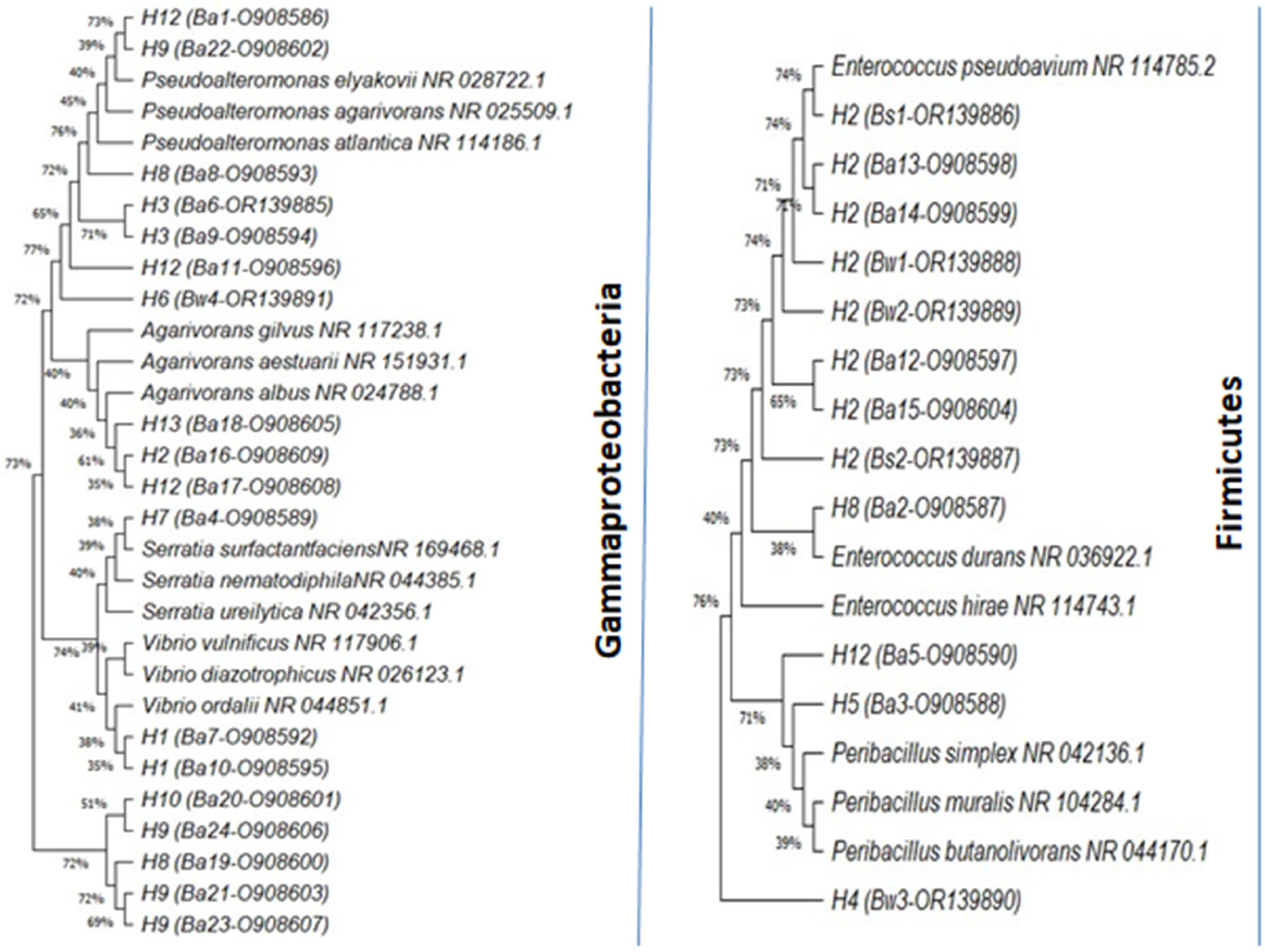
3.1. Molecular Identification of Isolates
3.2. Antibiotic Resistant Profile of CFF Associated Bacteria
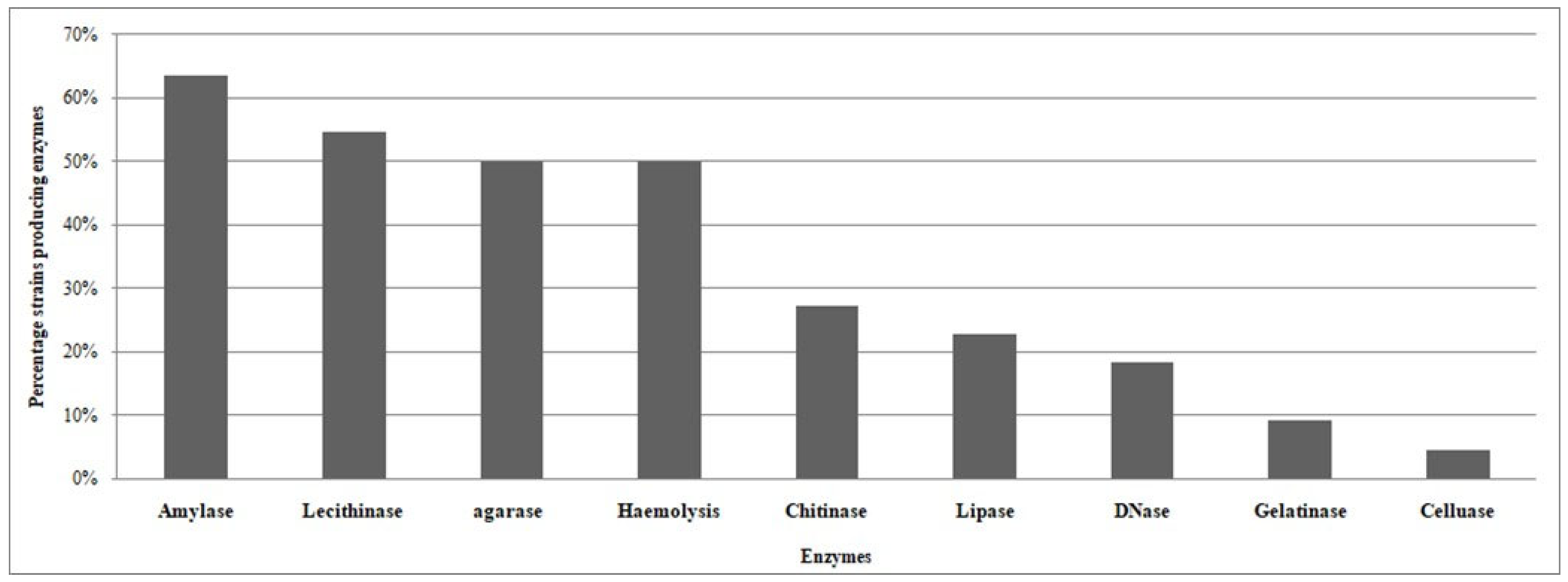
3.3. Screening of Qualitative Enzyme Production of CFF Associated Bacteria
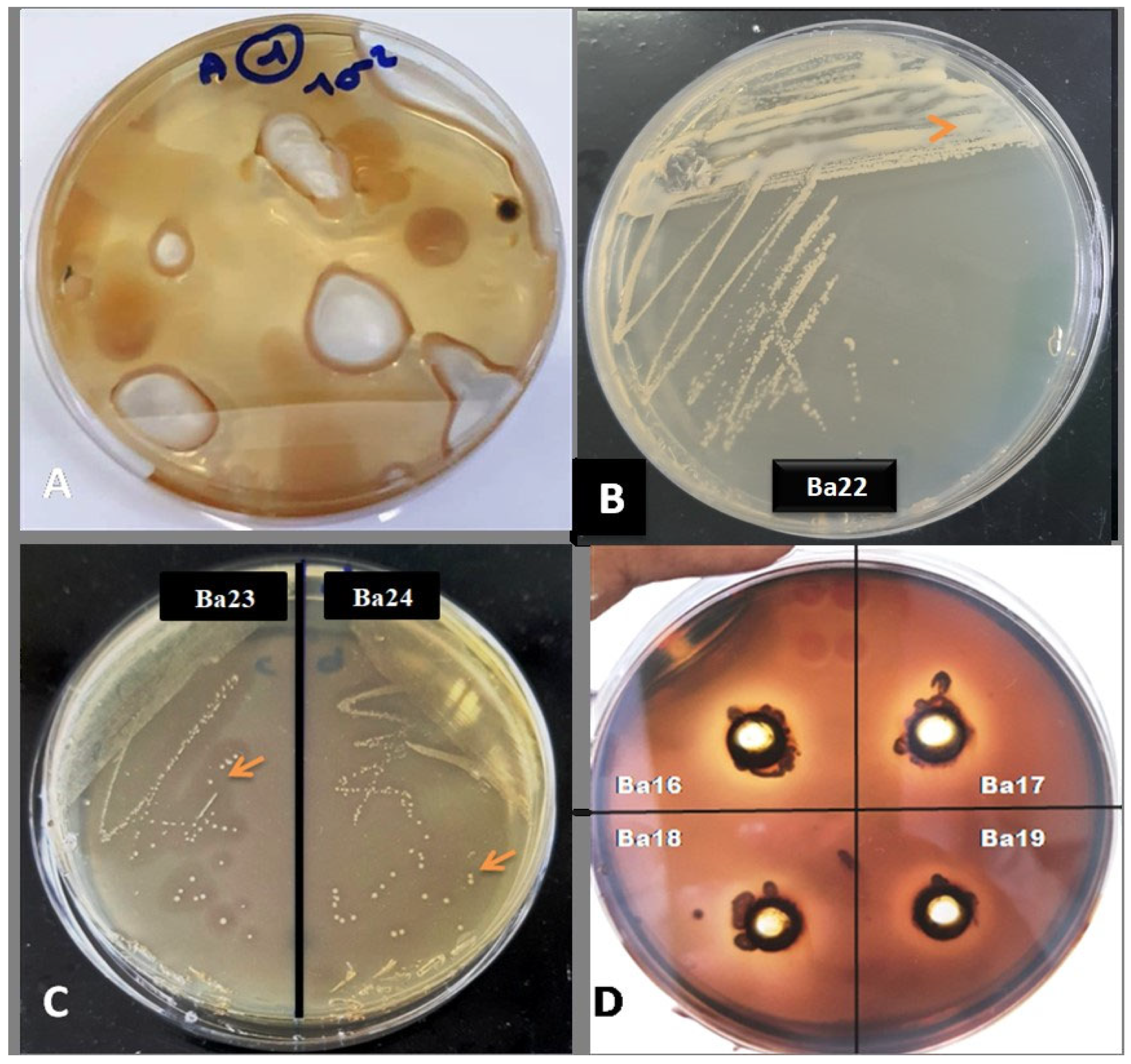
3.4. Qualitative Assays of Agarase Enzyme Production
3.5. Antimicrobial Activity of CFF Associated Bacteria
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goecke, F.; Labes, A.; Wiese, J.; Imhoff, J.F. Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 2010, 409, 267–300. [Google Scholar] [CrossRef]
- Zozaya-Valdés, E.; Roth-Schulze, A.J.; Thomas, T. Effects of temperature stress and aquarium conditions on the red macroalga Delisea pulchra and its associated microbial community. Front. Microbiol. 2016, 7, 161. [Google Scholar] [CrossRef]
- Srinivasan, R.; Kannappan, A.; Shi, C.; Lin, X. Marine Bacterial Secondary Metabolites: A Treasure House for Structurally Unique and Effective Antimicrobial Compounds. Mar. Drugs 2021, 19, 530. [Google Scholar] [CrossRef]
- Karthick, P.; Mohanraju, R. Antimicrobial Potential of Epiphytic Bacteria Associated with Seaweeds of Little Andaman, India. Front. Microbiol. 2018, 9, 611. [Google Scholar] [CrossRef]
- Menaa, F.; Wijesinghe, P.A.U.I.; Thiripuranatha, G.; Uzair, B.; Iqbal, H.; Ali Khan, B.; Menaa, B. Ecological and Industrial Implications of Dynamic Seaweed-Associated Microbiota Interactions Mar. Drugs 2020, 18, 641. [Google Scholar] [CrossRef]
- Martin, M.; Portetelle, D.; Michel, G.; Vandenbol, M. Microorganisms living on macroalgae: Diversity, interactions, and biotechnological applications. Appl. Microbiol. Biotechnol. 2014, 98, 2917–2935. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Wu, C.; Zhao, J.; Yan, T.; Jiang, P. Community structure of bacteria associated with drifting Sargassumhorneri, the causative species of golden tide in the Yellow Sea. Front. Microbiol. 2019, 10, 1192. [Google Scholar] [CrossRef]
- Goecke, F.; Thiel, V.; Wiese, J.; Labes, A.; Imhoff, J.F. Algae as an important environment for bacteria—Phylogenetic relationships among new bacterial species isolated from algae. Phycologia 2013, 52, 14–24. [Google Scholar] [CrossRef]
- Armstrong, E.; Yan, L.; Boyd, K.G.; Wright, P.C.; Burgess, J.G. The symbiotic role of marine microbes on living surfaces. Hydrobiologia 2001, 461, 37–40. [Google Scholar] [CrossRef]
- Kelecom, A. Secondary metabolites from marine microorganisms. An. Acad. Bras. Cienc. 2002, 74, 151–170. [Google Scholar] [CrossRef]
- Burgess, J.G.; Boyd, K.G.; Armstrong, E.; Jiang, Z.; Yan, L.; Berggren, M. The development of a marine natural product-based antifouling paint. Biofouling 2003, 19, 197–205. [Google Scholar] [CrossRef]
- Ismail, A.; Ktari, L.; Ahmed, M.; Bolhuis, H.; Bouhaouala-Zahar, B.; Stal, L.J.; Boudabbous, A.; El Bour, M. Heterotrophic bacteria associated with the green alga Ulva rigida: Identification and antimicrobial potential. J. Appl. Phycol. 2018, 30, 2883–2899. [Google Scholar] [CrossRef]
- Chen, J.; Zang, Y.; Yang, Z.; Qu, T.; Sun, T.; Liang, S.; Zhu, M.; Wang, Y.; Tang, X. Composition and Functional Diversity of Epiphytic Bacterial and Fungal Communities on Marine Macrophytes in an Intertidal Zone. Front. Microbiol. 2022, 13, 839465. [Google Scholar] [CrossRef] [PubMed]
- Trowbridge, C.D.; Todd, C.D. Host-plant changes of marine specialist herbivores: Ascoglossan sea slugs on the introduced macroalgae. Ecol. Monogr. 2001, 71, 219–243. [Google Scholar] [CrossRef]
- Djellouli, A. Sur la présence de Codium fragile (Suringar) Hariot (Codiaceae, Ulvophyceae) en Tunisie. Bull. Société Linnéenne Provence 1987, 39, 103–105. [Google Scholar]
- Sghaier, Y.R.; Zakhama-Sraieb, R.; Mouelhi, S.; Vazquez, M.; Valle, C.; Ramos-Espla, A.A.; Astier, J.M.; Verlaque, M.; Charfi-Cheikhrouha, F. Review of alien marine macrophytes in Tunisia. Mediterr. Mar. Sci. 2016, 17, 109–123. [Google Scholar] [CrossRef]
- Cherif, W.; Ktari, L.; El Bour, M.; Boudabous, A.; Grignon-Dubois, M. Codium fragile subsp. fragile (Suringar) Hariot in Tunisia: Morphological data and status of knowledge. ALGAE 2016, 31, 129–136. [Google Scholar] [CrossRef][Green Version]
- Morrissey, K.L.; Iveša, L.; Delva, S.; D’Hondt, S.; Willems, A.; De Clerck, O. Impacts of environmental stress on resistance and resilience of algal-associated bacterial communities. Ecol. Evol. 2021, 11, 15004–15019. [Google Scholar] [CrossRef]
- Silva, P.C. The dichotomous species of Codium in Britain. J. Mar. Biol. Assoc. 1955, 34, 565–577. [Google Scholar] [CrossRef]
- Trowbridge, C.D. Ecology of the green macroalga Codium fragile (Suringar) Hariot 1889: Invasive and noninvasive subspecies. Oceanogr. Mar. Biol. 1998, 36, 1–64. [Google Scholar]
- Hubbard, C.B.; Garbary, D.J. Morphological variation of Codium fragile (Chlorophyta) in eastern Canada. Bot. Mar. 2002, 45, 476–485. [Google Scholar] [CrossRef]
- Lemos, M.L.; Toranzo, A.E.; Barja, J.L. Antibiotic activity of epiphytic bacteria isolated from intertidal seaweeds. Microb. Ecol. 1985, 11, 149–163. [Google Scholar] [CrossRef]
- Dalton, H.M.; Poulsen, L.K.; Halasz, P.; Angles, M.L.; Goodman, A.E.; Marshall, K.C. Substratum induced morphological changes in a marine bacterium and their relevance to bioflm structure. J. Bacteriol. 1994, 176, 6900–6906. [Google Scholar] [CrossRef]
- Hassen, W.; Neifar, M.; Cherif, H.; Mahjoubi, M.; Souissi, Y.; Raddadi, N.; Fava, F.; Cherif, A. Assessment of genetic diversity and bioremediation potential of Pseudomonas isolated from pesticide-contaminated artichoke farm soils. Biotech 2018, 8, 263. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Soussy, C.J. Antibiogram Committee of the French Society of Microbiology; Centre Hospitalier Universitaire Henri Mondor: Créteil, France, 2008. [Google Scholar]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Sierra, G. A simple method for the detection of lipolytic activity of microorganisms and some observations on the influence of the contact between cells and fatty substrates. Antonie Van Leeuwenhoek 1957, 23, 15–22. [Google Scholar] [CrossRef]
- Schreiber, L.; Kjeldsen, K.U.; Funch, P.; Jensen, J.; Obst, M.; LópezLegentil, S.; Schramm, A. Endozoicomonas are specifc, facultative symbionts of sea squirts. Front. Microbiol. 2016, 7, 1042. [Google Scholar] [CrossRef]
- Garrity, P.; Jones, G.; Krieg, D.; Ludwig, N.R.; Rainey, W.F.A.; Schleifer, K.H.; Whitman, W. (Eds.) Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: New York, NY, USA, 2009; Volume 3. [Google Scholar]
- Krishnan, A.; Kumar, G.; Loganathan, K.; Rao, B. Optimization, production and partial purifcation of extracellular αamylase from Bacillus sp. marini. Arch. Appl. Sci. Res. 2011, 3, 33–42. [Google Scholar]
- Satpute, S.; Bhawsar, B.D.; Dhakephalkar, P.; Chopade, B. Assessment of different screening methods for selecting biosurfactant producing marine bacteria. Indian J. Mar. Sci. 2008, 37, 243–250. [Google Scholar]
- Smith, H.L.; Goodner, K. Detection of bacterial gelatinases by gelatin-agar plate methods. J. Bacteriol. 1958, 76, 662–665. [Google Scholar] [CrossRef]
- Kaur, K.; Dattajirao, V.; Shrivastava, V.; Bhardwaj, U. Isolation and characterization of chitosan-producing bacteria from beaches of Chennai, India. Enzym. Res. 2012, 2012, 421683. [Google Scholar] [CrossRef]
- Sinha, P.; Bedi, S. Production of sugars from cellulosic wastes by enzymatic hydrolysis. Int. J. Curr. Res. 2018, 10, 72141–72144. [Google Scholar]
- Kandasamy, K.P.; Subramanian, R.K.; Srinivasan, R.; Ragunath, S.G.; Balaji, G.; Gracy, M.; Latha, K. Shewanellaalgae and Microbulbiferelongatus from marine macro-algae—Isolation and characterization of agarhydrolysing bacteria. Access Microbiol. 2020, 2, e000170. [Google Scholar] [CrossRef]
- Fleming, H.P.; Etchells, J.L.; Costilow, R.N. Microbiol inhibition by an isolate of Pediococcus from cucumber brines. Appl. Microbiol. 1975, 30, 1040–1042. [Google Scholar] [CrossRef]
- Ismail, A.; Ktari, L.; Ahmed, M.; Bolhuis, H.; Boudabbous, A.; Stal, L.J. Antimicrobial activities of bacteria associated with the brown alga Padina pavonica. Front. Microbiol. 2016, 7, 1072. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Ebers, J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Florez, J.Z.; Camus, C.; Hengst, M.B.; Buschmann, A.H. A functional perspective analysis of macroalgae and epiphytic bacterial community interaction. Front. Microbiol. 2017, 8, 2561. [Google Scholar] [CrossRef]
- Fisher, M.M.; Wilcox, L.W.; Graham, L.E. Molecular characterization of epiphytic bacterial communities on Charophycean green algae applied and environmental. Microbiology 1998, 64, 4384–4389. [Google Scholar]
- Le Pennec, G.; Gall, E.A. The microbiome of Codium tomentosum: Original state and in the presence of copper. World J. Microbiol. Biotechnol. 2019, 35, 167. [Google Scholar] [CrossRef]
- Head, W.D.; Carpenter, E.J. Nitrogen fixation associated with the marine macroalga Codium fragile. Limnol. Oceanogr. 1975, 20, 815–823. [Google Scholar] [CrossRef]
- Rosenberg, G.; Paerl, H.W. Nitrogenfxation by blue-green algae associated with siphonous green seaweed Codium decorticatum: Effects on ammonium uptake. Mar. Biol. 1981, 61, 151–158. [Google Scholar] [CrossRef]
- Singh, R.P.; Reddy, C.R.K. Seaweed-microbial interactions: Key functions of seaweed-associated bacteria. FEMS Microbiol. Ecol. 2014, 88, 213–230. [Google Scholar] [CrossRef]
- Polianciuc, S.I.; Gurzău, A.E.; Kiss, B.; Ştefan, M.G.; Loghin, F. Antibiotics in the environment: Causes and consequences. Med. Pharm. Rep. 2020, 93, 231–240. [Google Scholar] [CrossRef]
- Tahrani, L.; Van Loco, J.; Anthonissen, R.; Verschaeve, L.; Ben Mansour, H.; Reyns, T. Identification and risk assessment of human and veterinary antibiotics in the wastewater treatment plants and the adjacent sea in Tunisia. Water Sci. Technol. 2017, 76, 3000–3021. [Google Scholar] [CrossRef]
- Afsa, S.; Hamden, K.; Martin, P.A.L.; Mansour, H.B. Occurrence of 40 pharmaceutically active compounds in hospital and urban wastewaters and their contribution to Mahdia coastal seawater contamination. Environ. Sci. Pollut. Res. 2020, 27, 1941–1955. [Google Scholar] [CrossRef]
- Alibi, S.; Beltifa, A.; Hassen, W.; Jaziri, A.; Soussia, L.; Zbidi, F.; Ben Mansour, H. Coastal Surveillance and Water Quality monitoring in the Rejiche Sea-Tunisia. Water Environ. Res. 2021, 93, 2025–2033. [Google Scholar] [CrossRef] [PubMed]
- Jebara, A.; Hassen, W.; Ousleti, A.; Mabrouk, L.; Jaziri, A.; Di Bella, G.; Ben Mansour, H. Multidrug-resistant epi-endophytic bacterial community in Posidonia oceanica of Mahdia coast as biomonitoring factor for antibiotic contamination. Arch Microbiol. 2022, 204, 229. [Google Scholar] [CrossRef]
- De Souza, P.M.; de Oliveirae, P.M. Application of microbial α-amylase in industry—A review. Braz. J. Microbiol. 2010, 41, 850–861. [Google Scholar] [CrossRef]
- Fenice, M.; Giambattista, R.D.; Leuba, J.L.; Federici, F. Inactivation of Mucorplumbeus by the combined actions of chitinase and high hydrostatic pressure. Int. J. Food Microbiol. 1999, 52, 109–113. [Google Scholar] [CrossRef]
- Raveendran, C.; Naveenan, T.; Varatharajan, G.R. Optimization of alkaline cellulase production by the marine-derived fungus Chaetomium sp. using agricultural and industrial wastes as substrates. Bot. Mar. 2010, 53, 275–282. [Google Scholar] [CrossRef]
- Lavilla-Pitogo, C.R. Agar-digesting bacteria associated with ‘rotten thallus syndrome’ of Gracilaria sp. Aquaculture 1992, 102, 1–7. [Google Scholar] [CrossRef]
- Schroeder, D.C.; Jaffer, M.A.; Coyne, V.E. Investigation of the role of a beta (1–4) agarase produced by Pseudoalteromonas gracilis B9 in eliciting disease symptoms in the red alga Gracilaria gracilis. Microbiology 2003, 149, 2919–2929. [Google Scholar] [CrossRef][Green Version]
- Nikapitiya, C.; Oh, C.H.; Lee, Y.D.; Lee, S.K.; Whang, I.S.; Lee, J.H. Characterization of a glycoside hydrolase family 50 thermostable β-agaraseAgrA from marine bacteria Agarivorans sp. AG17. Fish. Aquat. Sci. 2010, 13, 36–48. [Google Scholar] [CrossRef]
- Chen, H.; Yan, X.; Zhu, P.; Lin, J. Antioxidant activity and hepatoprotective potential of agaro-oligosaccharides in vitro and in vivo. Nutr. J. 2006, 5, 31. [Google Scholar] [CrossRef]
- Chen, X.; Fu, X.; Huang, L.; Xu, J.; Gao, X. Agar oligosaccharides: A review of preparation, structures, bioactivities and application. Carbohydr. Polym. 2021, 265, 118076. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, X.; Mou, H.; Guan, H. Anti-oxidation of agar oligosaccharides produced by agarase from a marine bacterium. J. Appl. Phycol. 2004, 16, 333–340. [Google Scholar] [CrossRef]
- Kobayashi, R.; Takisada, M.; Suzuki, T.; Kirimura, K.; Usami, S. Neoagarobiose as a novel moisturizer with whitening effect. Biosci. Biotechnol. Biochem. 1997, 61, 162–163. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.M.; Craigie, J.S.; Xie, Z.K. Protoplast production from Porphyralinearis using a simplified agarase procedure capable of commercial application. J. Appl. Phycol. 1994, 6, 35–39. [Google Scholar] [CrossRef]
- Dipakkore, S.; Reddy, C.; Jha, B. Production and seeding of protoplasts of Porphyraokhaensis (Bangiales, Rhodophyta) in laboratory culture. J. Appl. Phycol. 2005, 17, 331–337. [Google Scholar] [CrossRef]
- Burmeistera, M.; Lehrachb, H. Isolation of large DNA fragments from agarose gels using agarase. Trend Genet. 1989, 5, 41. [Google Scholar] [CrossRef]
- Aanshi, K.; Bhupendra. Isolation, Characterization and Purification of an Extracellular Enzyme Agarase from Agarolytic bacteria. Int. J. Curr. Res. 2015, 7, 19345–19349. [Google Scholar]
- Rajeswari, S.; Jaiganesh, R.; Muthukumar, R.; Jaganathan, M.K. Isolation and Characterization of an Agarase Producing Bacteria from Marine Sediment. Int. J. ChemTech Res. 2016, 9, 437–446. [Google Scholar]
- Fu, X.T.; Kim, S.M. Agarase: Review of major sources, categories, purification method, enzyme characteristics and applications. Mar. Drugs 2010, 8, 200–218. [Google Scholar] [CrossRef]
- Yu, F.S.; Hui, C.Y.; Yen, L. The discoveery of agarolytic bacterium with agrarse gene containing plasmid, and some enzymology characteristics. Int. J. Appl. Sci. Eng. 2009, 7, 25–41. [Google Scholar]
- Du, Z.J.; Lv, G.Q.; Rooney, A.P.; Miao, T.T.; Xu, Q.Q.; Chen, G.J. Agarivorans gilvus sp. nov. isolated from seaweed. Int. J. Syst. Evol. Microbiol. 2011, 61, 493–496. [Google Scholar] [CrossRef][Green Version]
- Comba-González, N.B.; Ruiz-Toquica, J.S.; López-Kleine, L.; Montoya-Castaño, D. Epiphytic bacteria of macroalgae of the Genus Ulva and their potential in producing enzymes having biotechnological interest. J. Mar. Biol. Oceanogr. 2016, 5, 1–9. [Google Scholar] [CrossRef]
- Osoegawa, K.; Woon, P.Y.; Zhao, B.; Frengen, E.; Tateno, M.; Catanese, J.J.; de Jong, P.J. An improved approach for construction of bacterial artificial chromosome libraries. Genomics 1998, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.Y. Production and functional properties of agaro-oligosaccharides. In Proceedings of Symposium on Scientific Study and Industrialization of Health Food; Health Food Society of Taiwan: Taipai, China, 2001; pp. 60–81. [Google Scholar] [CrossRef]
- Boyd, K.G.; Adams, D.R.; Burgess, J.G. Antibacterial and repellent activities of marine bacteria associated with algal surfaces. Biofouling 1999, 14, 227–236. [Google Scholar] [CrossRef]
- Jafarzade, M.; Shayesteh, F.; Usup, G.; Ahmad, A. Influence of culture conditions and medium composition on the production of antibacterial compounds by marine Serratia sp. WPRA3. J. Microbiol. 2013, 51, 373–379. [Google Scholar] [CrossRef]
- Arivuselvam, R.; Dera, A.A.; Parween Ali, S.; Alraey, Y.; Saif, A.; Hani, U.; Arumugam Ramakrishnan, S.; Azeeze, M.S.T.A.; Rajeshkumar, R.; Susil, A. Isolation, Identification, and Antibacterial Properties of Prodigiosin, a Bioactive Product Produced by a New Serratiamarcescens JSSCPM1 Strain: Exploring the Biosynthetic Gene Clusters of Serratia Species for Biological Applications. Antibiotics 2023, 12, 1466. [Google Scholar] [CrossRef]
- Atencio, L.A.; Dal Grande, F.; Young, G.O.; Gavilán, R.; Guzmán, H.M.; Schmitt, I.; Mejía, L.C.; Gutiérrez, M. Antimicrobial-producing Pseudoalteromonas from the marine environment of Panama shows a high phylogenetic diversity and clonal structure. J. Basic Microbiol. 2018, 58, 747–769. [Google Scholar] [CrossRef]
- Skovhus, T.L.; Ramsing, N.B.; Holmström, C.; Kjelleberg, S. Real-Time Quantitative PCR for assessment of abundance of Pseudoalteromonas Species in marine samples. Appl. Environ. Microbiol. 2004, 70, 2373–2382. [Google Scholar] [CrossRef]
- Holmström, C.; Kjelleberg, S. Marine Pseudoalteromonas species are associated with higher organisms and producebiologically active extracellular agents. FEMS Microbiol. Ecol. 1999, 30, 285–293. [Google Scholar] [CrossRef]
- Isnansetyo, A.; Kamei, Y. MC21-A, a bactericidal antibiotic produced by a new marine bacterium, Pseudoalteromonas phenolicasp. nov. O-BC30(T), against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 480–488. [Google Scholar] [CrossRef]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns-An Update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Martínez Cruz, P.; Ibáñez, A.L.; Monroy Hermosillo, O.A.; Ramírez Saad, H.C. Use of Probiotics in Aquaculture. Int. Sch. Res. Not. 2012, 2012, 916845. [Google Scholar] [CrossRef]
- Hmani, I.; Ktari, L.; Ismail, A.; EL Bour, M. Biotechnological potential of Ulva ohnoi epiphytic bacteria: Enzyme production and antimicrobial activities. Front. Mar. Sci. 2023, 10, 1042527. [Google Scholar] [CrossRef]


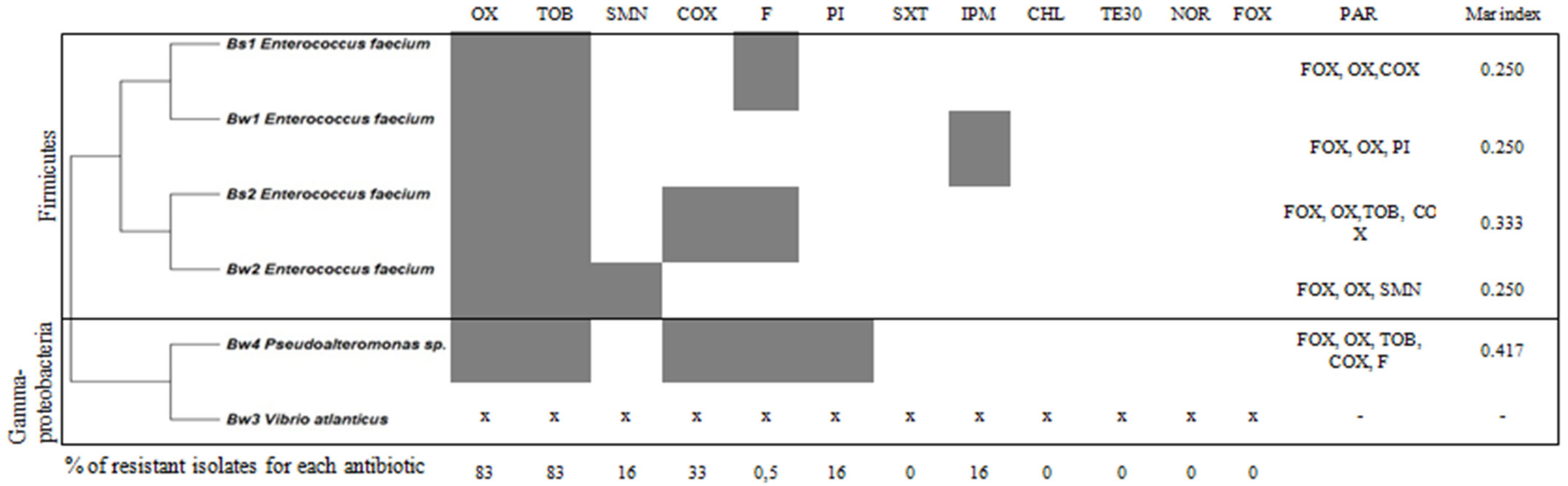
| Class | Abbreviation | Antibiotic | Amount (µg) |
|---|---|---|---|
| Phenicol | CHL | Chloramphenicol | 30 |
| Carbapenem | IPM | Imipenem | 10 |
| Diaminopyrimidines | SXT | Trimethoprim/Sulfamethoxazole | 1.25/23.75 |
| Quinolones | FOX | Cefoxitin | 30 |
| Quinolones | COX | Cefotaxime | 5 |
| Aminoglycoside | TOB | Tobramycin | 10 |
| Aminoglycoside | SMN | Streptomycin | 10 |
| Nitrofuran | F | Nitrofurantoin | 300 |
| Penicillin | OX | Oxacillin | 1 |
| TE30 | Tetracycline | 30 | |
| Pyridopyrimidine | PI | Pipemidicacid | 20 |
| Quinolone | NOR | Norfloxacin | 5 |
| ITS Haplotype | Isolates | Ox | Cat | Closestspecies in NCBI | Size (bp) | Identity (%) | Accession Number | Phylum |
|---|---|---|---|---|---|---|---|---|
| H1 | Ba10 | + | + | Vibrio anguillarum | 1050 | 99.62 | ON908595 | γp |
| H1 | Ba7 | + | + | Vibrio anguillarum | 1111 | 99.82 | ON908592 | γp |
| H2 | Ba12 | + | + | Enterococcus faecium | 1140 | 99.56 | ON908597 | F |
| H2 | Ba13 | + | + | Enterococcus faecium | 1136 | 99.74 | ON908598 | F |
| H2 | Ba14 | + | + | Enterococcus faecium | 1122 | 99.91 | ON908599 | F |
| H2 | Ba15 | + | + | Enterococcus faecium | 1005 | 99.80 | ON908604 | F |
| H2 | Ba16 | + | + | Agarivoranslitoreus | 1056 | 94.33 | ON908609 | γp |
| H2 | Bs1 | + | + | Enterococcus faecium | 1157 | 98.88 | OR139886 | F |
| H2 | Bs2 | + | + | Enterococcus faecium | 1128 | 99.65 | OR139887 | F |
| H2 | Bw1 | + | + | Enterococcus faecium | 1110 | 99.91 | OR139888 | F |
| H2 | Bw2 | + | + | Enterococcus faecium | 1145 | 99.74 | OR139889 | F |
| H3 | Ba6 | + | + | Pseudoalteromonas piscicida | 1124 | 99.82 | OR139885 | γp |
| H3 | Ba9 | + | + | Pseudoalteromonaspiscicida | 1086 | 99.72 | ON908594 | γp |
| H4 | Bw3 | + | + | Vibrio atlanticus | 1104 | 99.37 | OR139890 | γp |
| H5 | Ba3 | + | + | Peribacillusfrigoritolerans | 1140 | 99.22 | ON908588 | F |
| H6 | Bw4 | + | + | Pseudoalteromonas sp. | 988 | 90.26 | OR139891 | γp |
| H7 | Ba4 | + | + | Serratia marcescens | 1129 | 99.56 | ON908589 | γp |
| H8 | Ba8 | + | + | Pseudoalteromonasagarivorans | 1079 | 100 | ON908593 | γp |
| H8 | Ba19 | + | + | Pseudomonas khazarica | 1139 | 98.43 | ON908600 | γp |
| H8 | Ba2 | + | + | Enterococcus faecium | 1101 | 98.73 | ON908587 | F |
| H9 | Ba21 | + | + | Pseudomonas khazarica | 1104 | 98.46 | ON908603 | γp |
| H9 | Ba22 | + | − | Pseudoalteromonasagarivorans | 1129 | 99.47 | ON908602 | γp |
| H9 | Ba23 | + | + | Pseudomonas khazarica | 1072 | 98.42 | ON908607 | γp |
| H9 | Ba24 | + | + | Pseudomonas khazarica | 1109 | 97.93 | ON908606 | γp |
| H10 | Ba20 | + | − | Pseudomonas khazarica | 784 | 97.96 | ON908601 | γp |
| H11 | Ba17 | + | + | Agarivoransabus | 1056 | 94.33 | ON908608 | γp |
| H12 | Ba1 | + | + | Pseudoalteromonas spiralis | 1333 | 99.38 | ON908586 | γp |
| H12 | Ba5 | + | + | Peribacillusfrigoritolerans | 1117 | 99.91 | ON908590 | F |
| H12 | Ba11 | − | + | Pseudoalteromonasshioyasakiensis | 1011 | 98.81 | ON908596 | γp |
| H13 | Ba18 | + | + | Agarivorans sp. | 1009 | 92.54 | ON908605 | γp |
| REF | Isolates | Amylase | Lecithinase | Hemolysis | Chitinase | DNase | Lipase | Gelatinase | Cellulase | Agarase |
|---|---|---|---|---|---|---|---|---|---|---|
| CFF-associated bacteria | ||||||||||
| Ba1 | Pseudoalteromonasspiralis | − | − | + | − | − | + | − | − | − |
| Ba2 | Enterococcus aecium | + | + | + | − | − | + | − | − | − |
| Ba3 | Peribacillusfrigoritolerans | + | + | − | − | + | + | − | − | − |
| Ba4 | Serratia marcescens | + | − | + | − | − | − | − | − | − |
| Ba5 | Peribacillusfrigoritolerans | + | − | + | − | − | − | − | − | − |
| Ba8 | Pseudoalteromonasagarivorans | + | + | + | + | − | − | + | − | − |
| Ba9 | Pseudoalteromona spiscicida | + | − | + | − | + | − | − | − | − |
| Ba10 | Vibrio anguillarum | + | + | − | + | − | − | + | − | − |
| Ba11 | Pseudoalteromonasshioyasakiensis | + | − | − | − | − | − | − | − | − |
| Ba12 | Enterococcus faecium | x | x | x | x | x | x | x | x | − |
| Ba13 | Enterococcus faecium | + | + | + | − | − | − | − | − | + |
| Ba14 | Enterococcus faecium | + | − | + | + | − | − | − | − | + |
| Ba15 | Enterococcus faecium | − | − | − | − | − | − | − | − | + |
| Ba16 | Agarivoranslitoreus | − | − | − | − | − | − | − | − | + |
| Ba17 | Agarivoransabus | − | − | − | − | − | − | − | − | + |
| Ba18 | Agarivorans sp. | − | + | − | − | − | − | − | − | + |
| Ba19 | Pseudomonas khazarica | − | − | − | − | − | − | − | − | + |
| Ba20 | Pseudomonas khazarica | − | + | + | − | − | − | − | − | + |
| Ba21 | Pseudomonas khazarica | − | + | − | + | − | − | − | − | + |
| Ba22 | Pseudoalteromonasagarivorans | + | − | + | + | − | + | − | + | + |
| Ba23 | Pseudomonas khazarica | + | + | + | + | + | − | − | − | + |
| Ba24 | Pseudomonas khazarica | + | + | − | − | − | − | − | − | + |
| Sediment and surrounding water | ||||||||||
| Bs1 | Enterococcus faecium | − | − | − | − | − | − | − | − | − |
| Bs2 | Enterococcus faecium | + | − | − | + | − | − | − | − | − |
| Bw1 | Enterococcusfaecium | − | − | − | − | + | − | − | − | − |
| Bw2 | Enterococcus faecium | + | + | − | + | + | − | − | − | − |
| Bw3 | Vibrio atlanticus | + | − | − | − | − | − | − | − | − |
| Bw4 | Pseudoalteromonas sp. | + | − | + | − | + | − | − | − | − |
| Isolates | Agarolitic Index |
|---|---|
| Ba16 | 2.38 ± 0 |
| Ba17 | 2.13 ± 0 |
| Ba18 | 2.38 ± 0 |
| Ba19 | 1.13 ± 0 |
| Ba20 | 0.63 ± 0 |
| Ba21 | 1.13 ± 0 |
| Ba22 | 1.13 ± 0 |
| Ba23 | 0.71 ± 0.57 |
| Ba24 | 0.88 ± 0 |
| Isolates | Ref | Pathogenic bacteria | Yeast | |||||
|---|---|---|---|---|---|---|---|---|
| E.c | V.an | V.al | P.a | S.t | S.a | C.a | ||
| Pseudoalteromonas spiralis | Ba1 | 23 mm ±1.2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Peribacillus frigoritolerans | Ba3 | 10 mm ±1.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Serratia marcescens | Ba4 | 8 mm ±0.6 | 10 mm ±1 | 8 mm ±0.6 | 8 mm ±0.6 | 10 mm ±1 | 12 mm ±0.6 | 13 mm ±1 |
| Vibrio anguillarum | Ba7 | 13 mm ±0.6 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pseudoalteromonas agarivorans | Ba8 | 15 mm ±2.1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Vibrio anguillarum | Ba10 | 13 mm ±1.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pseudoalteromonas shioyasakiensis | Ba11 | 13 mm ±0.6 | 0 | 0 | 0 | 0 | 0 | 0 |
| Enterococcus faecium | Ba12 | 22 mm ±2.6 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pseudoalteromonas sp. | Bw4 | 22 mm ±2.1 | 0 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cherif, W.; Ktari, L.; Hassen, B.; Ismail, A.; El Bour, M. Epibiotic Bacteria Isolated from the Non-Indigenous Species Codium fragile ssp. fragile: Identification, Characterization, and Biotechnological Potential. Microorganisms 2024, 12, 1803. https://doi.org/10.3390/microorganisms12091803
Cherif W, Ktari L, Hassen B, Ismail A, El Bour M. Epibiotic Bacteria Isolated from the Non-Indigenous Species Codium fragile ssp. fragile: Identification, Characterization, and Biotechnological Potential. Microorganisms. 2024; 12(9):1803. https://doi.org/10.3390/microorganisms12091803
Chicago/Turabian StyleCherif, Wafa, Leila Ktari, Bilel Hassen, Amel Ismail, and Monia El Bour. 2024. "Epibiotic Bacteria Isolated from the Non-Indigenous Species Codium fragile ssp. fragile: Identification, Characterization, and Biotechnological Potential" Microorganisms 12, no. 9: 1803. https://doi.org/10.3390/microorganisms12091803
APA StyleCherif, W., Ktari, L., Hassen, B., Ismail, A., & El Bour, M. (2024). Epibiotic Bacteria Isolated from the Non-Indigenous Species Codium fragile ssp. fragile: Identification, Characterization, and Biotechnological Potential. Microorganisms, 12(9), 1803. https://doi.org/10.3390/microorganisms12091803






