Genome Mining of Pseudanabaena galeata CCNP1313 Indicates a New Scope in the Search for Antiproliferative and Antiviral Agents
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation
2.2. Isolation and Sequencing of Genomic DNA
2.3. Genome Assembling
2.4. Genome Annotation and Analysis
2.5. Genomic Data
2.6. LC-MS/MS Analysis
3. Results and Discussion
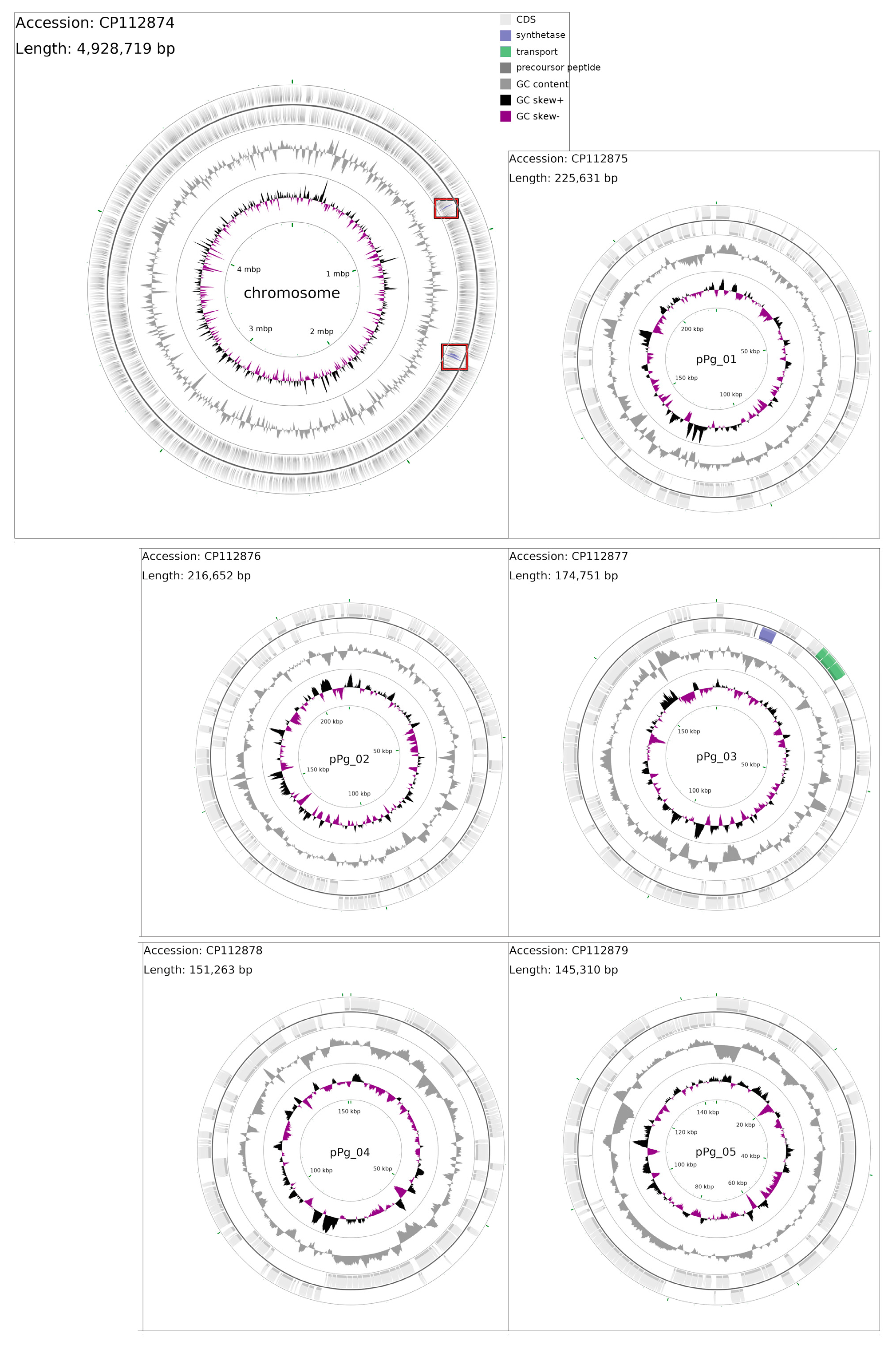
3.1. Analysis of the P. galeata CCNP1313 Genome
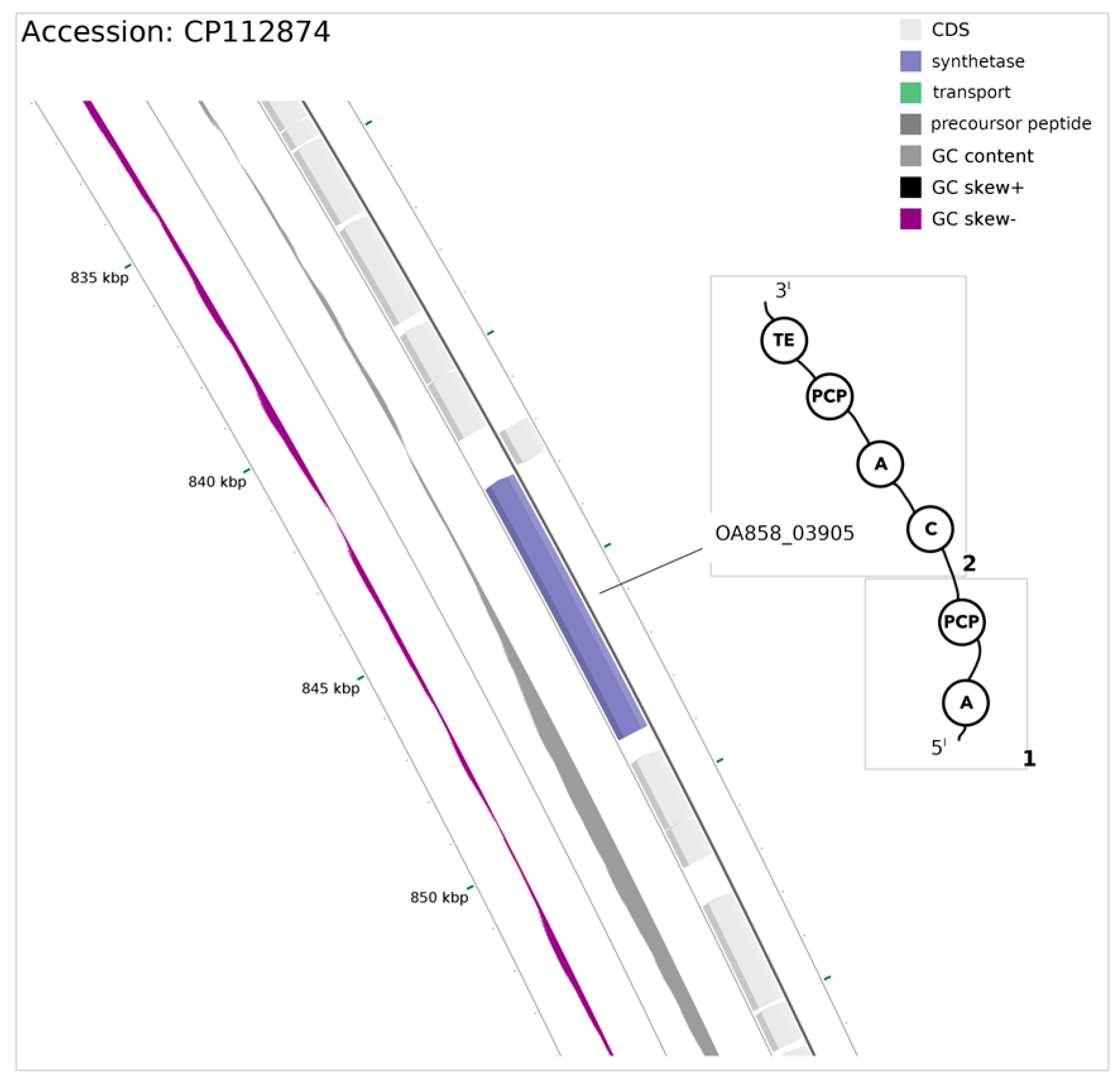
3.2. Non-Ribosomal Peptide Synthase (NRPS) Gene
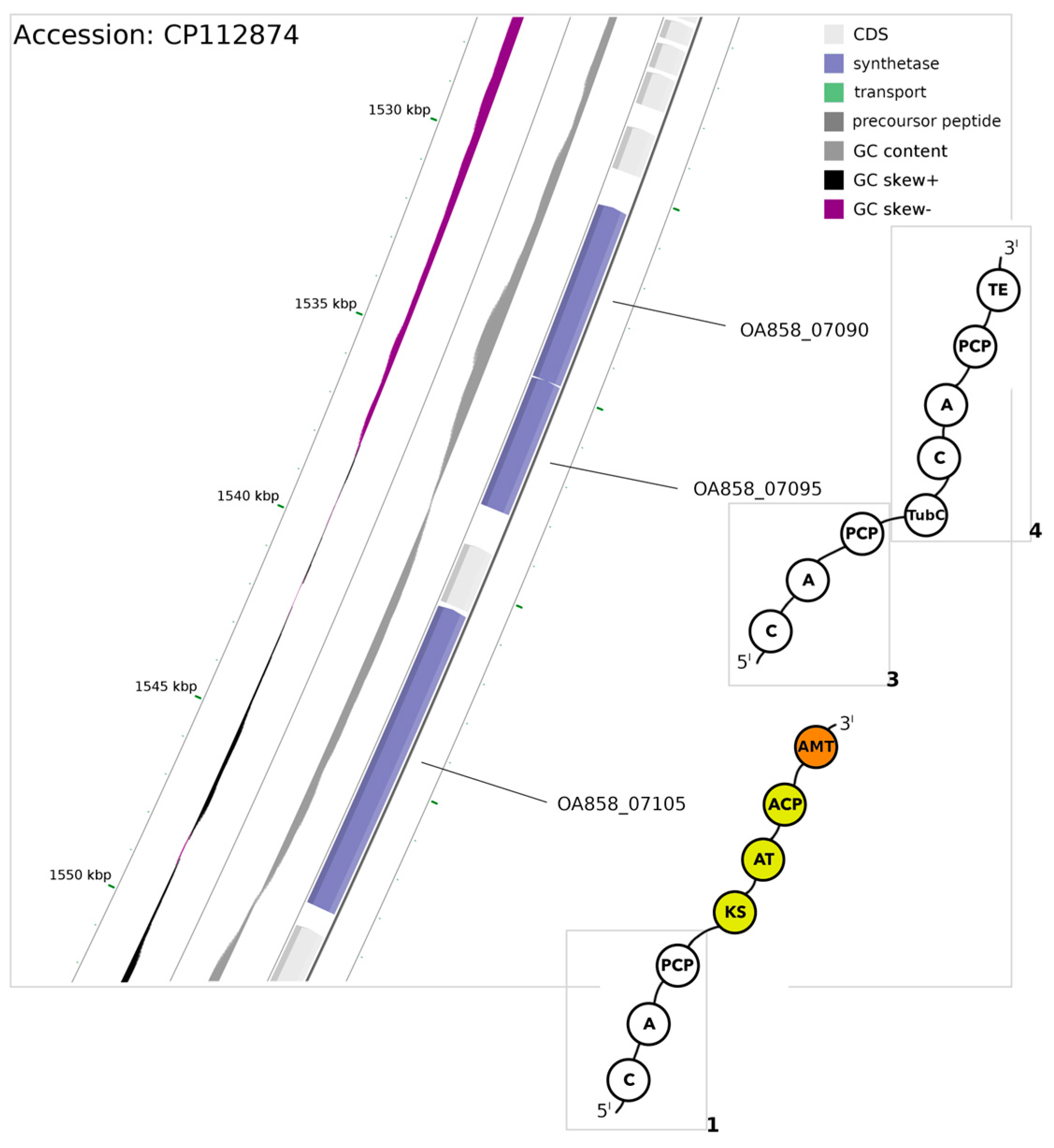
3.3. The Mixed PKS/NRPS System
3.4. Polyketide Synthase (PKS) Gene Clusters
3.5. Fatty Acid Synthases
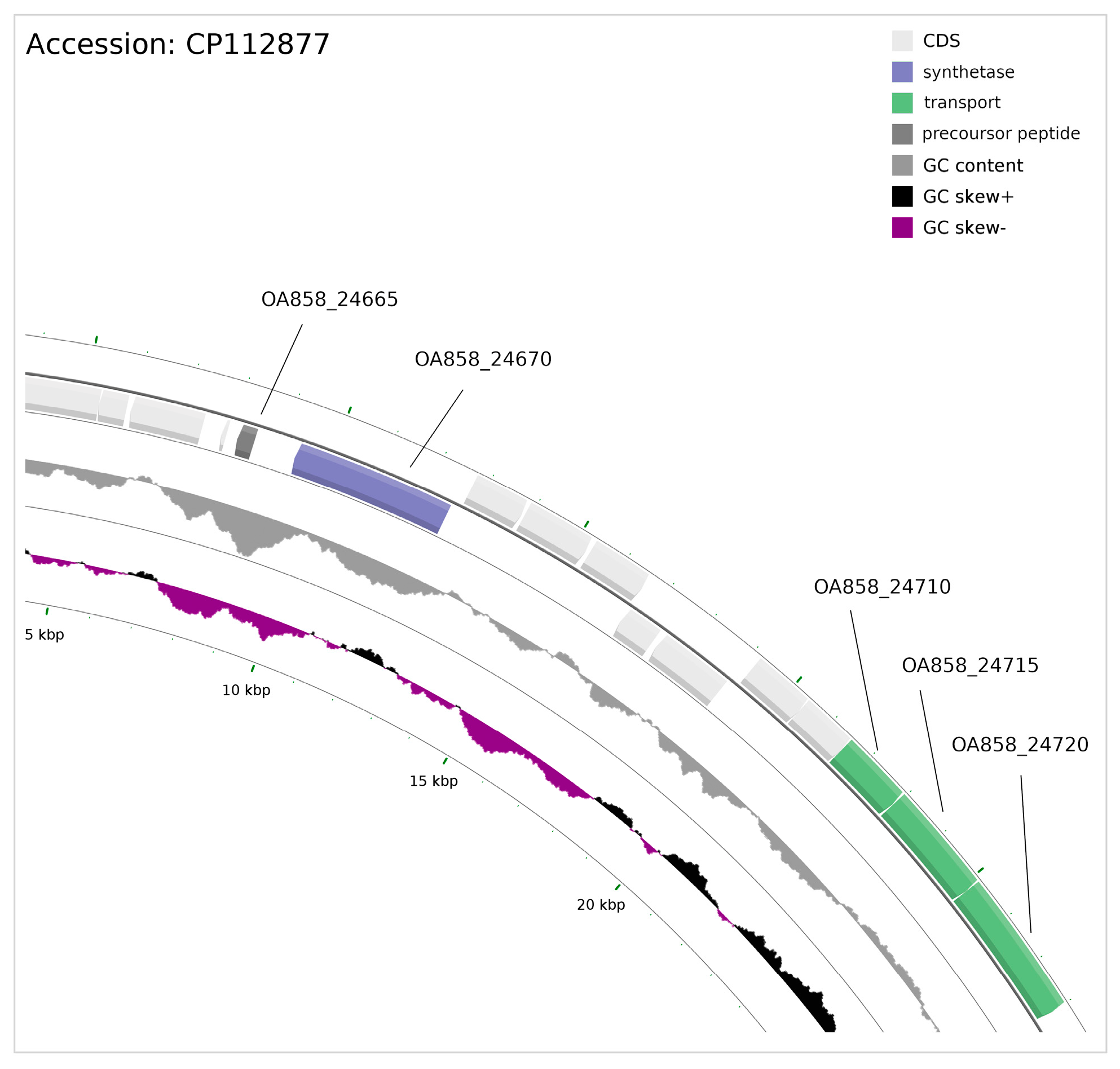
3.6. Ribosomally Synthesised and Post-Translationally Modified Peptides (RiPPs)
3.7. Peptides Previously Found in Mass Spectrometry (MS) Analysis
3.8. Cyanophage DNA Sequence in the P. galeata CCNP1313 Genome
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jones, A.C.; Monroe, E.A.; Eisman, E.B.; Gerwick, L.; Sherman, D.H.; Gerwick, W.H. The unique mechanistic transformations involved in the biosynthesis of modular natural products from marine cyanobacteria. Nat. Prod. Rep. 2010, 27, 1048. [Google Scholar] [CrossRef] [PubMed]
- Nandagopal, P.; Steven, A.N.; Chan, L.-W.; Rahmat, Z.; Jamaluddin, H.; Noh, N.I.M. Bioactive Metabolites Produced by Cyanobacteria for Growth Adaptation and Their Pharmacological Properties. Biology 2021, 10, 1061. [Google Scholar] [CrossRef] [PubMed]
- Demay, J.; Bernard, C.; Reinhardt, A.; Marie, B. Natural Products from Cyanobacteria: Focus on Beneficial Activities. Mar. Drugs 2019, 17, 320. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.T.; Phyo, M.Y. Marine Cyanobacteria: A Source of Lead Compounds and their Clinically-Relevant Molecular Targets. Molecules 2020, 25, 2197. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.R.; Pinto, E.; Torres, M.A.; Dörr, F.; Mazur-Marzec, H.; Szubert, K.; Tartaglione, L.; Dell’Aversano, C.; Miles, C.O.; Beach, D.G.; et al. CyanoMetDB, a comprehensive public database of secondary metabolites from cyanobacteria. Water Res. 2021, 196, 117017. [Google Scholar] [CrossRef] [PubMed]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural Product Biosynthetic Diversity and Comparative Genomics of the Cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Kehr, J.-C.; Gatte Picchi, D.; Dittmann, E. Natural product biosyntheses in cyanobacteria: A treasure trove of unique enzymes. Beilstein J. Org. Chem. 2011, 7, 1622–1635. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.S.; Pearson, L.A.; Neilan, B.A. Genome Mining and Evolutionary Analysis Reveal Diverse Type III Polyketide Synthase Pathways in Cyanobacteria. Genome Biol. Evol. 2021, 13, evab056. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.C.; Eslami, S.M.; Hetrick, K.J.; Ackenhusen, S.E.; Mitchell, D.A.; van der Donk, W.A. Precursor peptide-targeted mining of more than one hundred thousand genomes expands the lanthipeptide natural product family. BMC Genom. 2020, 21, 387. [Google Scholar] [CrossRef]
- Méjean, A.; Ploux, O. Biosynthesis of Cylindrospermopsin in Cyanobacteria: Characterization of CyrJ the Sulfotransferase. J. Nat. Prod. 2021, 84, 408–416. [Google Scholar] [CrossRef]
- Shih, P.M.; Wu, D.; Latifi, A.; Axen, S.D.; Fewer, D.P.; Talla, E.; Calteau, A.; Cai, F.; de Marsac, N.T.; Rippka, R.; et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Popin, R.V.; Alvarenga, D.O.; Castelo-Branco, R.; Fewer, D.P.; Sivonen, K. Mining of Cyanobacterial Genomes Indicates Natural Product Biosynthetic Gene Clusters Located in Conjugative Plasmids. Front. Microbiol. 2021, 12, 684565. [Google Scholar] [CrossRef] [PubMed]
- Sivonen, K.; Leikoski, N.; Fewer, D.P.; Jokela, J. Cyanobactins—Ribosomal cyclic peptides produced by cyanobacteria. Appl. Microbiol. Biotechnol. 2010, 86, 1213–1225. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.; Vasconcelos, V. Cyanobactins from Cyanobacteria: Current Genetic and Chemical State of Knowledge. Mar. Drugs 2015, 13, 6910–6946. [Google Scholar] [CrossRef] [PubMed]
- Ziemert, N.; Ishida, K.; Weiz, A.; Hertweck, C.; Dittmann, E. Exploiting the Natural Diversity of Microviridin Gene Clusters for Discovery of Novel Tricyclic Depsipeptides. Appl. Environ. Microbiol. 2010, 76, 3568–3574. [Google Scholar] [CrossRef] [PubMed]
- Cubillos-Ruiz, A.; Berta-Thompson, J.W.; Becker, J.W.; van der Donk, W.A.; Chisholm, S.W. Evolutionary radiation of lanthipeptides in marine cyanobacteria. Proc. Natl. Acad. Sci. USA 2017, 114, E5424–E5433. [Google Scholar] [CrossRef]
- Felczykowska, A.; Pawlik, A.; Mazur-Marzec, H.; Toruńska-Sitarz, A.; Narajczyk, M.; Richert, M.; Węgrzyn, G.; Herman-Antosiewicz, A. Selective inhibition of cancer cells’ proliferation by compounds included in extracts from Baltic Sea cyanobacteria. Toxicon 2015, 108, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cegłowska, M.; Szubert, K.; Grygier, B.; Lenart, M.; Plewka, J.; Milewska, A.; Lis, K.; Szczepański, A.; Chykunova, Y.; Barreto-Duran, E.; et al. Pseudanabaena galeata CCNP1313—Biological Activity and Peptides Production. Toxins 2022, 14, 330. [Google Scholar] [CrossRef]
- Nowak, R.M.; Jastrzębski, J.P.; Kuśmirek, W.; Sałamatin, R.; Rydzanicz, M.; Sobczyk-Kopcioł, A.; Sulima-Celińska, A.; Paukszto, Ł.; Makowczenko, K.G.; Płoski, R.; et al. Hybrid de novo whole-genome assembly and annotation of the model tapeworm Hymenolepis diminuta. Sci. Data 2019, 6, 302. [Google Scholar] [CrossRef]
- Wilson, K. Preparation of Genomic DNA from Bacteria. Curr. Protoc. Mol. Biol. 2001, 56, 2–4. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- de Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Zimin, A.V.; Puiu, D.; Luo, M.-C.; Zhu, T.; Koren, S.; Marçais, G.; Yorke, J.A.; Dvořák, J.; Salzberg, S.L. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res. 2017, 27, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-mapper: A web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. Protein Structure and Function Prediction Using I-TASSER. Curr. Protoc. Bioinform. 2015, 52, 5.8.1–5.8.15. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Arantes, A.S.; Stothard, P. Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genom. 2012, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Kassab, E.; Fuchs, M.; Haack, M.; Mehlmer, N.; Brueck, T.B. Engineering Escherichia coli FAB system using synthetic plant genes for the production of long chain fatty acids. Microb. Cell Factories 2019, 18, 163. [Google Scholar] [CrossRef] [PubMed]
- Stachelhaus, T.; Mootz, H.D.; Marahiel, M.A. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 1999, 6, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L.; Ravel, J.; Townsend, C.A. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 2000, 7, 211–224. [Google Scholar] [CrossRef]
- Ishikawa, F.; Nohara, M.; Takashima, K.; Tanabe, G. Probing the Compatibility of an Enzyme-Linked Immunosorbent Assay toward the Reprogramming of Nonribosomal Peptide Synthetase Adenylation Domains. ChemBioChem 2020, 21, 3056–3061. [Google Scholar] [CrossRef]
- Rausch, C. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 2005, 33, 5799–5808. [Google Scholar] [CrossRef]
- Alonzo, D.A.; Schmeing, T.M. Biosynthesis of depsipeptides, or Depsi: The peptides with varied generations. Protein Sci. 2020, 29, 2316–2347. [Google Scholar] [CrossRef] [PubMed]
- Graz, M.; Hunt, A.; Jamie, H.; Grant, G.; Milne, P. Antimicrobial activity of selected cyclic dipeptides. Die Pharm. 1999, 54, 772–775. [Google Scholar]
- Jorda, R.; Magar, P.; Hendrychová, D.; Pauk, K.; Dibus, M.; Pilařová, E.; Imramovský, A.; Kryštof, V. Novel modified leucine and phenylalanine dipeptides modulate viability and attachment of cancer cells. Eur. J. Med. Chem. 2020, 188, 112036. [Google Scholar] [CrossRef] [PubMed]
- Sivanathan, S.; Scherkenbeck, J. Cyclodepsipeptides: A Rich Source of Biologically Active Compounds for Drug Research. Molecules 2014, 19, 12368–12420. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.-S.; Nishimoto, K.; Motoyama, T.; Shimizu, T.; Hino, T.; Dohmae, N.; Nagano, S.; Osada, H. Unique features of the ketosynthase domain in a nonribosomal peptide synthetase–polyketide synthase hybrid enzyme, tenuazonic acid synthetase 1. J. Biol. Chem. 2020, 295, 11602–11612. [Google Scholar] [CrossRef] [PubMed]
- Cogan, D.P.; Zhang, K.; Li, X.; Li, S.; Pintilie, G.D.; Roh, S.-H.; Craik, C.S.; Chiu, W.; Khosla, C. Mapping the catalytic conformations of an assembly-line polyketide synthase module. Science 2021, 374, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Koper, K.; Han, S.-W.; Pastor, D.C.; Yoshikuni, Y.; Maeda, H.A. Evolutionary origin and functional diversification of aminotransferases. J. Biol. Chem. 2022, 298, 102122. [Google Scholar] [CrossRef]
- Kudo, F.; Miyanaga, A.; Eguchi, T. Biosynthesis of natural products containing β-amino acids. Nat. Prod. Rep. 2014, 31, 1056–1073. [Google Scholar] [CrossRef] [PubMed]
- Duitman, E.H.; Hamoen, L.W.; Rembold, M.; Venema, G.; Seitz, H.; Saenger, W.; Bernhard, F.; Reinhardt, R.; Schmidt, M.; Ullrich, C.; et al. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: A multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 1999, 96, 13294–13299. [Google Scholar] [CrossRef]
- Tillett, D.; Dittmann, E.; Erhard, M.; von Döhren, H.; Börner, T.; Neilan, B.A. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: An integrated peptide–polyketide synthetase system. Chem. Biol. 2000, 7, 753–764. [Google Scholar] [CrossRef]
- Chan, Y.A.; Podevels, A.M.; Kevany, B.M.; Thomas, M.G. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 2009, 26, 90–114. [Google Scholar] [CrossRef]
- Sabatini, M.; Comba, S.; Altabe, S.; Recio-Balsells, A.I.; Labadie, G.R.; Takano, E.; Gramajo, H.; Arabolaza, A. Biochemical characterization of the minimal domains of an iterative eukaryotic polyketide synthase. FEBS J. 2018, 285, 4494–4511. [Google Scholar] [CrossRef]
- Reeves, C.D.; Murli, S.; Ashley, G.W.; Piagentini, M.; Hutchinson, C.R.; McDaniel, R. Alteration of the Substrate Specificity of a Modular Polyketide Synthase Acyltransferase Domain through Site-Specific Mutations. Biochemistry 2001, 40, 15464–15470. [Google Scholar] [CrossRef] [PubMed]
- Petković, H.; Sandmann, A.; Challis, I.R.; Hecht, H.-J.; Silakowski, B.; Low, L.; Beeston, N.; Kuščer, E.; Garcia-Bernardo, J.; Leadlay, P.F.; et al. Substrate specificity of the acyl transferase domains of EpoC from the epothilone polyketide synthase. Org. Biomol. Chem. 2008, 6, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, M.C.; Neilan, B.A. Characterization of the Nodularin Synthetase Gene Cluster and Proposed Theory of the Evolution of Cyanobacterial Hepatotoxins. Appl. Environ. Microbiol. 2004, 70, 6353–6362. [Google Scholar] [CrossRef]
- Eusébio, N.; Castelo-Branco, R.; Sousa, D.; Preto, M.; D’agostino, P.; Gulder, T.A.M.; Leão, P.N. Discovery and Heterologous Expression of Microginins from Microcystis aeruginosa LEGE 91341. ACS Synth. Biol. 2022, 11, 3493–3503. [Google Scholar] [CrossRef]
- Nanson, J.D.; Himiari, Z.; Swarbrick, C.M.D.; Forwood, J.K. Structural Characterisation of the Beta-Ketoacyl-Acyl Carrier Protein Synthases, FabF and FabH, of Yersinia pestis. Sci. Rep. 2015, 5, 14797. [Google Scholar] [CrossRef] [PubMed]
- Jez, J.M.; Noel, J.P. Mechanism of Chalcone Synthase. J. Biol. Chem. 2000, 275, 39640–39646. [Google Scholar] [CrossRef]
- Jez, J.M.; Bowman, M.E.; Noel, J.P. Structure-Guided Programming of Polyketide Chain-Length Determination in Chalcone Synthase. Biochemistry 2001, 40, 14829–14838. [Google Scholar] [CrossRef]
- Meslet-Cladière, L.; Delage, L.; Leroux, C.J.-J.; Goulitquer, S.; Leblanc, C.; Creis, E.; Gall, E.A.; Stiger-Pouvreau, V.; Czjzek, M.; Potin, P. Structure/function analysis of a type iii polyketide synthase in the brown alga Ectocarpus siliculosus reveals a biochemical pathway in phlorotannin monomer biosynthesis. Plant Cell 2013, 25, 3089–3103. [Google Scholar] [CrossRef]
- Moche, M.; Dehesh, K.; Edwards, P.; Lindqvist, Y. The crystal structure of beta-ketoacyl-acyl carrier protein synthase II from Synechocystis sp. at 1.54 A resolution and its relationship to other condensing enzymes. J. Mol. Biol. 2001, 305, 491–503. [Google Scholar] [CrossRef]
- Martinez, M.A.; Zaballa, M.-E.; Schaeffer, F.; Bellinzoni, M.; Albanesi, D.; Schujman, G.E.; Vila, A.J.; Alzari, P.M.; de Mendoza, D. A novel role of malonyl-ACP in lipid homeostasis. Biochemistry 2010, 49, 3161–3167. [Google Scholar] [CrossRef]
- Oman, T.J.; van der Donk, W.A. Insights into the Mode of Action of the Two-Peptide Lantibiotic Haloduracin. ACS Chem. Biol. 2009, 4, 865–874. [Google Scholar] [CrossRef]
- van der Donk, W.A.; Nair, S.K. Structure and mechanism of lanthipeptide biosynthetic enzymes. Curr. Opin. Struct. Biol. 2014, 29, 58–66. [Google Scholar] [CrossRef]
- Li, B.; van der Donk, W.A. Identification of Essential Catalytic Residues of the Cyclase NisC Involved in the Biosynthesis of Nisin. J. Biol. Chem. 2007, 282, 21169–21175. [Google Scholar] [CrossRef]
- Havarstein, L.S.; Diep, D.B.; Nes, I.F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 1995, 16, 229–240. [Google Scholar] [CrossRef]
- Haft, D.H.; Basu, M.K.; Mitchell, D.A. Expansion of ribosomally produced natural products: A nitrile hydratase- and Nif11-related precursor family. BMC Biol. 2010, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Welker, M.; von Döhren, H. Cyanobacterial peptides—Nature’s own combinatorial biosynthesis. FEMS Microbiol. Rev. 2006, 30, 530–563. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Patton, G.C.; van der Donk, W.A. Mutants of the Zinc Ligands of Lacticin 481 Synthetase Retain Dehydration Activity but Have Impaired Cyclization Activity. Biochemistry 2007, 46, 6268–6276. [Google Scholar] [CrossRef] [PubMed]
- You, Y.O.; van der Donk, W.A. Mechanistic Investigations of the Dehydration Reaction of Lacticin 481 Synthetase Using Site-Directed Mutagenesis. Biochemistry 2007, 46, 5991–6000. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.-H.; Tang, W.; Lukk, T.; Yu, Y.; Nair, S.K.; van der Donk, W.A. The enterococcal cytolysin synthetase has an unanticipated lipid kinase fold. eLife 2015, 4, e07607. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Li, W.; Lai, Z.; Akhtar, M.U.; Dong, N.; Shan, A.; Ma, D. Effect of terminal arrangement of tryptophan on biological activity of symmetric α-helix-forming peptides. Chem. Biol. Drug Des. 2019, 94, 2051–2063. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M. Lysinoalanine in Food and in Antimicrobial Proteins. Adv. Exp. Med. Biol. 1999, 459, 145–159. [Google Scholar] [CrossRef]
- An, L.; Cogan, D.P.; Navo, C.D.; Jiménez-Osés, G.; Nair, S.K.; van der Donk, W.A. Substrate-assisted enzymatic formation of lysinoalanine in duramycin. Nat. Chem. Biol. 2018, 14, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Ökesli, A.; Cooper, L.E.; Fogle, E.J.; van der Donk, W.A. Nine post-translational modifications during the biosynthesis of cinnamycin. J. Am. Chem. Soc. 2011, 133, 13753–13760. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Pickens, L.B.; Tang, Y.; Chooi, Y.-H. Metabolic Engineering for the Production of Natural Products. Annu. Rev. Chem. Biomol. Eng. 2011, 2, 211–236. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]


| Locus Tag | Proposed aa Activated | Residue Position According to GrsA Phe Numbering | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | ||
| OA858_03905 [module 1] | hydroxy/carboxy-acid | S | I | F | H | S | G | I | C | M |
| OA858_03905 [module 2] | Arg | D | A | E | D | I | G | T | V | V |
| Locus Tag | Proposed aa Activated | Residue Position According to GrsA Phe Numbering | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | ||
| OA858_07105 [module 1] | Val | D | A | L | W | L | G | G | T | F |
| OA858__07095 [module 3] | Val/Trp/Phe | D | A | F | W | L | A | G | T | F |
| OA858__07090 [module 4] | Thr/Val | D | V | E | N | I | G | G | V | T |
| Protein Sequence ID | Hypothetical Product | Topology | Pattern Name | Pattern | Strand | Start | End | Matched |
|---|---|---|---|---|---|---|---|---|
| WGS74979.1 | sulfate permease (plasmid) | TM (inside) | PG638 | RMGF[LI] | + | 124 | 128 | RMGFL |
| WGS75066.1 | phosphate ABC transporter permease PstA (plasmid) | TM (helix) | GP598 | A[LI]V[LI][LI]A | − | 18 | 23 | AIVLIA |
| WGS74721.1 | AAA family ATPase (plasmid) | globular | GP598 | A[LI]V[LI][LI]A | − | 1066 | 1071 | ALVLLA |
| WGS72582.1 | solanesyl diphosphate synthase | globular | GP598 | A[LI]V[LI][LI]A | + | 55 | 60 | ALVLLA |
| WGS73177.1 | glycosyltransferase family 2 protein | TM (inside) | PG638 | RMGF[LI] | + | 325 | 329 | RMGFL |
| WGS73241.1 | hypothetical protein | globular | GP655 | A[LI]V[LI][LI]AG | − | 99 | 105 | AIVILAG |
| GP598 | A[LI]V[LI][LI]A | − | 100 | 105 | AIVILA | |||
| WGS74578.1 | glutamine-hydrolyzing carbamoyl-phosphate synthase small subunit | globular | GP598 | A[LI]V[LI][LI]A | − | 7 | 12 | ALVLIA |
| WGS74238.1 | DNA-directed RNA polymerase subunit gamma | globular | PG638 | RMGF[LI] | + | 101 | 105 | RMGFI |
| WGS70420.1 | hypothetical protein | globular | GP598 | A[LI]V[LI][LI]A | − | 28 | 33 | AIVIIA |
| WGS71453.1 | cation-translocating P-type ATPase | TM (inside) | GP655 | A[LI]V[LI][LI]AG | − | 435 | 441 | ALVLLAG |
| GP598 | A[LI]V[LI][LI]A | − | 436 | 441 | ALVLLA | |||
| WGS71843.1 | photosystem I core protein PsaB | TM (outside) | GP767 | YAAF[LI][LI]A | + | 729 | 735 | YAAFLIA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grabski, M.; Gawor, J.; Cegłowska, M.; Gromadka, R.; Mazur-Marzec, H.; Węgrzyn, G. Genome Mining of Pseudanabaena galeata CCNP1313 Indicates a New Scope in the Search for Antiproliferative and Antiviral Agents. Microorganisms 2024, 12, 1628. https://doi.org/10.3390/microorganisms12081628
Grabski M, Gawor J, Cegłowska M, Gromadka R, Mazur-Marzec H, Węgrzyn G. Genome Mining of Pseudanabaena galeata CCNP1313 Indicates a New Scope in the Search for Antiproliferative and Antiviral Agents. Microorganisms. 2024; 12(8):1628. https://doi.org/10.3390/microorganisms12081628
Chicago/Turabian StyleGrabski, Michał, Jan Gawor, Marta Cegłowska, Robert Gromadka, Hanna Mazur-Marzec, and Grzegorz Węgrzyn. 2024. "Genome Mining of Pseudanabaena galeata CCNP1313 Indicates a New Scope in the Search for Antiproliferative and Antiviral Agents" Microorganisms 12, no. 8: 1628. https://doi.org/10.3390/microorganisms12081628
APA StyleGrabski, M., Gawor, J., Cegłowska, M., Gromadka, R., Mazur-Marzec, H., & Węgrzyn, G. (2024). Genome Mining of Pseudanabaena galeata CCNP1313 Indicates a New Scope in the Search for Antiproliferative and Antiviral Agents. Microorganisms, 12(8), 1628. https://doi.org/10.3390/microorganisms12081628









