Exploring the Frozen Armory: Antiphage Defense Systems in Cold-Adapted Bacteria with a Focus on CRISPR-Cas Systems
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Dataset of 938 Cold-Adapted Bacterial Genomes
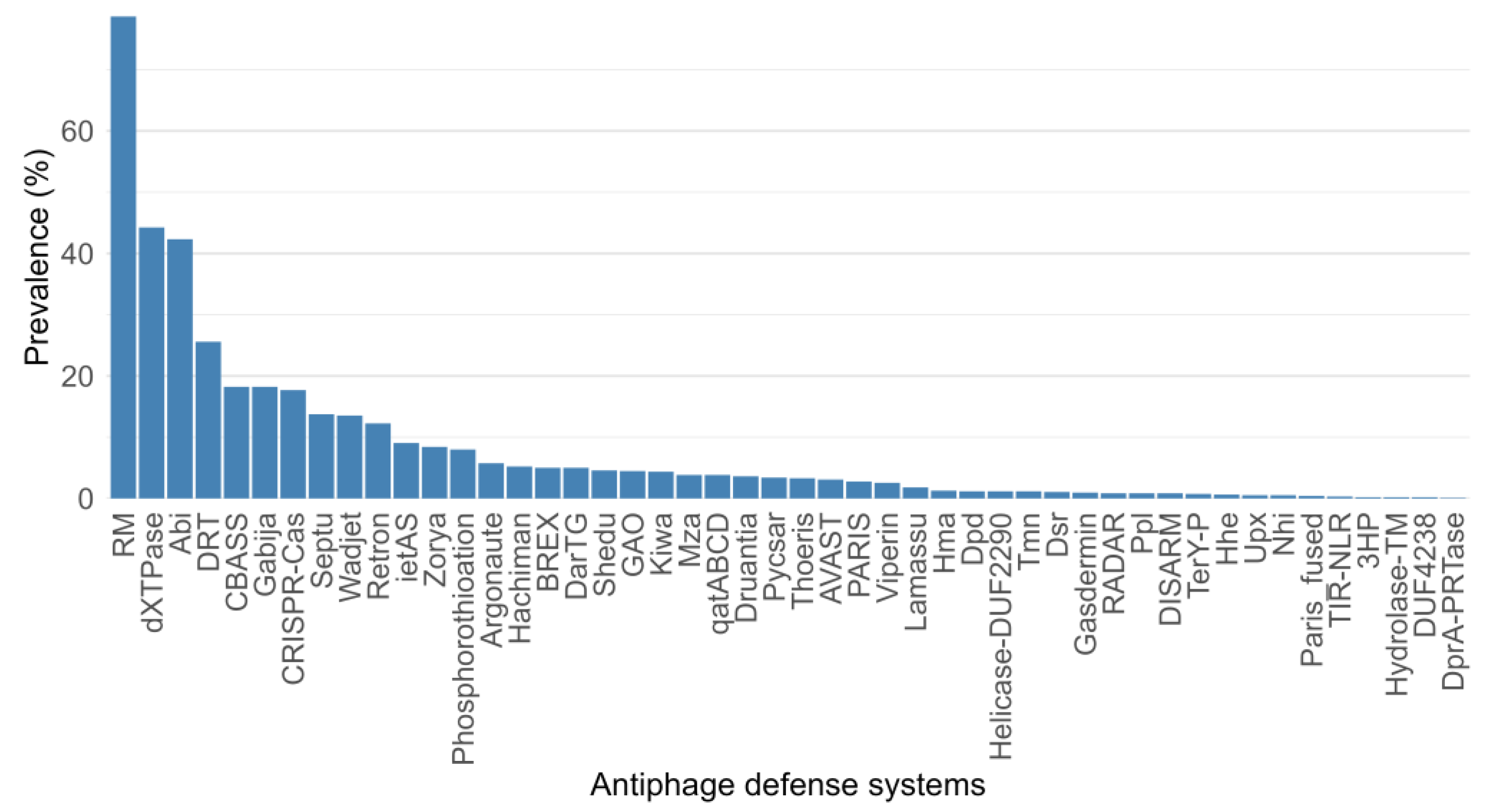
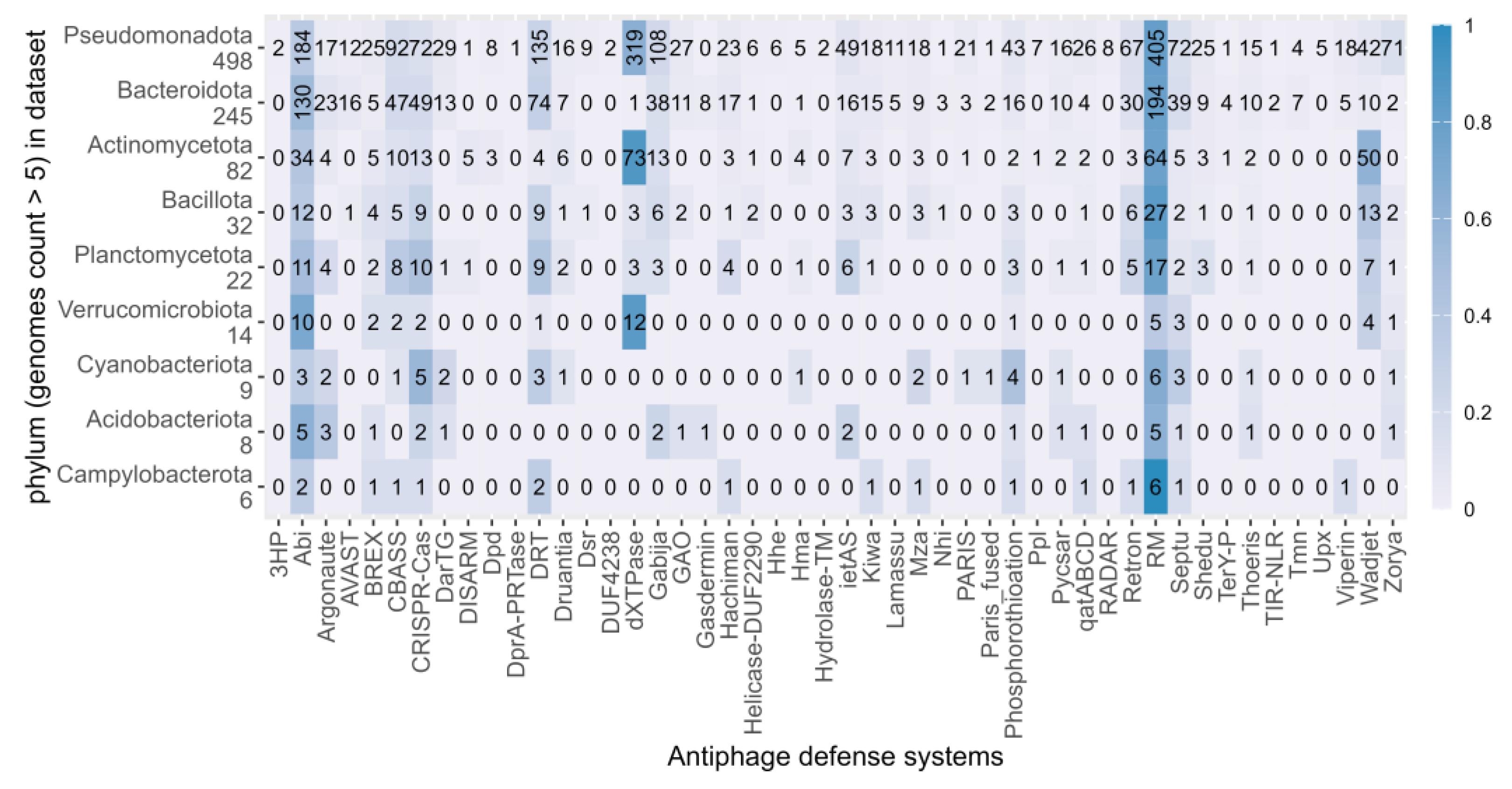
3.2. Abundance and Distribution of Antiphage Defense Systems in Cold-Adapted Bacteria
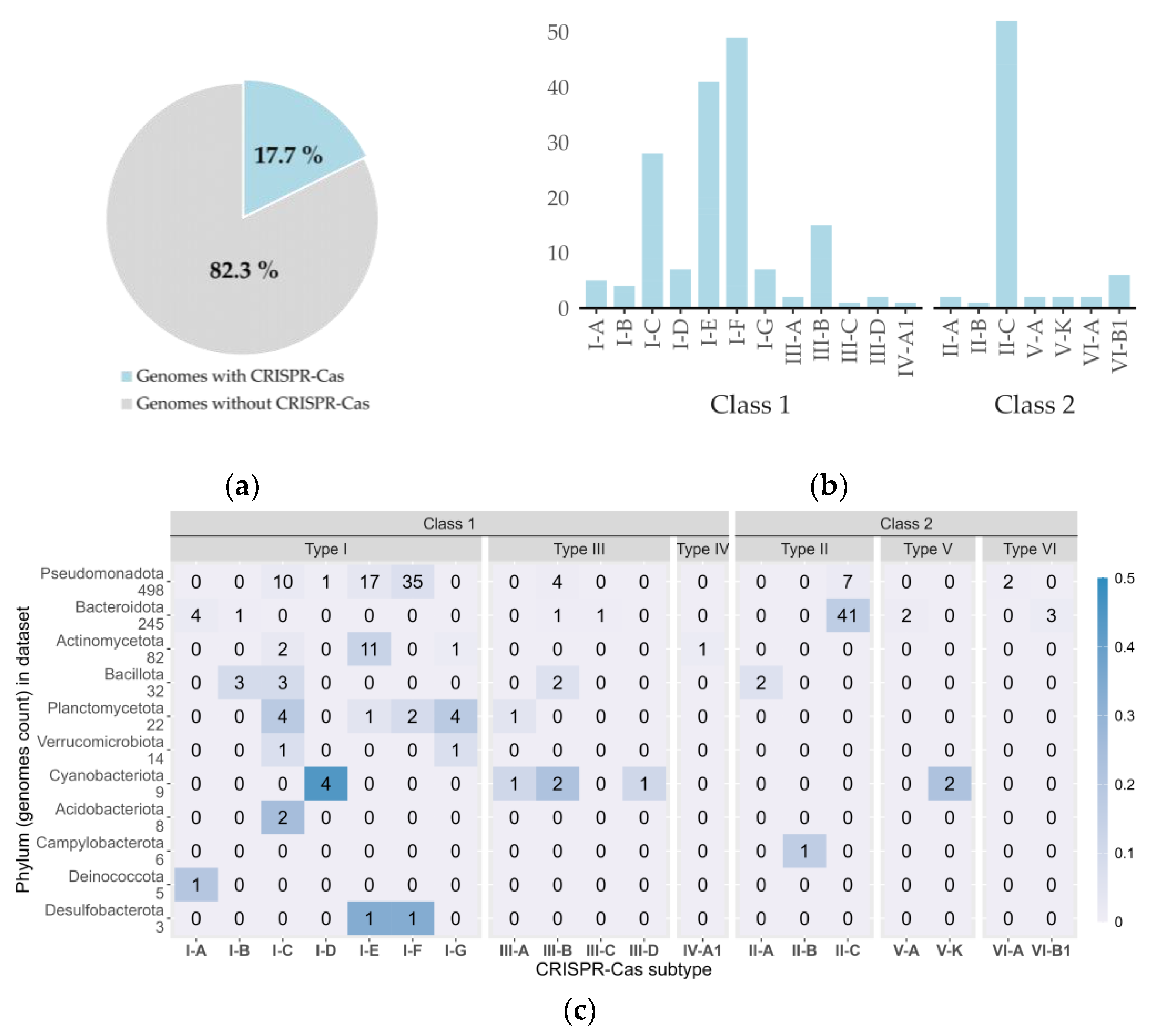
3.3. CRISPR-Cas Systems in Cold-Adapted Bacteria
3.4. Candidates for Cold-Active Genome Editing and Genome Engineering
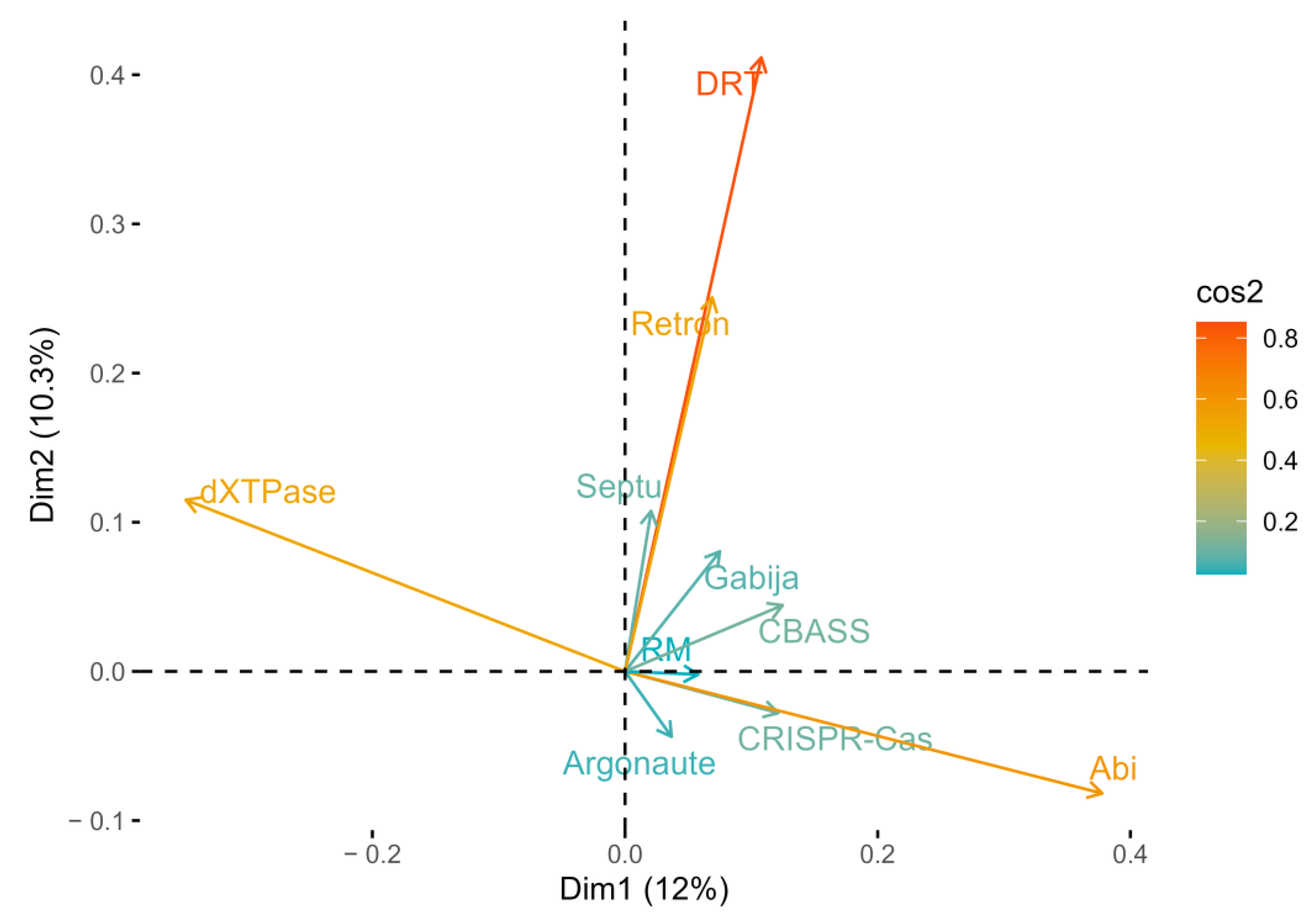
3.5. Patterns in Antiphage Defense System Occurrence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167. [Google Scholar] [CrossRef] [PubMed]
- Casanueva, A.; Tuffin, M.; Cary, C.; Cowan, D.A. Molecular adaptations to psychrophily: The impact of ‘omic’ technologies. Trends Microbiol. 2010, 18, 374–381. [Google Scholar] [CrossRef] [PubMed]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, S.; Collins, T.; Marx, J.-C.; Feller, G.; Gerday, C.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Filippova, S.N.; Surgucheva, N.A.; Sorokin, V.V.; Akimov, V.N.; Karnysheva, E.A.; Brushkov, A.V.; Andersen, D.; Gal’chenko, V.F. Bacteriophages in Arctic and Antarctic low-temperature systems. Microbiology 2016, 85, 359–366. [Google Scholar] [CrossRef]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Senčilo, A.; Luhtanen, A.-M.; Saarijärvi, M.; Bamford, D.H.; Roine, E. Cold-active bacteriophages from the Baltic Sea ice have diverse genomes and virus–host interactions. Environ. Microbiol. 2015, 17, 3628–3641. [Google Scholar] [CrossRef]
- Rostøl, J.T.; Marraffini, L. (Ph)ighting Phages: How Bacteria Resist Their Parasites. Cell Host Microbe 2019, 25, 184–194. [Google Scholar] [CrossRef]
- Bernheim, A.; Sorek, R. The pan-immune system of bacteria: Antiviral defence as a community resource. Nat. Rev. Microbiol. 2020, 18, 113–119. [Google Scholar] [CrossRef]
- Dy, R.L.; Richter, C.; Salmond, G.P.; Fineran, P.C. Remarkable Mechanisms in Microbes to Resist Phage Infections. Annu. Rev. Virol. 2014, 1, 307–331. [Google Scholar] [CrossRef] [PubMed]
- Kronheim, S.; Daniel-Ivad, M.; Duan, Z.; Hwang, S.; Wong, A.I.; Mantel, I.; Nodwell, J.R.; Maxwell, K.L. A chemical defence against phage infection. Nature 2018, 564, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Lopatina, A.; Tal, N.; Sorek, R. Abortive Infection: Bacterial Suicide as an Antiviral Immune Strategy. Annu. Rev. Virol. 2020, 7, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, H.; Tong, Y. Unveil the Secret of the Bacteria and Phage Arms Race. Int. J. Mol. Sci. 2023, 24, 4363. [Google Scholar] [CrossRef] [PubMed]
- Vasu, K.; Nagaraja, V. Diverse Functions of Restriction-Modification Systems in Addition to Cellular Defense. Microbiol. Mol. Biol. Rev. 2013, 77, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- van der Oost, J.; Jore, M.M.; Westra, E.R.; Lundgren, M.; Brouns, S.J. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009, 34, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Deveau, H.; Garneau, J.E.; Moineau, S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010, 64, 475–493. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef]
- Barrangou, R. The roles of CRISPR–Cas systems in adaptive immunity and beyond. Curr. Opin. Immunol. 2015, 32, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Cury, J.; Bernheim, A. CRISPR-Cas and restriction-modification team up to achieve long-term immunity. Trends Microbiol. 2022, 30, 513–514. [Google Scholar] [CrossRef] [PubMed]
- Tesson, F.; Bernheim, A. Synergy and regulation of antiphage systems: Toward the existence of a bacterial immune system? Curr. Opin. Microbiol. 2023, 71, 102238. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Doudna, J.A. CRISPR technology: A decade of genome editing is only the beginning. Science 2023, 379, eadd8643. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Classification and Nomenclature of CRISPR-Cas Systems: Where from Here? Cris. J. 2018, 1, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Nishiga, M.; Liu, C.; Qi, L.S.; Wu, J.C. The use of new CRISPR tools in cardiovascular research and medicine. Nat. Rev. Cardiol. 2022, 19, 505–521. [Google Scholar] [CrossRef]
- Hillary, V.E.; Ceasar, S.A. A Review on the Mechanism and Applications of CRISPR/Cas9/Cas12/Cas13/Cas14 Proteins Utilized for Genome Engineering. Mol. Biotechnol. 2023, 65, 311–325. [Google Scholar] [CrossRef]
- Liu, W.; Zuo, S.; Shao, Y.; Bi, K.; Zhao, J.; Huang, L.; Xu, Z.; Lian, J. Retron-mediated multiplex genome editing and continuous evolution in Escherichia coli. Nucleic Acids Res. 2023, 51, 8293–8307. [Google Scholar] [CrossRef] [PubMed]
- Graver, B.A.; Chakravarty, N.; Solomon, K.V. Prokaryotic Argonautes for in vivo biotechnology and molecular diagnostics. Trends Biotechnol. 2023, 42, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Payne, L.J.; Todeschini, T.C.; Wu, Y.; Perry, B.J.; Ronson, C.W.; Fineran, P.C.; Nobrega, F.L.; Jackson, S.A. Identification and classification of antiviral defence systems in bacteria and archaea with PADLOC reveals new system types. Nucleic Acids Res. 2021, 49, 10868–10878. [Google Scholar] [CrossRef] [PubMed]
- Georjon, H.; Bernheim, A. The highly diverse antiphage defence systems of bacteria. Nat. Rev. Microbiol. 2023, 21, 686–700. [Google Scholar] [CrossRef]
- Vassallo, C.N.; Doering, C.R.; Littlehale, M.L.; Teodoro, G.I.C.; Laub, M.T. A functional selection reveals previously undetected anti-phage defence systems in the E. coli pangenome. Nat. Microbiol. 2022, 7, 1568–1579. [Google Scholar] [CrossRef] [PubMed]
- Weissman, J.L.; Laljani, R.M.R.; Fagan, W.F.; Johnson, P.L.F. Visualization and prediction of CRISPR incidence in microbial trait-space to identify drivers of antiviral immune strategy. ISME J. 2019, 13, 2589–2602. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.R.; Liu, Z.L.; Niu, D.K. Precipitous Increase of Bacterial CRISPR-Cas Abundance at Around 45 °C. Front. Microbiol. 2022, 13, 773114. [Google Scholar] [CrossRef] [PubMed]
- Nogi, Y. Taxonomy of Psychrophiles. In Extremophiles Handbook; Horikoshi, K., Ed.; Springer: Tokyo, Japan, 2011; pp. 778–787. [Google Scholar]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Mori, T.; Yamagishi, K.; Takeyama, H. SAG-QC: Quality control of single amplified genome information by subtracting non-target sequences based on sequence compositions. BMC Bioinform. 2017, 18, 152. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Russel, J.; Pinilla-Redondo, R.; Mayo-Muñoz, D.; Shah, S.A.; Sørensen, S.J. CRISPRCasTyper: Automated Identification, Annotation, and Classification of CRISPR-Cas Loci. CRISPR J. 2020, 3, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, L.; Luo, X.; Chen, M.; Tang, W.; Zhan, L.; Dai, Z.; Lam, T.T.; Guan, Y.; Yu, G. Ggtree: A serialized data object for visualization of a phylogenetic tree and annotation data. iMeta 2022, 1, e56. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses, 1.0.7; CRAN (Comprehensive R Archive Network). 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 16 August 2023).
- Tal, N.; Millman, A.; Stokar-Avihail, A.; Fedorenko, T.; Leavitt, A.; Melamed, S.; Yirmiya, E.; Avraham, C.; Brandis, A.; Mehlman, T.; et al. Bacteria deplete deoxynucleotides to defend against bacteriophage infection. Nat. Microbiol. 2022, 7, 1200–1209. [Google Scholar] [CrossRef] [PubMed]
- González-Delgado, A.; Mestre, M.R.; Martínez-Abarca, F.; Toro, N. Prokaryotic reverse transcriptases: From retroelements to specialized defense systems. FEMS Microbiol. Rev. 2021, 45, fuab025. [Google Scholar] [CrossRef]
- Mestre, M.R.; Gao, L.A.; Shah, S.A.; López-Beltrán, A.; González-Delgado, A.; Martínez-Abarca, F.; Iranzo, J.; Redrejo-Rodríguez, M.; Zhang, F.; Toro, N. UG/Abi: A highly diverse family of prokaryotic reverse transcriptases associated with defense functions. Nucleic Acids Res. 2022, 50, 6084–6101. [Google Scholar] [CrossRef] [PubMed]
- Tesson, F.; Hervé, A.; Mordret, E.; Touchon, M.; d’Humières, C.; Cury, J.; Bernheim, A. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat. Commun. 2022, 13, 2561. [Google Scholar] [CrossRef]
- Zaayman, M.; Wheatley, R.M. Fitness costs of CRISPR-Cas systems in bacteria. Microbiology 2022, 168, 001209. [Google Scholar] [CrossRef]
- Dieser, M.; Smith, H.J.; Ramaraj, T.; Foreman, C.M. Janthinobacterium CG23_2: Comparative Genome Analysis Reveals Enhanced Environmental Sensing and Transcriptional Regulation for Adaptation to Life in an Antarctic Supraglacial Stream. Microorganisms 2019, 7, 454. [Google Scholar] [CrossRef]
- Cooper, Z.S.; Rapp, J.Z.; Shoemaker, A.M.D.; Anderson, R.E.; Zhong, Z.P.; Deming, J.W. Evolutionary Divergence of Marinobacter Strains in Cryopeg Brines as Revealed by Pangenomics. Front. Microbiol. 2022, 13, 879116. [Google Scholar] [CrossRef] [PubMed]
- Bosi, E.; Fondi, M.; Orlandini, V.; Perrin, E.; Maida, I.; de Pascale, D.; Tutino, M.L.; Parrilli, E.; Lo Giudice, A.; Filloux, A.; et al. The pangenome of (Antarctic) Pseudoalteromonas bacteria: Evolutionary and functional insights. BMC Genom. 2017, 18, 93. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Powell, S.M.; Wilson, R.; Bowman, J.P. Extensive gene acquisition in the extremely psychrophilic bacterial species Psychroflexus torquis and the link to sea-ice ecosystem specialism. Genome Biol. Evol. 2014, 6, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Waldvogel, A.M.; Pfenninger, M. Temperature dependence of spontaneous mutation rates. Genome Res. 2021, 31, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Pucci, F.; Rooman, M. Physical and molecular bases of protein thermal stability and cold adaptation. Curr. Opin. Struct. Biol. 2017, 42, 117–128. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; He, R.; Wang, L.; Wang, Y.; Zeng, W.; Zhang, Z.; Wang, F.; Ma, L. A programmable pAgo nuclease with RNA target preference from the psychrotolerant bacterium Mucilaginibacter paludis. Nucleic Acids Res. 2022, 50, 5226–5238. [Google Scholar] [CrossRef]
- Millman, A.; Bernheim, A.; Stokar-Avihail, A.; Fedorenko, T.; Voichek, M.; Leavitt, A.; Oppenheimer-Shaanan, Y.; Sorek, R. Bacterial Retrons Function In Anti-Phage Defense. Cell 2020, 183, 1551–1561.e1512. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sandsdalen, G.D.; Kumar, A.; Hjerde, E. Exploring the Frozen Armory: Antiphage Defense Systems in Cold-Adapted Bacteria with a Focus on CRISPR-Cas Systems. Microorganisms 2024, 12, 1028. https://doi.org/10.3390/microorganisms12051028
Sandsdalen GD, Kumar A, Hjerde E. Exploring the Frozen Armory: Antiphage Defense Systems in Cold-Adapted Bacteria with a Focus on CRISPR-Cas Systems. Microorganisms. 2024; 12(5):1028. https://doi.org/10.3390/microorganisms12051028
Chicago/Turabian StyleSandsdalen, Greta Daae, Animesh Kumar, and Erik Hjerde. 2024. "Exploring the Frozen Armory: Antiphage Defense Systems in Cold-Adapted Bacteria with a Focus on CRISPR-Cas Systems" Microorganisms 12, no. 5: 1028. https://doi.org/10.3390/microorganisms12051028
APA StyleSandsdalen, G. D., Kumar, A., & Hjerde, E. (2024). Exploring the Frozen Armory: Antiphage Defense Systems in Cold-Adapted Bacteria with a Focus on CRISPR-Cas Systems. Microorganisms, 12(5), 1028. https://doi.org/10.3390/microorganisms12051028






