A Review on Colistin Resistance: An Antibiotic of Last Resort
Abstract
1. Introduction
2. Importance of Colistin
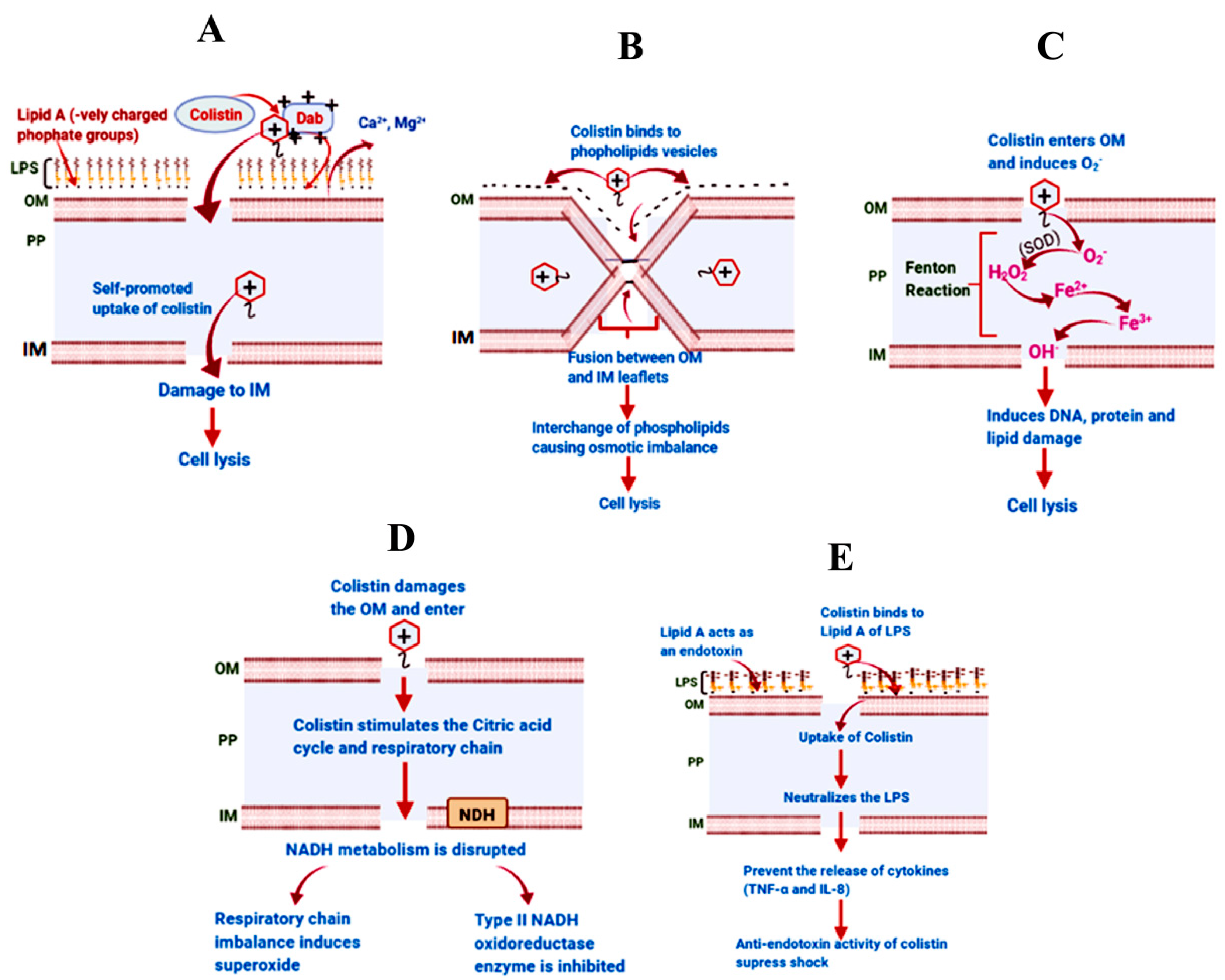
3. Mechanism of Action of Colistin against Gram-Negative Bacteria
3.1. The Classical Membrane Lysis Pathway
3.2. Vesicle–Vesicle Contact Pathway
3.3. Hydroxyl Radical Death Pathway
3.4. Respiratory Enzyme Inhibition Pathway
3.5. Anti-Endotoxin Activity of Colistin
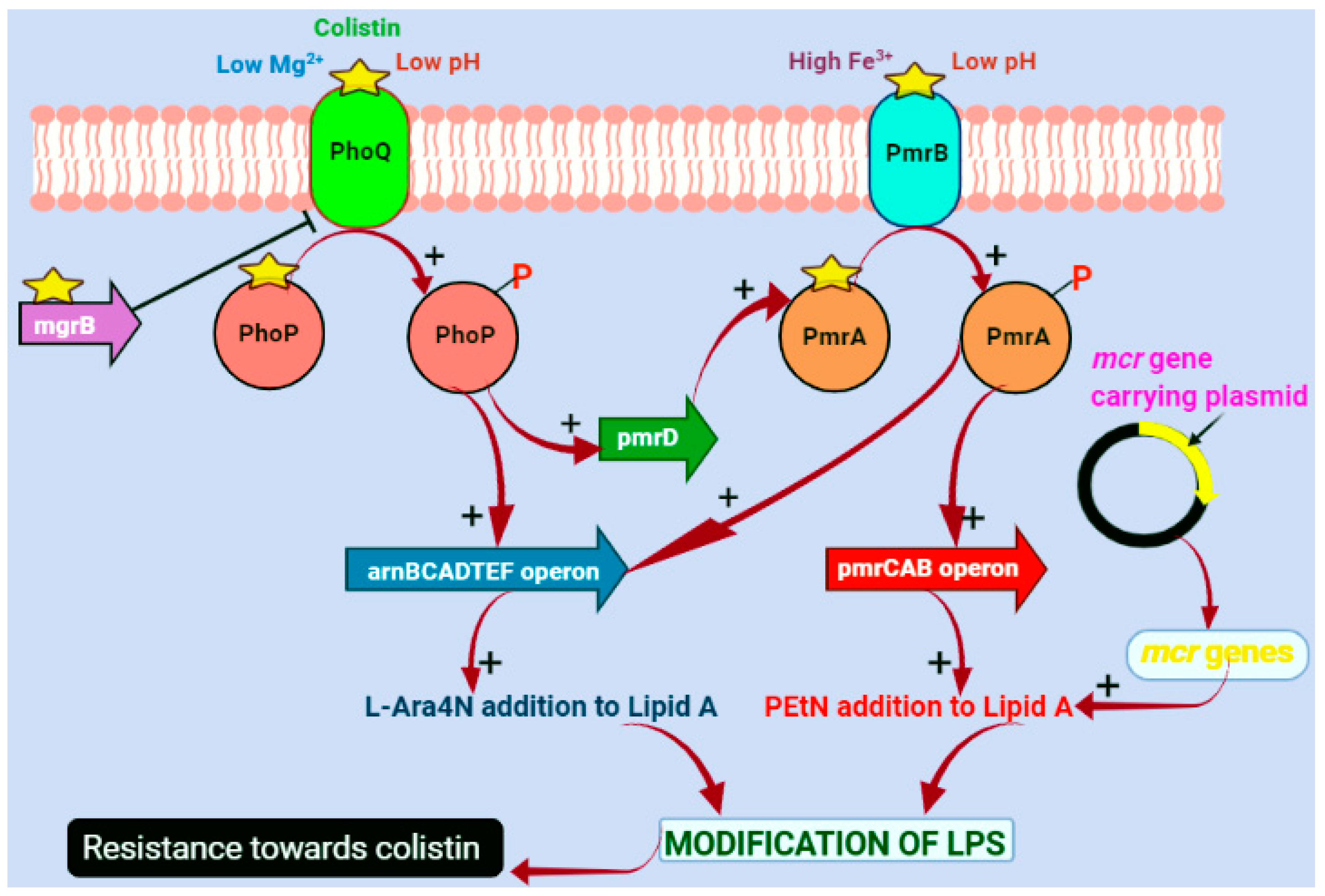
4. Molecular Mechanisms of Colistin Resistance
4.1. Chromosomal-Mediated Colistin Resistance
4.2. Plasmid-Mediated Colistin Resistance
5. Dissemination of Colistin Resistance (HGT: Transformation, Transduction, Conjugation)
6. Global Prevalence of mcr-Mediated Colistin Resistance (Environmental and Clinical Isolates)
6.1. The mcr-1 Gene
6.2. The mcr-2 Gene
6.3. The mcr-3 Gene
6.4. The mcr-4 Gene
6.5. The mcr-5 Gene
6.6. The mcr-6 Gene
6.7. The mcr-7 Gene
6.8. The mcr-8 Gene
6.9. The mcr-9 Gene
6.10. The mcr-10 Gene
7. Phenotypic and Molecular Detection Methods of Colistin-Resistant Gram-Negative Bacteria
7.1. Screening Resistant Isolates Using Selective Media
- ➢
- Super Polymyxin: It is a widely utilized screening medium, developed by ElitechMicrobio in Signes, France. It contains a lower concentration of colistin, specifically 3.5 μg/mL, which enables the straightforward identification of isolates resistant to colistin that have low minimum inhibitory concentration (MIC) values, whether the resistance is intrinsic or acquired [137,138]. In addition to colistin, SuperPolymyxin contains 5 μg/mL of amphotericin B, 10 μg/mL of daptomycin, and EMB powder. The EMB powder produces a distinct metallic green reflection and suppresses the growth of Gram-positive bacteria with selectivity.
- ➢
- CHROMagar COL-APSE: It is an agar-based selective medium developed subsequent to SuperPolymyxin, offering a broader target spectrum [139]. This medium incorporates colistin sulfate and oxazolidinone antibiotics. Its chromogenic properties enable the efficient differentiation of Enterobacteriaceae as well as colistin-resistant Gram-negative non-fermenters.
- ➢
- LBJMR (Lucie-Bardet-Jean-Marc-Rolain) Medium: This recently developed medium offers versatility by detecting colistin-resistant Enterobacteriaceae, Gram-negative non-fermenters, and vancomycin-resistant enterococci (VRE) [140]. Enhanced sensitivity and specificity were observed with a purple agar base supplemented with bromocresol purple and glucose compared to other combinations. Yellow colonies of Enterococci and Enterobacteriaceae, varying in size, are observable against the purple background. Additionally, this medium can facilitate the detection of pathogens in patients diagnosed with cystic fibrosis.
7.2. Determination of MIC
- The Sensititre system developed by Thermo Fisher Scientific (Waltham, MA, USA) has a customizable plate layout with 96 wells containing antibiotics. All the steps including inoculation, incubation, and reading can be automated [143].
- The Vitek 2 system developed by BioM’erieux, Marcy l’Etoile, France uses 64 wells containing dehydrated antibiotics and other reagents and is semi-automated [143].
- BD Phoenix developed by Becton Dickinson, Le Pont de Claix, France uses an 84-well plate and an oxidation-reduction indicator to determine the antimicrobial susceptibility in 6–16 h.
7.3. Matrix-Assisted Laser Desorption-Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
7.4. Rapid Polymyxin Nordmann Poirel (RPNP) Test for Enterobacteriaceae
7.5. Inhibition of MCR-1 Activity
7.6. Polymyxin Drop Test
8. Molecular Methods of Detection
8.1. Whole Genome Sequencing (WGS) and Polymerase Chain Reaction (PCR)
8.2. Loop-Mediated Isothermal Amplification (LAMP)
8.3. Microarray
8.4. Ribotyping (16S-23S ITS)
8.5. Multilocus Sequence Typing (MLST)
8.6. Infrared Biotyping (IRBT)
9. Judicial Use of Colistin
10. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Terreni, M.; Taccani, M.; Pregnolato, M. New antibiotics for multidrug-resistant bacterial strains: Latest research developments and future perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef] [PubMed]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q.M.R. Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Front. Med. 2021, 8, 677720. [Google Scholar] [CrossRef] [PubMed]
- Hassan, J.; El-Gemayel, L.; Bashour, I.; Kassem, I.I. On the edge of a precipice: The global emergence and dissemination of plasmid-borne mcr genes that confer resistance to colistin, a last-resort antibiotic. In Antibiotics and Antimicrobial Resistance Genes in the Environment; Elsevier: Amsterdam, The Netherlands, 2020; pp. 155–182. [Google Scholar]
- Rubens, R.S.; Arruda, I.d.S.A.; Almeida, R.M.; Nóbrega, Y.K.d.M.; Carneiro, M.d.S.; Dalmolin, T.V. Challenges in the Detection of Polymyxin Resistance: From Today to the Future. Microorganisms 2024, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure—Activity relationships of polymyxin antibiotics. J. Med. Chem. 2010, 53, 1898–1916. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Li, J. Pharmacology of polymyxins: New insights into an ‘old’ class of antibiotics. Future Microbiol. 2013, 8, 711–724. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Diene, S.M.; Kempf, M.; Berrazeg, M.; Bakour, S.; Gupta, S.K.; Thongmalayvong, B.; Akkhavong, K.; Somphavong, S.; Paboriboune, P. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: An epidemiological and molecular study. Int. J. Antimicrob. Agents 2014, 44, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.A.; Vargas, M.A.; Regueiro, V.; Llompart, C.M.; Albertí, S.; Bengoechea, J.A. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 2004, 72, 7107–7114. [Google Scholar] [CrossRef] [PubMed]
- Bengoechea, J.A.; Skurnik, M. Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol. Microbiol. 2000, 37, 67–80. [Google Scholar] [CrossRef]
- Nation, R.L.; Li, J. Colistin in the 21st century. Curr. Opin. Infect. Dis. 2009, 22, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Landman, D.; Georgescu, C.; Martin, D.A.; Quale, J. Polymyxins revisited. Clin. Microbiol. Rev. 2008, 21, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Bae, I.K.; Lee, H.; Jeong, S.H.; Yong, D.; Lee, K. In vivo emergence of colistin resistance in Acinetobacter baumannii clinical isolates of sequence type 357 during colistin treatment. Diagn. Microbiol. Infect. Dis. 2014, 79, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Rafailidis, P.I.; Ioannidou, E.; Alexiou, V.G.; Matthaiou, D.K.; Karageorgopoulos, D.E.; Kapaskelis, A.; Nikita, D.; Michalopoulos, A. Colistin therapy for microbiologically documented multidrug-resistant Gram-negative bacterial infections: A retrospective cohort study of 258 patients. Int. J. Antimicrob. Agents 2010, 35, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Xavier, B.B.; Lammens, C.; Ruhal, R.; Kumar-Singh, S.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016, 21, 30280. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.; Hadjadj, L.; Rolain, J.-M.; Olaitan, A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents 2016, 48, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Afify, F.A.; Shata, A.H.; Aboelnaga, N.; Osama, D.; Elsayed, S.W.; Saif, N.A.; Mouftah, S.F.; Shawky, S.M.; Mohamed, A.A.; Loay, O. Emergence of carbapenem resistant gram-negative pathogens with high rate of colistin resistance in Egypt: A cross sectional study to assess resistance trends during the COVID-19 pandemic. J. Genet. Eng. Biotechnol. 2024, 22, 100351. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Michalopoulos, A. Polymyxins: Old antibiotics are back. Lancet 2006, 367, 633–634. [Google Scholar] [CrossRef]
- Biswas, S.; Brunel, J.-M.; Dubus, J.-C.; Reynaud-Gaubert, M.; Rolain, J.-M. Colistin: An update on the antibiotic of the 21st century. Expert Rev. Anti-Infect. Ther. 2012, 10, 917–934. [Google Scholar] [CrossRef]
- Stein, A.; Raoult, D. Colistin: An antimicrobial for the 21st century? Clin. Infect. Dis. 2002, 35, 901–902. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.; Giske, C.G.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Bialvaei, A.Z.; Samadi Kafil, H. Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 2015, 31, 707–721. [Google Scholar] [CrossRef]
- Srinivas, K.; Rao, T.; Kiranmayi, C.; Kavitha, L. Antibiogram of Campylobacter jejuni and Campylobacter coli isolated from animals, foods of animal origin and humans in areas surrounding Gannavaram, Andhra Pradesh. Int. J. Sci. Environ. Technol 2019, 8, 233–246. [Google Scholar]
- Gurjar, M. Colistin for lung infection: An update. J. Intensive Care 2015, 3, 3. [Google Scholar] [CrossRef]
- Katsunuma, Y.; Hanazumi, M.; Fujisaki, H.; Minato, H.; Hashimoto, Y.; Yonemochi, C. Associations between the use of antimicrobial agents for growth promotion and the occurrence of antimicrobial-resistant Escherichia coli and enterococci in the feces of livestock and livestock farmers in Japan. J. Gen. Appl. Microbiol. 2007, 53, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Beaudry, F.; Letellier, A. Resistance to colistin: What is the fate for this antibiotic in pig production? Int. J. Antimicrob. Agents 2016, 48, 119–126. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed Ahmed, M.A.E.-G.; Zhong, L.-L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.-B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Kasiakou, S.K.; Saravolatz, L.D. Colistin: The revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Bergen, P.J.; Li, J.; Rayner, C.R.; Nation, R.L. Colistin methanesulfonate is an inactive prodrug of colistin against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2006, 50, 1953–1958. [Google Scholar] [CrossRef]
- Nishino, K.; Latifi, T.; Groisman, E.A. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2006, 59, 126–141. [Google Scholar] [CrossRef]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Qin, W.; Lin, J.; Fang, S.; Qiu, J. Antibacterial mechanisms of polymyxin and bacterial resistance. BioMed Res. Int. 2015, 2015, 679109. [Google Scholar] [CrossRef]
- Kanazawa, K.; Sato, Y.; Ohki, K.; Okimura, K.; Uchida, Y.; Shindo, M.; Sakura, N. Contribution of each amino acid residue in polymyxin B3 to antimicrobial and lipopolysaccharide binding activity. Chem. Pharm. Bull. 2009, 57, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.-P.S.; Hancock, R.E. The relationship between peptide structure and antibacterial activity. Peptides 2003, 24, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- Pristovsek, P.; Kidric, J. The search for molecular determinants of LPS inhibition by proteins and peptides. Curr. Top. Med. Chem. 2004, 4, 1185–1201. [Google Scholar] [CrossRef]
- Clausell, A.; Garcia-Subirats, M.; Pujol, M.; Busquets, M.A.; Rabanal, F.; Cajal, Y. Gram-negative outer and inner membrane models: Insertion of cyclic cationic lipopeptides. J. Phys. Chem. B 2007, 111, 551–563. [Google Scholar] [CrossRef]
- Cajal, Y.; Rogers, J.; Berg, O.G.; Jain, M.K. Intermembrane molecular contacts by polymyxin B mediate exchange of phospholipids. Biochemistry 1996, 35, 299–308. [Google Scholar] [CrossRef]
- Sampson, T.R.; Liu, X.; Schroeder, M.R.; Kraft, C.S.; Burd, E.M.; Weiss, D.S. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob. Agents Chemother. 2012, 56, 5642–5649. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Johnson, N.; Cizmas, L.; McDonald, T.J.; Kim, H. A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere 2016, 150, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Deris, Z.Z.; Swarbrick, J.D.; Roberts, K.D.; Azad, M.A.; Akter, J.; Horne, A.S.; Nation, R.L.; Rogers, K.L.; Thompson, P.E.; Velkov, T. Probing the penetration of antimicrobial polymyxin lipopeptides into gram-negative bacteria. Bioconjug. Chem. 2014, 25, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, K.; Ohnishi, T.; Kaback, H.R. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 1987, 26, 7732–7737. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhu, Y.; Fu, J.; Qiu, J.; Yin, J. Enhanced NADH metabolism involves colistin-induced killing of Bacillus subtilis and Paenibacillus polymyxa. Molecules 2019, 24, 387. [Google Scholar] [CrossRef] [PubMed]
- Şentürk, S. Evaluation of the anti-endotoxic effects of polymyxin-E (colistin) in dogs with naturally occurred endotoxic shock. J. Vet. Pharmacol. Ther. 2005, 28, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Baeuerlein, A.; Ackermann, S.; Parlesak, A. Transepithelial activation of human leukocytes by probiotics and commensal bacteria: Role of Enterobacteriaceae-type endotoxin. Microbiol. Immunol. 2009, 53, 241–250. [Google Scholar] [CrossRef]
- Andrade, F.F.; Silva, D.; Rodrigues, A.; Pina-Vaz, C. Colistin update on its mechanism of action and resistance, present and future challenges. Microorganisms 2020, 8, 1716. [Google Scholar] [CrossRef]
- Kaye, K.S.; Pogue, J.M.; Tran, T.B.; Nation, R.L.; Li, J. Agents of last resort: Polymyxin resistance. Infect. Dis. Clin. 2016, 30, 391–414. [Google Scholar] [CrossRef]
- McPhee, J.B.; Bains, M.; Winsor, G.; Lewenza, S.; Kwasnicka, A.; Brazas, M.D.; Brinkman, F.S.; Hancock, R. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 3995–4006. [Google Scholar] [CrossRef] [PubMed]
- Gunn, J.S. The Salmonella PmrAB regulon: Lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 2008, 16, 284–290. [Google Scholar] [CrossRef]
- Kato, A.; Chen, H.D.; Latifi, T.; Groisman, E.A. Reciprocal control between a bacterium’s regulatory system and the modification status of its lipopolysaccharide. Mol. Cell 2012, 47, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.D.; Groisman, E.A. The biology of the PmrA/PmrB two-component system: The major regulator of lipopolysaccharide modifications. Annu. Rev. Microbiol. 2013, 67, 83–112. [Google Scholar] [CrossRef] [PubMed]
- Rubin, E.J.; Herrera, C.M.; Crofts, A.A.; Trent, M.S. PmrD is required for modifications to Escherichia coli endotoxin that promote antimicrobial resistance. Antimicrob. Agents Chemother. 2015, 59, 2051–2061. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Groisman, E.A. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004, 18, 2302–2313. [Google Scholar] [CrossRef]
- Véscovi, E.G.; Soncini, F.C.; Groisman, E.A. Mg2+ as an extracellular signal: Environmental regulation of Salmonella virulence. Cell 1996, 84, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Soncini, F.C.; Groisman, E.A. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 1996, 178, 6796–6801. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Lee, E.-J.; Huang, H.; Groisman, E.A. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science 2006, 314, 1607–1609. [Google Scholar] [CrossRef]
- Yeo, W.-S.; Zwir, I.; Huang, H.V.; Shin, D.; Kato, A.; Groisman, E.A. Intrinsic negative feedback governs activation surge in two-component regulatory systems. Mol. Cell 2012, 45, 409–421. [Google Scholar] [CrossRef][Green Version]
- Schweizer, H.P. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: Unanswered questions. Genet. Mol. Res. 2003, 2, 48–62. [Google Scholar] [PubMed]
- Pamp, S.J.; Gjermansen, M.; Johansen, H.K.; Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 2008, 68, 223–240. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St Michael, F.; Cox, A.D.; et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, J.H.; Harper, M.; Adler, B.; Nation, R.L.; Li, J.; Boyce, J.D. Insertion sequence IS Aba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3022–3024. [Google Scholar] [CrossRef]
- Berglund, B. Acquired resistance to colistin via chromosomal and plasmid-mediated mechanisms in Klebsiella pneumoniae. Infect. Microbes Dis. 2019, 1, 10–19. [Google Scholar] [CrossRef]
- Zurfluh, K.; Tasara, T.; Poirel, L.; Nordmann, P.; Stephan, R. Draft genome sequence of Escherichia coli S51, a chicken isolate harboring a chromosomally encoded mcr-1 gene. Genome Announc. 2016, 4, e00796-16. [Google Scholar] [CrossRef] [PubMed]
- Sherman, E.X.; Hufnagel, D.A.; Weiss, D.S. MCR-1 confers cross-resistance to lysozyme. Lancet Infect. Dis. 2016, 16, 1226–1227. [Google Scholar] [CrossRef][Green Version]
- Kai, J.; Wang, S. Recent progress on elucidating the molecular mechanism of plasmid-mediated colistin resistance and drug design. Int. Microbiol. 2020, 23, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Bastidas-Caldes, C.; de Waard, J.H.; Salgado, M.S.; Villacís, M.J.; Coral-Almeida, M.; Yamamoto, Y.; Calvopiña, M. Worldwide prevalence of mcr-mediated colistin-resistance Escherichia coli in isolates of clinical samples, healthy humans, and livestock—A systematic review and meta-analysis. Pathogens 2022, 11, 659. [Google Scholar] [CrossRef] [PubMed]
- Dadashi, M.; Sameni, F.; Bostanshirin, N.; Yaslianifard, S.; Khosravi-Dehaghi, N.; Nasiri, M.J.; Goudarzi, M.; Hashemi, A.; Hajikhani, B. Global prevalence and molecular epidemiology of mcr-mediated colistin resistance in Escherichia coli clinical isolates: A systematic review. J. Glob. Antimicrob. Resist. 2022, 29, 444–461. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, H.; Liu, Y.-H.; Feng, Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018, 26, 794–808. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tian, G.-B.; Zhang, R.; Shen, Y.; Tyrrell, J.M.; Huang, X.; Zhou, H.; Lei, L.; Li, H.-Y.; Doi, Y. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: An epidemiological and clinical study. Lancet Infect. Dis. 2017, 17, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, T.R.; Ishikawa, K.; Takeda, K.; Oh, J.-S.; Kondo, H.; Hashizume, H.; Tanaka, H.; Knecht, S.D.; Bilén, S.G.; Hori, M. Dynamic analysis of reactive oxygen nitrogen species in plasma-activated culture medium by UV absorption spectroscopy. J. Appl. Phys. 2017, 122, 213301. [Google Scholar] [CrossRef]
- Neumann, B.; Rackwitz, W.; Hunfeld, K.-P.; Fuchs, S.; Werner, G.; Pfeifer, Y. Genome sequences of two clinical Escherichia coli isolates harboring the novel colistin-resistance gene variants mcr-1.26 and mcr-1.27. Gut Pathog. 2020, 12, 40. [Google Scholar] [CrossRef]
- Partridge, S.R.; Di Pilato, V.; Doi, Y.; Feldgarden, M.; Haft, D.H.; Klimke, W.; Kumar-Singh, S.; Liu, J.-H.; Malhotra-Kumar, S.; Prasad, A. Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J. Antimicrob. Chemother. 2018, 73, 2625–2630. [Google Scholar] [CrossRef] [PubMed]
- AbuOun, M.; Stubberfield, E.J.; Duggett, N.A.; Kirchner, M.; Dormer, L.; Nunez-Garcia, J.; Randall, L.P.; Lemma, F.; Crook, D.W.; Teale, C. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 2017, 72, 2745–2749. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Li, H.; Shen, Y.; Liu, Z.; Wang, S.; Shen, Z.; Zhang, R.; Walsh, T.R.; Shen, J.; Wang, Y. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. mBio 2017, 8, e00543-17. [Google Scholar] [CrossRef]
- Yang, Y.-Q.; Li, Y.-X.; Lei, C.-W.; Zhang, A.-Y.; Wang, H.-N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1791–1795. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzotti, G.; Magistrali, C.F. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017, 22, 30589. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio 2019, 10, e00853-19. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Zhou, Y.; Li, J.; Yin, W.; Wang, S.; Zhang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Kempf, M.; Bakour, S.; Flaudrops, C.; Berrazeg, M.; Brunel, J.M.; Drissi, M.; Mesli, E.; Touati, A.; Rolain, J.M. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS ONE 2012, 7, e31676. [Google Scholar] [CrossRef] [PubMed]
- Tse, H.; Yuen, K.-Y. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 2016, 16, 145–146. [Google Scholar] [CrossRef] [PubMed]
- Campos, J.; Cristino, L.; Peixe, L.; Antunes, P. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1, 4,[5], 12: I:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill. 2016, 21, 30270. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Treier, A.; Schmitt, K.; Stephan, R. Mobile fosfomycin resistance genes in Enterobacteriaceae—An increasing threat. Microbiologyopen 2020, 9, e1135. [Google Scholar] [CrossRef] [PubMed]
- Lima, T.; Domingues, S.; Da Silva, G.J. Plasmid-mediated colistin resistance in Salmonella enterica: A review. Microorganisms 2019, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Jeannot, K.; Bolard, A.; Plesiat, P. Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 2017, 49, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y. Transferability of MCR-1/2 polymyxin resistance: Complex dissemination and genetic mechanism. ACS Infect. Dis. 2018, 4, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Snesrud, E.; He, S.; Chandler, M.; Dekker, J.P.; Hickman, A.B.; McGann, P.; Dyda, F. A model for transposition of the colistin resistance gene mcr-1 by IS Apl1. Antimicrob. Agents Chemother. 2016, 60, 6973–6976. [Google Scholar] [CrossRef]
- Quiroga, C.; Nastro, M.; Di Conza, J. Current scenario of plasmid-mediated colistin resistance in Latin America. Rev. Argent. Microbiol. 2019, 51, 93–100. [Google Scholar] [CrossRef]
- Faccone, D.; Moredo, F.A.; Giacoboni, G.I.; Albornoz, E.; Alarcón, L.; Nievas, V.F.; Corso, A. Multidrug-resistant Escherichia coli harbouring mcr-1 and bla(CTX-M) genes isolated from swine in Argentina. J. Glob. Antimicrob. Resist. 2019, 18, 160–162. [Google Scholar] [CrossRef]
- Higashino, H.R.; Marchi, A.P.; Martins, R.C.R.; Batista, M.V.; Perdigão Neto, L.V.; Lima, V.A.C.d.C.; Rossi, F.; Guimarães, T.; Levin, A.S.; Rocha, V. Colistin-resistant Klebsiella pneumoniae co-harboring KPC and MCR-1 in a hematopoietic stem cell transplantation unit. Bone Marrow Transplant. 2019, 54, 1118–1120. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Shi, Y.L.; Peng, Y.; Xu, C.; Zhang, C.; Chen, Y.; Luo, X.Q.; Li, Q.M.; Zhao, C.L.; Lei, J.; et al. BL02, a phage against carbapenem- and polymyxin-B resistant Klebsiella pneumoniae, isolated from sewage: A preclinical study. Virus Res. 2023, 331, 199126. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.S.; Elshafiee, E.A.; Khalefa, H.S.; Kadry, M.; Hamza, D.A. Evidence of colistin resistance genes (mcr-1 and mcr-2) in wild birds and its public health implication in Egypt. Antimicrob. Resist. Infect. Control 2019, 8, 197. [Google Scholar] [CrossRef] [PubMed]
- Nang, S.C.; Li, J.; Velkov, T. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 2019, 45, 131–161. [Google Scholar] [CrossRef] [PubMed]
- Volland, H.; Dortet, L.; Bernabeu, S.; Boutal, H.; Haenni, M.; Madec, J.-Y.; Robin, F.; Beyrouthy, R.; Naas, T.; Simon, S. Development and multicentric validation of a lateral flow immunoassay for rapid detection of MCR-1-producing Enterobacteriaceae. J. Clin. Microbiol. 2019, 57, e01454-18. [Google Scholar] [CrossRef]
- Elbediwi, M.; Li, Y.; Paudyal, N.; Pan, H.; Li, X.; Xie, S.; Rajkovic, A.; Feng, Y.; Fang, W.; Rankin, S.C. Global burden of colistin-resistant bacteria: Mobilized colistin resistance genes study (1980–2018). Microorganisms 2019, 7, 461. [Google Scholar] [CrossRef] [PubMed]
- Calero-Cáceres, W.; Balcázar, J.L. Evolution and dissemination of mobile colistin resistance genes: Limitations and challenges in Latin American countries. Lancet Microbe 2023, 4, e567–e568. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Poirel, L.; Kieffer, N.; Châtre, P.; Saras, E.; Métayer, V.; Dumoulin, R.; Nordmann, P.; Madec, J.-Y. Co-occurrence of extended spectrum β lactamase and MCR-1 encoding genes on plasmids. Lancet Infect. Dis. 2016, 16, 281–282. [Google Scholar] [CrossRef] [PubMed]
- Juhász, E.; Iván, M.; Pintér, E.; Pongrácz, J.; Kristóf, K. Colistin resistance among blood culture isolates at a tertiary care centre in Hungary. J. Glob. Antimicrob. Resist. 2017, 11, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Yin, W.; Shen, Z.; Wang, Y.; Shen, J.; Walsh, T.R. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 2020, 75, 3087–3095. [Google Scholar] [CrossRef]
- Park, J.Y.; Heo, S.T.; Kwon, K.T.; Lee, K.J.; Choi, J.A. MCR1 and KPC2 co-producing Klebsiella pneumoniae bacteremia: First case in Korea. Infect. Chemother. 2019, 51, 399–404. [Google Scholar] [CrossRef] [PubMed]
- McGann, P.; Snesrud, E.; Maybank, R.; Corey, B.; Ong, A.C.; Clifford, R.; Hinkle, M.; Whitman, T.; Lesho, E.; Schaecher, K.E. Escherichia coli harboring mcr-1 and bla CTX-M on a novel IncF plasmid: First report of mcr-1 in the United States. Antimicrob. Agents Chemother. 2016, 60, 4420–4421. [Google Scholar] [CrossRef] [PubMed]
- Hasman, H.; Hammerum, A.M.; Hansen, F.; Hendriksen, R.S.; Olesen, B.; Agersø, Y.; Zankari, E.; Leekitcharoenphon, P.; Stegger, M.; Kaas, R.S. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill. 2015, 20, 30085. [Google Scholar] [CrossRef] [PubMed]
- Lei, T.; Zhang, J.; Jiang, F.; He, M.; Zeng, H.; Chen, M.; Wu, S.; Wang, J.; Ding, Y.; Wu, Q. First detection of the plasmid-mediated colistin resistance gene mcr-1 in virulent Vibrio parahaemolyticus. Int. J. Food Microbiol. 2019, 308, 108290. [Google Scholar] [CrossRef] [PubMed]
- Wise, M.G.; Estabrook, M.A.; Sahm, D.F.; Stone, G.G.; Kazmierczak, K.M. Prevalence of mcr-type genes among colistin-resistant Enterobacteriaceae collected in 2014–2016 as part of the INFORM global surveillance program. PLoS ONE 2018, 13, e0195281. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Graells, C.; De Keersmaecker, S.C.; Vanneste, K.; Pochet, B.; Vermeersch, K.; Roosens, N.; Dierick, K.; Botteldoorn, N. Detection of plasmid-mediated colistin resistance, mcr-1 and mcr-2 genes, in Salmonella spp. isolated from food at retail in Belgium from 2012 to 2015. Foodborne Pathog. Dis. 2018, 15, 114–117. [Google Scholar] [CrossRef]
- Dutta, A.; Barua, H.; Jalal, M.; Dhar, P.; Biswas, S.; Biswas, P. An investigation of plasmid-mediated colistin resistance mechanism, MCR in Escherichia coli of human, veterinary and environmental origin in Bangladesh. Int. J. Infect. Dis. 2018, 73, 54. [Google Scholar] [CrossRef]
- Mitra, S.; Basu, S.; Rath, S.; Sahu, S.K. Colistin resistance in Gram-negative ocular infections: Prevalence, clinical outcome and antibiotic susceptibility patterns. Int. Ophthalmol. 2020, 40, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, L.; Wang, J.; Yassin, A.K.; Butaye, P.; Kelly, P.; Gong, J.; Guo, W.; Li, J.; Li, M.; et al. Molecular detection of colistin resistance genes (mcr-1, mcr-2 and mcr-3) in nasal/oropharyngeal and anal/cloacal swabs from pigs and poultry. Sci. Rep. 2018, 8, 3705. [Google Scholar] [CrossRef] [PubMed]
- Al-Kadmy, I.M.; Ibrahim, S.A.; Al-Saryi, N.; Aziz, S.N.; Besinis, A.; Hetta, H.F. Prevalence of genes involved in colistin resistance in Acinetobacter baumannii: First report from Iraq. Microb. Drug Resist. 2020, 26, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Kawahara, R.; Harada, K.; Teruya, S.; Nakayama, T.; Motooka, D.; Nakamura, S.; Nguyen, P.D.; Kumeda, Y.; Van Dang, C. The presence of colistin resistance gene mcr-1 and-3 in ESBL producing Escherichia coli isolated from food in Ho Chi Minh City, Vietnam. FEMS Microbiol. Lett. 2018, 365, fny100. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, P.; Yang, X.; Wang, Z.; Fanning, S.; Wang, J.; Du, P.; Bai, L. Identification of a novel hybrid plasmid coproducing MCR-1 and MCR-3 variant from an Escherichia coli strain. J. Antimicrob. Chemother. 2019, 74, 1517–1520. [Google Scholar] [CrossRef]
- Xiang, R.; Liu, B.H.; Zhang, A.Y.; Lei, C.W.; Ye, X.L.; Yang, Y.X.; Chen, Y.P.; Wang, H.N. Colocation of the Polymyxin Resistance Gene mcr-1 and a Variant of mcr-3 on a Plasmid in an Escherichia coli Isolate from a Chicken Farm. Antimicrob. Agents Chemother. 2018, 62, e00501-18. [Google Scholar] [CrossRef]
- Wang, X.; Zhai, W.; Wang, S.; Shen, Z.; Wang, Y.; Zhang, Q. A novel transposon, Tn 6518, mediated transfer of mcr-3 variant in ESBL-producing Aeromonas veronii. Infect. Drug Resist. 2020, 13, 893–899. [Google Scholar] [CrossRef]
- Sun, R.-Y.; Ke, B.-X.; Fang, L.-X.; Guo, W.-Y.; Li, X.-P.; Yu, Y.; Zheng, S.-L.; Jiang, Y.-W.; He, D.-M.; Sun, J. Global clonal spread of mcr-3-carrying MDR ST34 Salmonella enterica serotype Typhimurium and monophasic 1, 4,[5], 12: I:- variants from clinical isolates. J. Antimicrob. Chemother. 2020, 75, 1756–1765. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, L.; Vidal, A.; Seminati, C.; Tello, M.; Redondo, N.; Darwich, L.; Martín, M. Antimicrobial resistance profile and prevalence of extended-spectrum beta-lactamases (ESBL), AmpC beta-lactamases and colistin resistance (mcr) genes in Escherichia coli from swine between 1999 and 2018. Porc. Health Manag. 2020, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, L.; Wang, J.; Butaye, P.; Huang, K.; Qiu, H.; Zhang, X.; Gong, W.; Wang, C. Molecular detection of colistin resistance genes (mcr-1 to mcr-5) in human vaginal swabs. BMC Res. Notes 2018, 11, 1–4. [Google Scholar] [CrossRef]
- García, V.; García-Meniño, I.; Mora, A.; Flament-Simon, S.C.; Díaz-Jiménez, D.; Blanco, J.E.; Alonso, M.P.; Blanco, J. Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 Enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006–2017). Int. J. Antimicrob. Agents 2018, 52, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Avila, L.U.; Loyola-Cruz, M.A.; Hernández-Cortez, C.; Bello-López, J.M.; Castro-Escarpulli, G. Colistin Resistance in Aeromonas spp. Int. J. Mol. Sci. 2021, 22, 5974. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Sun, C.; Hulth, A.; Li, J.; Nilsson, L.E.; Zhou, Y.; Börjesson, S.; Bi, Z.; Bi, Z.; Sun, Q.; et al. Mobile colistin resistance gene mcr-5 in porcine Aeromonas hydrophila. J. Antimicrob. Chemother. 2018, 73, 1777–1780. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, J.; Wang, J.; Butaye, P.; Kelly, P.; Li, M.; Yang, F.; Gong, J.; Yassin, A.K.; Guo, W. Newly identified colistin resistance genes, mcr-4 and mcr-5, from upper and lower alimentary tract of pigs and poultry in China. PLoS ONE 2018, 13, e0193957. [Google Scholar] [CrossRef]
- Nesporova, K.; Jamborova, I.; Valcek, A.; Medvecky, M.; Literak, I.; Dolejska, M. Various conjugative plasmids carrying the mcr-5 gene in Escherichia coli isolates from healthy chickens in Paraguay. J. Antimicrob. Chemother. 2019, 74, 3394–3397. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, L.D.R.; Furlan, J.P.R.; Ramos, M.S.; Gallo, I.F.L.; de Freitas, L.V.P.; Stehling, E.G. Co-occurrence of mcr-1, mcr-3, mcr-7 and clinically relevant antimicrobial resistance genes in environmental and fecal samples. Arch. Microbiol. 2020, 202, 1795–1800. [Google Scholar] [CrossRef] [PubMed]
- Kneis, D.; Berendonk, T.U.; Heß, S. High prevalence of colistin resistance genes in German municipal wastewater. Sci. Total Environ. 2019, 694, 133454. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Feng, Y.; Liu, L.; Yao, Z.; Zong, Z. A cluster of colistin-and carbapenem-resistant Klebsiella pneumoniae carrying bla NDM-1 and mcr-8.2. J. Infect. Dis. 2020, 221 (Suppl. S2), S237–S242. [Google Scholar] [CrossRef]
- Farzana, R.; Jones, L.S.; Barratt, A.; Rahman, M.A.; Sands, K.; Portal, E.; Boostrom, I.; Espina, L.; Pervin, M.; Uddin, A.N. Emergence of mobile colistin resistance (mcr-8) in a highly successful Klebsiella pneumoniae sequence type 15 clone from clinical infections in Bangladesh. mSphere 2020, 5, e00023-20. [Google Scholar] [CrossRef]
- Hadjadj, L.; Baron, S.A.; Olaitan, A.O.; Morand, S.; Rolain, J.-M. Co-occurrence of variants of mcr-3 and mcr-8 genes in a Klebsiella pneumoniae isolate from Laos. Front. Microbiol. 2019, 10, 2720. [Google Scholar] [CrossRef]
- Li, Y.; Dai, X.; Zeng, J.; Gao, Y.; Zhang, Z.; Zhang, L. Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci. Rep. 2020, 10, 8113. [Google Scholar] [CrossRef]
- Lin, M.; Yang, Y.; Yang, Y.; Chen, G.; He, R.; Wu, Y.; Zhong, L.-L.; El-Sayed Ahmed, M.A.E.-G.; Feng, S.; Shen, C. Co-occurrence of mcr-9 and bla NDM-1 in Enterobacter cloacae isolated from a patient with bloodstream infection. Infect. Drug Resist. 2020, 13, 1397–1402. [Google Scholar] [CrossRef]
- Xu, L.; Wan, F.; Fu, H.; Tang, B.; Ruan, Z.; Xiao, Y.; Luo, Q. Emergence of Colistin Resistance Gene mcr-10 in Enterobacterales Isolates Recovered from Fecal Samples of Chickens, Slaughterhouse Workers, and a Nearby Resident. Microbiol. Spectr. 2022, 10, e0041822. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.; Lao, G.; Zhou, Z.; Liu, Z.; Cai, J.; Sun, Q. Identification of Mobile Colistin Resistance Gene mcr-10 in Disinfectant and Antibiotic Resistant Escherichia coli from Disinfected Tableware. Antibiotics 2022, 11, 883. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Cui, Y.; Quan, J.; Zhao, D.; Han, X.; Shi, Q.; Wang, Q.; Jiang, Y.; Du, X.; Li, X. High prevalence of colistin resistance and mcr-9/10 genes in Enterobacter spp. in a tertiary hospital over a decade. Int. J. Antimicrob. Agents 2022, 59, 106573. [Google Scholar] [CrossRef]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, N.; He, Z.; Dai, X.; Zhao, F.; Li, Y.; Xiong, W.; Zeng, Z. Bavachin Rejuvenates Sensitivity of Colistin against Colistin-Resistant Gram-Negative Bacteria. Int. J. Mol. Sci. 2024, 25, 2349. [Google Scholar] [CrossRef] [PubMed]
- Gashaw, M.; Gudina, E.K.; Ali, S.; Gabriele, L.; Seeholzer, T.; Alemu, B.; Froeschl, G.; Kroidl, A.; Wieser, A. Molecular characterization of carbapenem-resistance in Gram-negative isolates obtained from clinical samples at Jimma Medical Center, Ethiopia. Front. Microbiol. 2024, 15, 1336387. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Jayol, A.; Poirel, L. A universal culture medium for screening polymyxin-resistant Gram-negative isolates. J. Clin. Microbiol. 2016, 54, 1395–1399. [Google Scholar] [CrossRef]
- Nordmann, P.; Jayol, A.; Poirel, L. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg. Infect. Dis. 2016, 22, 1038. [Google Scholar] [CrossRef]
- Abdul Momin, M.H.F.; Bean, D.C.; Hendriksen, R.S.; Haenni, M.; Phee, L.M.; Wareham, D.W. CHROMagar COL-APSE: A selective bacterial culture medium for the isolation and differentiation of colistin-resistant Gram-negative pathogens. J. Med. Microbiol. 2017, 66, 1554–1561. [Google Scholar] [CrossRef] [PubMed]
- Bardet, L.; Le Page, S.; Leangapichart, T.; Rolain, J.-M. LBJMR medium: A new polyvalent culture medium for isolating and selecting vancomycin and colistin-resistant bacteria. BMC Microbiol. 2017, 17, 220. [Google Scholar] [CrossRef] [PubMed]
- Jayol, A.; Nordmann, P.; Brink, A.; Villegas, M.V.; Dubois, V.; Poirel, L. High-Level Resistance to Colistin Mediated by Various Mutations in the crrB Gene among Carbapenemase-Producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2017, 61, e01423-17. [Google Scholar] [CrossRef]
- Jayol, A.; Nordmann, P.; Lehours, P.; Poirel, L.; Dubois, V. Comparison of methods for detection of plasmid-mediated and chromosomally encoded colistin resistance in Enterobacteriaceae. Clin. Microbiol. Infect. 2018, 24, 175–179. [Google Scholar] [CrossRef]
- Bardet, L.; Rolain, J.-M. Development of new tools to detect colistin-resistance among Enterobacteriaceae strains. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 3095249. [Google Scholar] [CrossRef] [PubMed]
- Osei Sekyere, J.; Govinden, U.; Essack, S. Review of established and innovative detection methods for carbapenemase-producing Gram-negative bacteria. J. Appl. Microbiol. 2015, 119, 1219–1233. [Google Scholar] [CrossRef] [PubMed]
- Hrabák, J.; Studentová, V.; Walková, R.; Zemlicková, H.; Jakubu, V.; Chudácková, E.; Gniadkowski, M.; Pfeifer, Y.; Perry, J.D.; Wilkinson, K.; et al. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2012, 50, 2441–2443. [Google Scholar] [CrossRef] [PubMed]
- Burckhardt, I.; Zimmermann, S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 2011, 49, 3321–3324. [Google Scholar] [CrossRef] [PubMed]
- Hooff, G.P.; van Kampen, J.J.; Meesters, R.J.; van Belkum, A.; Goessens, W.H.; Luider, T.M. Characterization of β-lactamase enzyme activity in bacterial lysates using MALDI-mass spectrometry. J. Proteome Res. 2012, 11, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Hoyos-Mallecot, Y.; Riazzo, C.; Miranda-Casas, C.; Rojo-Martín, M.; Gutiérrez-Fernández, J.; Navarro-Marí, J. Rapid detection and identification of strains carrying carbapenemases directly from positive blood cultures using MALDI-TOF MS. J. Microbiol. Methods 2014, 105, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Lartigue, M.F. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for bacterial strain characterization. Infect. Genet. Evol. 2013, 13, 230–235. [Google Scholar] [CrossRef]
- Lee, W.; Chung, H.-S.; Lee, Y.; Yong, D.; Jeong, S.H.; Lee, K.; Chong, Y. Comparison of matrix-assisted laser desorption ionization–time-of-flight mass spectrometry assay with conventional methods for detection of IMP-6, VIM-2, NDM-1, SIM-1, KPC-1, OXA-23, and OXA-51 carbapenemase-producing Acinetobacter spp. Pseudomonas aeruginosa, and Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 2013, 77, 227–230. [Google Scholar] [CrossRef]
- Osei Sekyere, J.; Govinden, U.; Bester, L.; Essack, S. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: Emerging resistance mechanisms and detection methods. J. Appl. Microbiol. 2016, 121, 601–617. [Google Scholar] [CrossRef] [PubMed]
- Dortet, L.; Bonnin, R.A.; Pennisi, I.; Gauthier, L.; Jousset, A.B.; Dabos, L.; Furniss, R.C.D.; Mavridou, D.A.; Bogaerts, P.; Glupczynski, Y. Rapid detection and discrimination of chromosome-and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: The MALDIxin test. J. Antimicrob. Chemother. 2018, 73, 3359–3367. [Google Scholar] [CrossRef] [PubMed]
- Osei Sekyere, J. Mcr colistin resistance gene: A systematic review of current diagnostics and detection methods. Microbiologyopen 2019, 8, e00682. [Google Scholar] [CrossRef] [PubMed]
- Caniaux, I.; Van Belkum, A.; Zambardi, G.; Poirel, L.; Gros, M. MCR: Modern colistin resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Guo, J.; Cheng, Q.; Yang, Z.; Chan, E.W.C.; Chen, S.; Hao, Q. Crystal Structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for Colistin Resistance. Sci. Rep. 2016, 6, 38793. [Google Scholar] [CrossRef] [PubMed]
- Hinchliffe, P.; Yang, Q.E.; Portal, E.; Young, T.; Li, H.; Tooke, C.L.; Carvalho, M.J.; Paterson, N.G.; Brem, J.; Niumsup, P.R. Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci. Rep. 2017, 7, 39392. [Google Scholar] [CrossRef] [PubMed]
- Coppi, M.; Cannatelli, A.; Antonelli, A.; Baccani, I.; Di Pilato, V.; Sennati, S.; Giani, T.; Rossolini, G.M. A simple phenotypic method for screening of MCR-1-mediated colistin resistance. Clin. Microbiol. Infect. 2018, 24, 201.e1–201.e3. [Google Scholar] [CrossRef] [PubMed]
- Esposito, F.; Fernandes, M.R.; Lopes, R.; Muñoz, M.; Sabino, C.P.; Cunha, M.P.; Silva, K.C.; Cayô, R.; Martins, W.M.; Moreno, A.M. Detection of colistin-resistant MCR-1-positive Escherichia coli by use of assays based on inhibition by EDTA and zeta potential. J. Clin. Microbiol. 2017, 55, 3454–3465. [Google Scholar] [CrossRef] [PubMed]
- Pasteran, F.; Danze, D.; Menocal, A.; Cabrera, C.; Castillo, I.; Albornoz, E.; Lucero, C.; Rapoport, M.; Ceriana, P.; Corso, A. Simple phenotypic tests to improve accuracy in screening chromosomal and plasmid-mediated colistin resistance in Gram-negative bacilli. J. Clin. Microbiol. 2020, 59, e01701-20. [Google Scholar] [CrossRef]
- Perez, L.R.R.; Carniel, E.; Narvaez, G.A.; Dias, C.G. Evaluation of a polymyxin drop test for polymyxin resistance detection among non-fermentative gram-negative rods and enterobacterales resistant to carbapenems. APMIS 2021, 129, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Nijhuis, R.; Veldman, K.; Schelfaut, J.; Van Essen-Zandbergen, A.; Wessels, E.; Claas, E.; Gooskens, J. Detection of the plasmid-mediated colistin-resistance gene mcr-1 in clinical isolates and stool specimens obtained from hospitalized patients using a newly developed real-time PCR assay. J. Antimicrob. Chemother. 2016, 71, 2344–2346. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Feng, Y.; Xu, W.; Feng, J.; Yan, C.; Fu, T.; Zhao, H.; Cui, J.; Gan, L.; Liu, S.; et al. Rapid Detection of Multi-Resistance Strains Carrying mcr-1 Gene Using Recombinase-Aided Amplification Directly on Clinical Samples. Front. Microbiol. 2022, 13, 852488. [Google Scholar] [CrossRef] [PubMed]
- Chabou, S.; Leangapichart, T.; Okdah, L.; Le Page, S.; Hadjadj, L.; Rolain, J.-M. Real-time quantitative PCR assay with Taqman® probe for rapid detection of MCR-1 plasmid-mediated colistin resistance. New Microbes New Infect. 2016, 13, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Bontron, S.; Poirel, L.; Nordmann, P. Real-time PCR for detection of plasmid-mediated polymyxin resistance (mcr-1) from cultured bacteria and stools. J. Antimicrob. Chemother. 2016, 71, 2318–2320. [Google Scholar] [CrossRef] [PubMed]
- Donà, V.; Bernasconi, O.J.; Kasraian, S.; Tinguely, R.; Endimiani, A. A SYBR® Green-based real-time PCR method for improved detection of mcr-1-mediated colistin resistance in human stool samples. J. Glob. Antimicrob. Resist. 2017, 9, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, G.; Davis, K.E.; Poljak, Z.; Friendship, R.; Mulvey, M.R.; Deckert, A.E.; Reid-Smith, R.J.; Boerlin, P. A method to detect Escherichia coli carrying the colistin-resistance genes mcr-1 and mcr-2 using a single real-time polymerase chain reaction and its application to chicken cecal and porcine fecal samples. Can. J. Vet. Res. 2018, 82, 312–315. [Google Scholar] [PubMed]
- Li, J.; Shi, X.; Wang, Y.; Wang, S. A multiplex SYBR green real-time PCR assay for the detection of three colistin resistance genes from cultured bacteria, feces, and environment samples. Front. Microbiol. 2017, 8, 302964. [Google Scholar] [CrossRef] [PubMed]
- Tolosi, R.; Apostolakos, I.; Laconi, A.; Carraro, L.; Grilli, G.; Cagnardi, P.; Piccirillo, A. Rapid detection and quantification of plasmid-mediated colistin resistance genes (mcr-1 to mcr-5) by real-time PCR in bacterial and environmental samples. J. Appl. Microbiol. 2020, 129, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018, 23, 17-00672. [Google Scholar] [CrossRef] [PubMed]
- Lescat, M.; Poirel, L.; Nordmann, P. Rapid multiplex polymerase chain reaction for detection of mcr-1 to mcr-5 genes. Diagn. Microbiol. Infect. Dis. 2018, 92, 267–269. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Zou, D.; Huang, S.; Lei, H.; Yang, Z.; Su, Y.; He, X.; Zhao, Q.; Wang, Y.; Liu, W.; Huang, L. Sensitive and Rapid Detection of the Plasmid-Encoded Colistin-Resistance Gene mcr-1 in Enterobacteriaceae Isolates by Loop-Mediated Isothermal Amplification. Front. Microbiol. 2017, 8, 2356. [Google Scholar] [CrossRef] [PubMed]
- Imirzalioglu, C.; Falgenhauer, L.; Schmiedel, J.; Waezsada, S.E.; Gwozdzinski, K.; Roschanski, N.; Roesler, U.; Kreienbrock, L.; Schiffmann, A.P.; Irrgang, A.; et al. Evaluation of a Loop-Mediated Isothermal Amplification-Based Assay for the Rapid Detection of Plasmid-Encoded Colistin Resistance Gene mcr-1 in Enterobacteriaceae Isolates. Antimicrob. Agents Chemother. 2017, 61, e02326-16. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.L.; Zhou, Q.; Tan, C.Y.; Roberts, A.P.; El-Sayed Ahmed, M.A.E.; Chen, G.; Dai, M.; Yang, F.; Xia, Y.; Liao, K.; et al. Multiplex loop-mediated isothermal amplification (multi-LAMP) assay for rapid detection of mcr-1 to mcr-5 in colistin-resistant bacteria. Infect. Drug Resist. 2019, 12, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi, O.J.; Principe, L.; Tinguely, R.; Karczmarek, A.; Perreten, V.; Luzzaro, F.; Endimiani, A. Evaluation of a new commercial microarray platform for the simultaneous detection of β-lactamase and mcr-1 and mcr-2 genes in Enterobacteriaceae. J. Clin. Microbiol. 2017, 55, 3138–3141. [Google Scholar] [CrossRef] [PubMed]
- Genovese, C.; La Fauci, V.; D’Amato, S.; Squeri, A.; Anzalone, C.; Costa, G.B.; Fedele, F.; Squeri, R. Molecular epidemiology of antimicrobial resistant microorganisms in the 21th century: A review of the literature. Acta Bio Medica Atenei Parmensis 2020, 91, 256. [Google Scholar]
- Kumar, S.; Anwer, R.; Azzi, A. Molecular typing methods & resistance mechanisms of MDR Klebsiella pneumoniae. AIMS Microbiol. 2023, 9, 112–130. [Google Scholar]
- Liu, S.; Wang, X.; Ge, J.; Wu, X.; Zhao, Q.; Li, Y.M.; Wang, R. Analysis of carbapenemase-resistant genotypes of highly virulent Klebsiella pneumoniae and clinical infection characteristics of different MLST types. Evid.-Based Complement. Altern. Med. eCAM 2021, 2021, 9838103. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Patil, P.P.; Singhal, L.; Ray, P.; Patil, P.B.; Gautam, V. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates reveals the emergence of bla(OXA-23) and bla(NDM-1) encoding international clones in India. Infect. Genet. Evol. 2019, 75, 103986. [Google Scholar] [CrossRef]
- Chen, C.-m.; Wang, M.; Li, X.-p.; Li, P.-l.; Tian, J.-j.; Zhang, K.; Luo, C. Homology analysis between clinically isolated extraintestinal and enteral Klebsiella pneumoniae among neonates. BMC Microbiol. 2021, 21, 25. [Google Scholar]
- Wang, C.-H.; Ma, L.; Huang, L.-Y.; Yeh, K.-M.; Lin, J.-C.; Siu, L.K.; Chang, F.-Y. Molecular epidemiology and resistance patterns of blaOXA-48 Klebsiella pneumoniae and Escherichia coli: A nationwide multicenter study in Taiwan. J. Microbiol. Immunol. Infect. 2021, 54, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-Z.; Li, X.-P.; Zhang, Y.-J.; Zhong, W.-C.; Liu, Y.-H.; Liao, X.-P.; Sun, J.; Zhou, Y.-F. Molecular characteristic of mcr-1 gene in Escherichia coli from aquatic products in Guangdong, China. J. Glob. Antimicrob. Resist. 2024, 36, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, L.; Wang, X.; Li, J.; Tang, B. Evaluation of IR Biotyper for Lactiplantibacillus plantarum Typing and Its Application Potential in Probiotic Preliminary Screening. Front. Microbiol. 2022, 13, 823120. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhou, H.; Lu, J.; Sun, Q.; Liu, C.; Zeng, Y.; Zhang, R. Evaluation of the IR Biotyper for Klebsiella pneumoniae typing and its potentials in hospital hygiene management. Microb. Biotechnol. 2021, 14, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Tobin, L.A.; Jarocki, V.M.; Kenyon, J.; Drigo, B.; Donner, E.; Djordjevic, S.P.; Hamidian, M. Genomic analysis of diverse environmental Acinetobacter isolates identifies plasmids, antibiotic resistance genes, and capsular polysaccharides shared with clinical strains. Appl. Environ. Microbiol. 2024, 90, e0165423. [Google Scholar] [CrossRef] [PubMed]
- Barathan, M.; Ng, S.-L.; Lokanathan, Y.; Ng, M.H.; Law, J.X. Unseen Weapons: Bacterial Extracellular Vesicles and the Spread of Antibiotic Resistance in Aquatic Environments. Int. J. Mol. Sci. 2024, 25, 3080. [Google Scholar] [CrossRef] [PubMed]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef] [PubMed]
- Sambaza, S.S.; Naicker, N. Contribution of wastewater to antimicrobial resistance: A review article. J. Glob. Antimicrob. Resist. 2023, 34, 23–29. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Liguori, K.; Keenum, I.; Davis, B.C.; Calarco, J.; Milligan, E.; Harwood, V.J.; Pruden, A. Antimicrobial resistance monitoring of water environments: A framework for standardized methods and quality control. Environ. Sci. Technol. 2022, 56, 9149–9160. [Google Scholar] [CrossRef]
- Yuan, T.; Pian, Y. Hospital wastewater as hotspots for pathogenic microorganisms spread into aquatic environment: A review. Front. Environ. Sci. 2023, 10, 1091734. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018, 42, fux053. [Google Scholar] [CrossRef] [PubMed]
- Ehalt Macedo, H.; Lehner, B.; Nicell, J.; Grill, G.; Li, J.; Limtong, A.; Shakya, R. Distribution and characteristics of wastewater treatment plants within the global river network. Earth Syst. Sci. Data 2022, 14, 559–577. [Google Scholar] [CrossRef]
- Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 2021, 19, e06651. [Google Scholar] [PubMed]
- Velazquez-Meza, M.E.; Galarde-López, M.; Carrillo-Quiróz, B.; Alpuche-Aranda, C.M. Antimicrobial resistance: One Health approach. Vet. World 2022, 15, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Cella, E.; Giovanetti, M.; Benedetti, F.; Scarpa, F.; Johnston, C.; Borsetti, A.; Ceccarelli, G.; Azarian, T.; Zella, D.; Ciccozzi, M. Joining Forces against Antibiotic Resistance: The One Health Solution. Pathogens 2023, 12, 1074. [Google Scholar] [CrossRef]
- Ahmad, N.; Joji, R.M.; Shahid, M. Evolution and implementation of One Health to control the dissemination of antibiotic-resistant bacteria and resistance genes: A review. Front. Cell. Infect. Microbiol. 2023, 12, 1065796. [Google Scholar] [CrossRef] [PubMed]
- Adebisi, Y.A. Balancing the risks and benefits of antibiotic use in a globalized world: The ethics of antimicrobial resistance. Glob. Health 2023, 19, 27. [Google Scholar] [CrossRef] [PubMed]
- Almutairi, M.M. Synergistic activities of colistin combined with other antimicrobial agents against colistin-resistant Acinetobacter baumannii clinical isolates. PLoS ONE 2022, 17, e0270908. [Google Scholar] [CrossRef] [PubMed]
- Lertsrisatit, Y.; Santimaleeworagun, W.; Thunyaharn, S.; Traipattanakul, J. In vitro activity of colistin mono-and combination therapy against colistin-resistant Acinetobacter baumannii, mechanism of resistance, and clinical outcomes of patients infected with colistin-resistant A. baumannii at a Thai university hospital. Infect. Drug Resist. 2017, 10, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Brennan-Krohn, T.; Pironti, A.; Kirby, J.E. Synergistic Activity of Colistin-Containing Combinations against Colistin-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2018, 62, e00873-18. [Google Scholar] [CrossRef]
- Srisakul, S.; Wannigama, D.L.; Higgins, P.G.; Hurst, C.; Abe, S.; Hongsing, P.; Saethang, T.; Luk-In, S.; Liao, T.; Kueakulpattana, N.; et al. Overcoming addition of phosphoethanolamine to lipid A mediated colistin resistance in Acinetobacter baumannii clinical isolates with colistin-sulbactam combination therapy. Sci. Rep. 2022, 12, 11390. [Google Scholar] [CrossRef] [PubMed]
- MacNair, C.R.; Stokes, J.M.; Carfrae, L.A.; Fiebig-Comyn, A.A.; Coombes, B.K.; Mulvey, M.R.; Brown, E.D. Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 2018, 9, 458. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mondal, A.H.; Khare, K.; Saxena, P.; Debnath, P.; Mukhopadhyay, K.; Yadav, D. A Review on Colistin Resistance: An Antibiotic of Last Resort. Microorganisms 2024, 12, 772. https://doi.org/10.3390/microorganisms12040772
Mondal AH, Khare K, Saxena P, Debnath P, Mukhopadhyay K, Yadav D. A Review on Colistin Resistance: An Antibiotic of Last Resort. Microorganisms. 2024; 12(4):772. https://doi.org/10.3390/microorganisms12040772
Chicago/Turabian StyleMondal, Aftab Hossain, Kriti Khare, Prachika Saxena, Parbati Debnath, Kasturi Mukhopadhyay, and Dhananjay Yadav. 2024. "A Review on Colistin Resistance: An Antibiotic of Last Resort" Microorganisms 12, no. 4: 772. https://doi.org/10.3390/microorganisms12040772
APA StyleMondal, A. H., Khare, K., Saxena, P., Debnath, P., Mukhopadhyay, K., & Yadav, D. (2024). A Review on Colistin Resistance: An Antibiotic of Last Resort. Microorganisms, 12(4), 772. https://doi.org/10.3390/microorganisms12040772






