Abstract
Antimicrobial-resistant Klebsiella pneumoniae is one of the predominant pathogens in healthcare settings. However, the prevalence and resistome of this organism within residential aged care facilities (RACFs), which are potential hotspots for antimicrobial resistance, remain unexplored. Here, we provide a phenotypic and molecular characterization of antimicrobial-resistant K. pneumoniae isolated from RACFs. K. pneumoniae was isolated from urine, faecal and wastewater samples and facility swabs. The antimicrobial susceptibility profiles of all the isolates were determined and the genomic basis for resistance was explored with whole-genome sequencing on a subset of isolates. A total of 147 K. pneumoniae were isolated, displaying resistance against multiple antimicrobials. Genotypic analysis revealed the presence of beta-lactamases and the ciprofloxacin-resistance determinant QnrB4 but failed to confirm the basis for the observed cephalosporin resistance. Clonal spread of the multidrug-resistant, widely disseminated sequence types 323 and 661 was observed. This study was the first to examine the resistome of K. pneumoniae isolates from RACFs and demonstrated a complexity between genotypic and phenotypic antimicrobial resistance. The intra-facility dissemination and persistence of multidrug-resistant clones is concerning, given that residents are particularly vulnerable to antimicrobial resistant infections, and it highlights the need for continued surveillance and interventions to reduce the risk of outbreaks.
1. Introduction
Antimicrobial resistance (AMR) is an accelerating global emergency [1]. AMR is annually associated with 4.95 million deaths, and directly causes 1.27 million deaths globally—which is estimated to be more than HIV/AIDS or malaria [2]. Without more effective intervention, AMR may become the single largest cause of global mortality by 2050 [3]. Species of the Klebsiella pneumoniae complex, which includes K. variicola [4], are amongst the most prevalent global AMR pathogens, responsible for 193,000 annual deaths while contributing to an additional 600,000 [2]. Within Australia alone, K. pneumoniae complex bloodstream infections have an associated 13.4% 30-day all-cause mortality [5]. K. pneumoniae complex isolates are intrinsically resistant to many antimicrobials, and they are increasingly resistant to the antimicrobials relied on as treatment options, such as cephalosporins, fluroquinolones and carbapenems [5]. Carbapenem-resistant and third-generation cephalosporin-resistant K. pneumoniae each currently cause between 50,000 and 100,000 deaths annually [2]. Additionally, fluroquinolone resistance, often plasmid-mediated, is increasingly identified in clinical isolates, further complicating potential treatment regimens [6,7].
The genome of K. pneumoniae is highly variable, with more than half comprising accessory genes [8,9,10]. This genomic plasticity facilitates Klebsiella spp. both acquiring and disseminating resistance genes, via plasmids and other mobile genetic elements, which are responsible for much of the observed resistance [8,9,10]. Both intrinsic and acquired resistance mechanisms within K. pneumoniae have been studied extensively, with the presence of resistance determinants correlating to a resistant phenotype [8,10,11]. However, emerging, and inexplicable antimicrobial resistance within Klebsiella spp. has been identified [12,13,14,15].
A known contributor in the development of AMR is the frequent use of antibiotics, which in residential aged care facilities (RACFs) is well documented [16,17]. Residents of RACFs are older, frequently immunocompromised and prone to bacterial infections necessitating antimicrobial treatment. There is well-documented inappropriate extended use of antimicrobial prophylaxis in this population [18]. As in many institutions, there is also a high use of biocidal cleaning agents [16,19,20]. Additionally, close living conditions and frequent transfers of residents to other healthcare settings could promote AMR development and dissemination [21,22,23]. Current surveillance characterizes RACFs as significant reservoirs of antimicrobial-resistant organisms [16,24,25,26,27], with rates of AMR similar to those in hospitals [5]. Reported outbreaks of drug-resistant K. pneumoniae from long-term care facilities indicate a potential presence of AMR K. pneumoniae within RACFs [28,29]. However, no study has yet assessed the prevalence nor the resistome and antimicrobial susceptibility profiles of the WHO priority pathogen K. pneumoniae complex isolates within this setting.
This study reports on the surveillance of K. pneumoniae complex collected from RACFs in Adelaide, Australia. The comprehensive sampling regime included residents, the facility and the wastewater generated by the facility. The phenotypic antimicrobial resistance profiles of isolates were characterized and compared with genotypic analyses of resistance determinants. This pilot surveillance study aimed to provide, to our knowledge, the first comprehensive report on the resistome of Klebsiella pneumoniae complex isolates from RACFs and on the prevalence of resistant K. pneumoniae in aged care facilities, and it contributes to a more comprehensive understanding of resistance determinants within this species.
2. Materials and Methods
2.1. Sampling
Faecal, urine, facility and wastewater samples were collected from within aged care facilities (‘Facility 1’, 170 beds; ‘Facility 2’, 70 beds; ‘Facility 3’, 58 beds) and a retirement cohort (‘Retirement’, 38 beds total) in Adelaide, Australia. Faecal and urine samples were collected by a registered nurse.
For the collection of facility environmental swabs, which was performed across all facilities, sterile cotton-tipped applicators moistened in sterile saline (0.9% w/v) were wiped over touchpoints, and bathroom sinks, including drains, within each residence. Swabs were transported to the laboratory on ice, re-suspended in 200 µL peptone water with 20% v/v glycerol as cryoprotectant and stored at −80 °C until processing.
Wastewater sampling was performed at 3-month intervals from October 2019 to February 2021 and was not performed at Facility 3 due to physical lack of access. In each instance, wastewater samples of ~200 mL were collected hourly, for ten hours, and pooled for analysis.
All samples were collected on-site and held at 4 °C during transit to the host laboratory immediately after collection for same-day processing.
2.2. Isolation and Identification of K. pneumoniae
Faecal, facility and wastewater samples were plated onto rehydrated Brilliance™ E. coli/coliform selective agar (dehydrated, CM1046B, Thermo Scientific™, Melbourne, Australia) supplemented with either 1 mg/L ceftazidime (Sigma, Melbourne, Australia), 0.1 mg/L ciprofloxacin (Sigma, Australia) or no supplementation, whilst urine samples were plated onto rehydrated Brilliance™ UTI agar (dehydrated, CM0949B, Thermo Scientific™, Australia). Non-selective rehydrated Columbia agar base (dehydrated, NCM0038A, Neogen Australia, Bundamba, Australia) was used throughout the study to assess bacterial growth. Plates containing wastewater and facility samples were incubated at 25 °C for 48 h, whilst faecal and urine plates were incubated at 37 °C for 24 h.
Following colony purification, morphologically distinct colonies were identified to species and subspecies by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) (Bruker Daltonik GmbH, Bremen, Germany). Klebsiella spp. were stored in cryoprotective enrichment media, tryptic soy broth (dehydrated, CM0129, Thermo Scientific™, Australia) + 20% v/v glycerol, at −80 °C.
2.3. Antimicrobial Susceptibility Testing
K. pneumoniae complex isolates were subjected to antimicrobial susceptibility assays using the broth microdilution method as outlined in the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [30] guidelines. ‘Resistance’ was defined as a minimum inhibitory concentration (MIC) above the EUCAST breakpoint value [30]. Given the increasingly published links between antimicrobial resistance and biocide tolerance [31,32], tolerance to the biocides benzalkonium chloride, chlorhexidine and triclosan was also assessed using the same method and was determined by comparison with reported ECOFF values for K. pneumoniae [33]. Escherichia coli ATCC 25922 was used as a quality control throughout.
2.4. Efflux Pump Inhibition Assays
Assays quantifying the synergism between antimicrobials and efflux pump inhibitors were performed in accordance with methods widely described within the literature [32,34,35,36]. Bacterial growth was assessed in the presence of antimicrobials in combination with the RND-family efflux pump inhibitor phenylalanine-arginine β-naphthylamide (PAβN), to investigate if active efflux was responsible for the observed antimicrobial resistance.
2.5. Genomic DNA Isolation and Whole-Genome Sequencing
For whole-genome sequencing (WGS), DNA was extracted using an MN NucleoSpin®Microbial DNA kit (Machery-Nagel GmbH and Co.KG, Duren, Germany), following the manufacturer’s instructions. The quantity (ng/μL) and purity (absorbance 260 nm:280 nm) of DNA was assessed using a Cytation5 imaging reader (BioTek instruments, Winoosi, VT, USA). Extracted genomic DNA was visualised on 1% w/v agarose gel via electrophoresis to ensure no shearing of extracts. WGS was performed at SA Pathology Adelaide, Australia. The Nextera XT DNA library preparation kit (Illumina Inc., San Diego, CA, USA) was used to prepare sequenced libraries, as per the manufacturer’s instructions. Sequencing was performed on the Illumina NextSeq 550 platform with the NextSq 500/550 Mid-Output kit v2.5 for 300 cycles (Illumina Inc.).
2.6. Bioinformatic and Statistical Analyses
Raw paired-end sequencing reads were assembled and annotated using the TORMES pipeline version 1.3 [37]. Following the removal of sequence adaptors by Trimmomatic version 0.4 [38], sequence quality assessment was performed by Prinseq version 0.20.4 [39] and Sickle version 1.33 [40]. Draft genomes were then assembled by SPAdes version 3.15.4 [41] and annotated by Prokka version 1.14.5 [42]. Draft genomes were screened for antimicrobial resistance genes using ResFinder version 2.1 [43], the Antibiotic Resistance Gene-ANNOTation (ARG-ANNOT) database [44] and the comprehensive antibiotic resistance database (CARD) [45].
Identification of single-nucleotide polymorphisms (SNPs) within whole genomes was performed using CSIphylogeny V1.4 [46], in August 2023, with parameters as follows: minimum depth at SNP position at 10×, minimum distance between SNPs at 10 bp and minimum SNP quality score of 30. Isolates were classified as clones based on previous research into K. pneumoniae clonality and sample collection information [47,48,49]. Plasmid presence and replicons were assessed using PlasmidFinder 2.1 [50].
Downstream visualisation and annotation of phylogenetic data, which were generated within the TORMES pipeline, was achieved through the use of the Interactive Tree of Life v.6 (iTOL) [51]. Bar graphs were generated using GraphPad Prism v.9 [52]. Manual probing of nucleotide and amino acid sequences of key genes and proteins was performed by sequence aligning tools Clustal omega [53] and ESPript v3.0 [54] to identify the presence of mutations.
2.7. Data Availability
The collective WGSs of isolates used in this study were deposited to the NCBI database BioProject ‘PRJNA949397′ (Table 1).

Table 1.
NCBI database accession numbers for isolates within this study.
2.8. Ethics Approval
Ethics approval to collect clinical samples, which included faeces and urine, from RACF residents was granted by the University of South Australia’s human research ethics committee (Application ID: 201882).
3. Results
3.1. A Total of 147 K. pneumoniae Complex Isolates Were Identified
Faecal, urine and facility swabs were collected from 123 residents across three RACFs and from a retirement living cohort in Adelaide, Australia, while the wastewater from the facilities was also sampled as representative of all the residents and to capture clonal dissemination. A total of 147 K. pneumoniae complex isolates were identified from these samples (Table 2). Isolates were predominantly recovered from faecal samples, with 56% (n = 82) of isolates from this source. The 82 faecal and 2 urine K. pneumoniae complex isolates are representative of 40 participants, as some participants provided multiple samples. The next largest source was wastewater, with 40% (n = 59) of isolates derived from wastewater samples. Facility swabs yielded n = 4 (2%) of isolates, and urine samples yielded n = 2 (1%) of isolates.

Table 2.
The distribution of the 147 K. pneumoniae complex isolates by facility and sample type.
At the species level, 134 (91%) of K. pneumoniae complex isolates were K. pneumoniae and 13 (9%) were K. variicola. The number of isolates recovered was proportionate to the number of beds in each facility, with Facility 1 having both the greatest number of beds (n = 170) and greatest number of isolates (n = 97, 66 %).
3.2. K. pneumoniae Complex Resistant to Ceftazidime, Ciprofloxacin and Trimethoprim-Sulfamethoxazole was Isolated
Since ESBL producing Enterobacterales have been highlighted as a problem in RACFs in Australia [55,56], the residents were screened for carriage of ESBL producing K. pneumoniae using Brilliance™ E. coli/coliform selective agar supplemented with 1 mg/L ceftazidime. K. pneumoniae were isolated from 7 of the 102 participants from the RACFs using this media, relating to a 7% incidence of colonization, which is similar to the national average and correlates with a previous study on the incidence of other Enterobacterales in RACFs [56]. However, when the antimicrobial susceptibility of all the isolates was determined, incidence of ceftazidime and cefepime resistance was much higher at 46% and 16%, respectively (Table 3), most likely due to the clonal spread of ceftazidime-resistant K. pneumoniae that were also isolated from wastewater and the facility. Most of the cephalosporin-resistant isolates displayed high-level ceftazidime resistance with MIC values of >64 mg/L, values well above the resistance breakpoint of 4 mg/L. Only 11% of the ceftazidime isolates were from agar plates supplemented with ceftazidime, indicating that the ceftazidime resistance observed was not merely due to selection bias during isolation.

Table 3.
MIC distribution (mg/L) for 147 K. pneumoniae complex isolates recovered from three RACFs and one retirement living cohort.
Similarly, a high incidence of resistance was observed against ciprofloxacin, with 69% (n = 102) of isolates being ciprofloxacin-resistant (MIC ≥ 0.5 mg/L), with only 15% of the ciprofloxacin-resistant isolates being isolated from plates containing ciprofloxacin. Further, high-level ciprofloxacin resistance (MIC ≥ 64 mg/L) was observed for 5% (n = 8) of isolates. The incidence of ciprofloxacin resistance was followed by trimethoprim-sulfamethoxazole resistance, which was observed in 47% (n = 69) of isolates.
Carbapenem antimicrobials, primarily meropenem, are currently treatments relied on for AMR K. pneumoniae [5]. No isolates were resistant to the carbapenem meropenem; however, 3% (n = 4) of isolates were non-wild type with MIC values 1-2-fold above EUCAST breakpoints. One of the isolates was resistant to the last resort antibiotic colistin with an MIC of 32 µg/mL.
Overall, aminoglycoside resistance was relatively low compared to the instance of resistance to other antibiotics tested, with tobramycin and amikacin resistance observed in 10% (n = 15) and 5% (n = 8) of isolates, respectively.
Given the link between biocide tolerance and antimicrobial resistance [31,32], and the risk of background low levels of biocides causing AMR [53,54], the biocide sensitivity of isolates was also investigated (Table 4). No EUCAST breakpoints and ECOFF data are available for biocides. Therefore, our data were compared to published ECOFF values [33] to infer tolerance to biocides (Table 4). Three commonly used biocides were assessed in this study, namely, triclosan, chlorhexidine digluconate and benzalkonium chloride, all of which have been linked to the development of AMR [57,58]. Almost no tolerance to chlorhexidine was observed, with only 1.4% (n = 2) of isolates displaying an MIC above the ECOFF threshold of 64 mg/L. This contrasts with the tolerance to triclosan that was observed for a relatively high proportion (n = 67, 46%) of isolates. Tolerance to benzalkonium chloride was observed for n = 119 (96%) of isolates.

Table 4.
MIC distribution (mg/L) for 147 K. pneumoniae complex isolates for three commonly used biocides.
3.3. Multidrug Resistance (MDR) Was Prevalent within RACFs, Especially within Facility 1
Each facility harboured some isolates which were sensitive to all classes of antimicrobials, but most isolates (n = 115, 78%) displayed resistance to one or more classes of antimicrobial tested and were thus AMR organisms (Figure 1A). Of the 40 residents who provided faecal and urine samples, n = 35 (87.5%) were found to be carrying an AMR organism.
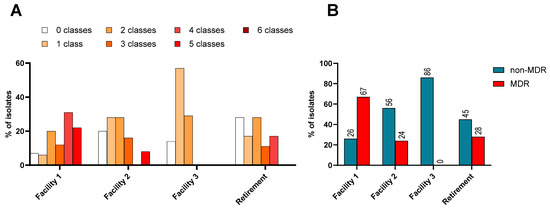
Figure 1.
(A): Percentage of isolates resistant to 0–6 different classes of antimicrobials, grouped by number of classes and differentiated by facility. (B): The distribution of non-MDR and MDR K. pneumoniae complex isolates from across the three RACFs and one retirement setting within this study.
The problem of K. pneumoniae complex is amplified when organisms display multidrug resistance (MDR), as treatment options are fewer. An MDR organism is defined as displaying resistance to antimicrobials in three or more different classes. Facility 1 had the highest proportion of MDR isolates at 72% (Figure 1B). This facility was also the only facility with isolates that were resistant to five classes of antibiotics tested and it had the lowest proportion of isolates without any resistance, which was 7% (Figure 1A).
3.4. Resistome Analysis of a Subset of K. pneumoniae Complex Could Not Identify the Genomic Basis for the Cephalosporin or Colistin Resistance
With antimicrobial susceptibility testing revealing a broad range of antimicrobial resistance phenotypes within the RACF K. pneumoniae complex isolates, whole-genome sequencing (WGS) was performed to elucidate the genetic bases of this resistance. A cohort of the 147 isolates were selected for sequencing, based on their resistance profiles (Table 5). Isolates displaying resistance to ceftazidime, cefepime and ciprofloxacin were targeted for WGS as relatively a high incidence of resistance was observed against these antimicrobials. Isolates both resistant and sensitive to the last-resort treatment option colistin were also included [59,60]. Additionally, an isolate sensitive to all the antibiotics tested (A031) was also included to allow for comparisons. All isolates selected were from Facility 1, the site with the highest proportions of MDR organisms observed.

Table 5.
EUCAST breakpoints, sequence types (STs) and minimum inhibitory concentrations (MICs) for antimicrobials used in this study and sample type information of sequenced K. pneumoniae complex isolates.
Resistome analysis revealed the presence of four beta-lactam-resistance determinants, blaDHA-1, blaSHV-1, blaSHV-27 and blaCTX-M-14, while no carbapenemases were identified (Figure 2). Isolate A031 was the only isolate carrying blaSHV-1, a cephalosporin-hydrolysing beta-lactamase which does not hydrolyse oxyimino cephalosporins such as ceftazidime [61]. Accordingly, isolate A031 was sensitive to ceftazidime. The other twelve sequenced isolates contained the AmpC beta-lactamase blaDHA-1 and a variant of blaSHV-1, blaSHV-27 and were resistant to ceftazidime. Previous studies have shown that both blaDHA-1 and blaSHV-27 can confer resistance to third-generation cephalosporins such as ceftazidime [61,62,63,64].
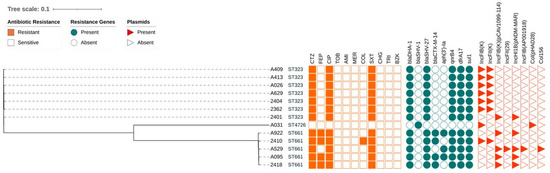
Figure 2.
Phylogenetic tree constructed from core-genome SNPs of K. pneumoniae complex isolates. Sequence types, antimicrobial susceptibility (orange squares), presence of resistance determinants (teal circles) and plasmid presence (red triangles) are annotated to the phylogenetic tree. Presence is indicated by a shaded field, absence indicated by an empty field. CTZ: ceftazidime; FEP: cefepime; CIP: ciprofloxacin; TOB: tobramycin; AMI: amikacin; MER: meropenem; COL: colistin; SXT: trimethoprim-sulfamethoxazole; CHG: chlorhexidine digluconate; TRI: triclosan; BZK: benzalkonium chloride.
Four isolates were resistant to the fourth-generation cephalosporin cefepime. The determinant for the cefepime resistance was not identified.
The blaSHV-27 gene is frequently associated with other ESBL genes in resistant organisms, such as the CTX-M-type [65,66], consistent with findings here. Four of the sequenced isolates with differing cephalosporin resistance phenotypes co-harboured both the blaSHV-27 gene and a blaCTX-M-14: isolates A922, 2410, A095 and 2418. The beta-lactamase blaCTX-M-14 is part of the CTX-M class A ESBLs, which also include CTX-M-15 [67]. Unlike CTX-M-15, which is highly active against cefotaxime and is shown to be increasing in its ability to hydrolyse ceftazidime [68,69], CTX-M-14 is not typically associated with ceftazidime or cefepime resistance [70]. Therefore, despite isolates A922, 2410, A095 and 2418 harbouring this gene and displaying a resistant phenotype, blaCTX-M-14 might not be the resistance determinant responsible for cefepime resistance. The results obtained and the resistance determinants identified in this study do not clearly explain the high levels of ceftazidime resistance detected in many of the K. pneumoniae isolates assessed here. In addition, none of these beta-lactamases are known to confer cefepime resistance, which is observed in isolates A922, A095, 2410 and 2418. The presence of the identified beta-lactamases blaSHV-1, blaSHV-27 and blaCTX-M-14 therefore does not correlate conclusively with the resistance phenotypes observed against the beta-lactam antimicrobials assessed in this study.
Fluoroquinolone resistance was assessed using ciprofloxacin, and widespread ciprofloxacin resistance was observed. The fluoroquinolone-resistance determinant QnrB4, an alternate target protein which binds to gyrase enzymes, thus offering protection to host organisms against the quinolone class drugs, was identified and correlated largely with the observed ciprofloxacin resistance here (Figure 2) [71]. Typically, carriage of the qnrB4 gene in isolation is correlated with only low-level resistance to ciprofloxacin [66,67], as was observed for the isolates that were sequenced in this study.
The observed trimethoprim-sulfamethoxazole resistance could be attributed to the presence of known resistance determinants. Trimethoprim-sulfamethoxazole-resistance determinants dfrA17 and sul1 (an integron-encoded dihydrofolate reductase and dihydropteroate synthase [72,73], respectively) were identified, and they were present in all isolates with a resistant phenotype (Figure 2).
3.5. Mutations in Genes Known to Confer Resistance to Antimicrobials
Despite the observed phenotypic resistance to cephalosporins and colistin, no well-known genetic resistance determinants were present, and those determinants that were found did not entirely correlate with resistance. Similarly, no genetic determinants were identified for the high-level resistance to ciprofloxacin observed for two of the isolates. Therefore, a manual probing for mutations in specific genes, which when mutated are known to confer resistance to antimicrobials, was performed across the sequenced isolates to identify mutations which could be responsible for the resistant phenotypes (Supplementary Table S1).
No mutations in the fluoroquinolone target proteins GyrA/B or ParE were observed in isolates with a ciprofloxacin-resistant phenotype. A ParC M304S substitution mutation was observed in six ciprofloxacin-resistant isolates but was absent in the other ciprofloxacin-resistant isolates, making the deduction of its role in ciprofloxacin resistance difficult to determine. Further, this mutation did not correlate with the ciprofloxacin MIC values observed. This mutation has also not been reported for other ciprofloxacin-resistant bacteria and needs further study to clarify its role.
Resistance to colistin within Klebsiella has been reported to be conferred by the presence of mutations in the phoP/Q, mgrB, eptA and eptB genes [67,68,69,70,71]. Therefore, these genes were probed for mutations. No mutations were observed in the transcriptional regulatory proteins PhoP and PhoQ for any of the sequenced isolates. The sole colistin resistance isolate, 2410, contained an M66I mutation in PrmA, T240M and T246A, mutations in PmrB and a T224M mutation in EptA. However, mutations were also present in the colistin-sensitive isolates A095, A529, 2418 and A922; hence, it could not be responsible for the observed colistin resistance. Both the PmrA and PmrB mutations identified in this study have also been determined in colistin-resistant K. pneumonia isolates from China [74]; however, their role in colistin resistance has not been experimentally verified. No mutations within the PhoPQ-negative regulator MgrB were observed.
In addition to probing for mutations which could confer colistin resistance, mutations which could account for the observed resistance to ceftazidime and cefepime were investigated in the sequenced isolates. The amino acid sequences of the blaCTX-M-14 beta-lactamases present in each cefepime-resistant isolate but also a cefepime-sensitive isolate were probed for mutations, as mutations here have been recognized as affecting the substrate specificity [75], but none were identified. Mutations in outer membrane porins OmpK35, 36 and 37 are known to correlate with resistance to some beta-lactam antibiotics (cephalosporin and carbapenem resistance) for K. pneumoniae [15,76,77,78,79,80,81]. As all sequenced isolates apart from A922 were resistant to ceftazidime, and some to cefepime, these porins were also examined for mutations which could affect porin function. Twelve of the thirteen isolates contained a K132E substitution mutation in OmpK35. As these 12 included the ceftazidime-sensitive isolate A922 and only 4 of these were cefepime-resistant, the presence of this mutation within 12 isolates is inconclusive. An identical suite of 15 substitution and insertion mutations were found in the OmpK37 porin for six of the 13 sequenced isolates, while another six of 14 isolates contained a single amino acid substitution in the OmpK37 porin (Supplementary Table S1). While these mutations were observed, no clear correlation in phenotypic resistance to either ceftazidime or cefepime could be drawn. Further, while the mutations in OmpK35 and 37 appear novel and unreported in the literature, many of the mutations in OmpK36 are reported and associated with only ceftazidime resistance [80]. The presence of these porin mutations may therefore be contributing to ceftazidime and/or cefepime resistance but this requires more research to be conclusive.
As RND efflux pumps are associated with a broad substrate range and are an archetypal resistance determinant within K. pneumoniae, these pumps and regulatory genes were also assessed for mutations [36,82,83]. To this end, the RND efflux pump components AcrA and AcrB, as well as the repressor for the system, AcrR, and the global regulator, MarR, were also screened, with no mutations found. Further, efflux pump inhibition using efflux pump inhibitor PAβN was performed and demonstrated no change in MIC values (data not shown). Whilst overexpression of these components may occur by other mechanisms, such as transient, induced up-regulation, these data demonstrate that no mutations in the repressors could be responsible for an overexpression of the AcrAB efflux pump within these isolates.
3.6. The Majority of the K. pneumoniae Complex Isolates were the MDR-Outbreak-Causing Sequence Type 323
The sequenced isolates from this study spanned three different sequence types: ST4726, ST323 and ST661. The majority were of ST323 (n = 7), an ST previously associated with hospitals rather than RACFs [55,84] and recognized as an MDR clonal type [85]. Disease outbreaks have been caused by ST323 K. pneumoniae isolates in Africa and Australia [55,86].
Five of the sequenced isolates were ST661, an ST which has been recorded during outbreaks and found to be typically carrying carbapenemases [87,88]. Whilst none of the ST661 isolates here produced carbapenemases, all were ceftazidime-resistant, and three were additionally resistant to cefepime.
One isolate, A031, was the previously undescribed ST4726.
3.7. Clonal Spread of K. pneumoniae within RACFs was Observed within Facility 1
Single nucleotide polymorphism (SNP) analysis was conducted between isolates of the same sequence type to determine clonality and assess the potential for clonal spread in Facility 1 [47,48,49]. SNP analysis was performed on ST323 isolates using ST323 isolate A409 as reference (Supplementary Table S2), and ST661 isolates were assessed based on ST661 isolate A922 (Supplementary Table S3). The range of SNPs across the ST323 isolates ranged from 8 to 40 and that of the ST661 isolates ranged from 15 to 37. Despite the number of SNPs showing minor variances across the isolates spanning the two STs, the maximum number of SNPs observed was still low enough to infer clonal relatedness across all isolates of the same ST.
Of the ST323 isolates, all of which were clones, the first to be identified was isolate 2404, yielded by a faecal sample from resident 61 in January 2019. Isolate 2362 was then identified in a wastewater sample from December 2019, before isolate 2401 was detected in a faecal sample from a different resident, resident 60, in the same facility in January 2020. A413 and A409 were then recovered from the sink within the bathroom of resident 60 in Facility 1 in September 2020, while isolate A629 was recovered from a faecal sample of resident 77B of Facility 1 in December 2020. The observed clonality of isolates belonging to ST323, despite originating from different sample types and different dates, demonstrates clonal spread within Facility 1. Further, genomic assessment revealed isolate 2401, the most recently isolated, to be carrying different resistance genes, and plasmids. Together, these findings indicate the persistence and spread of clones, and potential adaptation and accumulation of resistance determinants over time. Despite their clonality, and the identification of identical resistance determinants and plasmids (except for isolate 2401 which had different plasmids than the other clones), the resistance profile of these varied. For example, a difference in cefepime MIC was observed, with isolate A409 having a cefepime MIC of >64 mg/L, compared to A413 at 1 mg/L. Conversely, isolates 2404, 2401 and A413 were all sensitive to cefepime (MIC = 1 mg/L). Whilst the ST323 clones were all ciprofloxacin-resistant, isolate A413 had a much lower ciprofloxacin MIC of 4 mg/L, while the other clones had MICs up to >64 mg/L. These differences in antimicrobial susceptibility across these clones, which could not be attributed to resistance mechanisms identified here, point to the persistence and presence of unknown resistance determinants within the RACFs.
Of the ST661 isolates, isolate A922 was the first identified and was from a wastewater sample in December 2019. Isolates 2410 and 2418 were then isolated from faecal samples of resident 50 in Facility 1, followed by isolate A095 in a faecal sample from the same resident at a later date. Lastly, a room swab collected in the rooms of participant 70 yielded isolate A529. Intra-site dissemination was therefore observed for ST661 isolates, including colonization of a resident and establishing persistence within the rooms of the facility. While small differences in antimicrobial susceptibility were observed across these isolates, these were within 1- to 2-fold differences and were not considered to be significant.
4. Discussion
Species of the Klebsiella pneumoniae complex are opportunistic pathogens which can cause serious infections and are increasingly resistant to antimicrobials [89]. The impact on mortality and the AMR capacity of K. pneumoniae exemplify the problem of antimicrobial resistance and mandate surveillance efforts. Current K. pneumoniae surveillance for outbreaks focusses on sepsis within healthcare settings like hospitals [90,91,92,93] rather than in aged care facilities. However, RACFs are now firmly implicated in the development and spread of AMR organisms [16,24,25,26,27,94], yet there is a dearth of data on the occurrence of the priority pathogen MDR K. pneumoniae complex and resistome burden from RACFs. Therefore, the purpose of this study was to look at the prevalence and antimicrobial susceptibility profile of the K. pneumoniae complex in RACFs.
A total of 147 isolates were isolated in this study from three RACFs and one retirement village in Adelaide, Australia. The three RACFs were managed by the same care provider, one that operates multiple RACFs Australia-wide, and they were thereby broadly representative RACFs. The facilities provided almost identical amenities to their residents, and the cohorts of residents (age, duration of stay) were comparable. Each of the facilities selected offered a diverse range of residency options, which included permanent care (including end of life), transitional (between hospital and community), respite (between community and RACF) and transfer (between one RACF and another). The levels of care at each of the RACFs included respite, long-term and end-of-life care. Each facility assessed in this study provided the same type of care to their residents. Potential demographic variables (socio-economic status, sex) may have been present; however, this study focussed on the prevalence of AMR pathogens within these environments and did not seek to measure these variables. To account for this and allow standardisation of comparisons, percentages, rather than total numbers, were the focus of the data presented here.
In this study, we were concerned about the incidence and spread of ESBL-producing and ciprofloxacin-resistant K. pneumoniae in RACFs. The incidence of carriage of putative ESBL-producing K. pneumoniae of 7% was in good correlation with the incidence of ESBL-producing Enterobacterales in RACFs detected in another study in Australia [57]. However, in contrast to those Enterobacterales, genomic analysis of the ceftazidime-resistant isolates from our study revealed that these organisms did not carry ESBL genes. The precise mechanism of ceftazidime resistance could not be definitively assigned, and the relative high incidence of ceftazidime resistance was most probably the result of clonal spread of resistant isolates within a facility.
Single amino acid substitution mutations were observed in outer membrane porins OmpK35, OmpK36 and OmpK37 (Supplementary Table S1). While both novel and previously reported mutations were observed, these did not correlate with a specific resistance phenotype. Further, cefepime resistance was observed in 4 of the 13 sequenced isolates, in the absence of canonical cefepime resistance determinants. With cefepime increasingly relevant as a treatment option in the context of AMR infections, an elucidation of this resistance determinant is important. Whilst this study cannot draw causal relationships, this does provide a potential further avenue for exploration into porin mutations and ceftazidime resistance and shows a need for further exploration into cefepime resistance.
Similarly, the high incidence of phenotypic ciprofloxacin resistance cannot be accounted for solely by the selection for ciprofloxacin resistance as the majority of the ciprofloxacin-resistant isolates were from plates without ciprofloxacin selection. However, genomic analysis of a subset of ciprofloxacin-resistant isolates revealed yet again clonal spread of resistant isolates carrying the qnrB4 gene throughout the facility. Carriage of the qnrB4 gene correlated with the carriage of beta-lactamase genes and genes for resistance to trimethoprim-sulfamethoxazole. Hence, both selective plating and intra-facility spread contributed to the relatively high incidence of ciprofloxacin resistance in K. pneumoniae isolates observed in this study. In comparison, ciprofloxacin resistance in K. pneumoniae isolates from Australian sepsis surveillance programs is currently estimated to be 7.75% [5]. Greece, recognized as having the highest levels of ciprofloxacin resistance globally from sepsis cases, reports resistance prevalence at 66.9% [18]. Data on asymptomatic K. pneumoniae carriage from faecal samples reported ciprofloxacin resistance at 9% in Norway [95] and 18% from community settings in Taiwan [96], while a 33% incidence of pre-admission carriage of ciprofloxacin-resistant K. pneumoniae was reported for neonatal hospitals in Madagascar [97].
While data are lacking on ciprofloxacin-resistant K. pneumoniae from RACFs specifically, other AMR Enterobacterales, such as Escherichia coli, are often identified from residents in aged care homes or from older patients and display ciprofloxacin resistance [96,98,99]. This co-presence could allow for intra-species dissemination of plasmid-mediated quinolone resistance genes associated with low-level plasmid resistance, such as qnrA, qnrB and qnrS [100,101,102]. Mutations in key ciprofloxacin resistance genes, such as the parC mutations identified in this study, have been increasingly identified in ciprofloxacin-resistant K. pneumoniae isolates with a range of MIC values [103,104] and this study adds to this growing body of evidence.
A review of the medical records of the participants during the previous year revealed that all the residents had received antibiotics. Although ceftazidime and cefepime were not prescribed to residents in the RACFs, the first-generation cephalosporin cephalexin was frequently prescribed and could have contributed to the development of resistance to third- and fourth-generation cephalosporins. Alternatively, the records did not capture prescribing during hospital stays and it is possible that these antibiotics could have been administered then. Ciprofloxacin or norfloxacin were among the top 10 most frequently prescribed antibiotics among the participants during the sampling period.
Given the antimicrobial resistance observed here to first- and second-line treatment options, such as cephalosporins and ciprofloxacin, resistance to last-resort and salvage antimicrobials like colistin is particularly relevant. Colistin-resistant K. pneumoniae typically harbour the mobile colistin-resistance gene mcr or a variant thereof [105,106,107], or have mutations or downregulations of the Lipid A synthesis pathway [108,109]. The mcr gene was not identified in the colistin-resistant isolate, indicating the observed colistin resistance may be due to another factor, such as changes in Lipid A synthesis. However, the mutations that were observed in PhoQ, PmrA, PmrB and EptA did not correlate with the observed resistance profile. Taken summarily, reported mutations correlative with resistance were observed, unreported mutations correlative with sensitivity were observed, and these findings act as contributions to the surveillance on resistance and mutations, but cannot conclusively link mutations to resistance at this stage.
Another avenue requiring further exploration is the classification of clonality, defined by the number of SNPs across core genomes. Numerous studies have used SNP analysis to define clonality, with a varying number of SNPs used as the threshold between clonality and just species-level relatedness [47,48,49]. A study comparing SNPs from K. pneumoniae isolates from handwashing sinks found 21 SNPs between isolates and classified these as clones [49]. Other studies of K. pneumoniae clonality have ranged from classifying isolates with 6 SNPs up to 75 SNPs as clones [110,111,112,113]. In our study, the highest numbers of SNPs observed between isolates classified as clones were within the range supported by the literature and isolates of the same ST were therefore classified as clones. Accordingly, we observed clonal spread of MDR K. pneumoniae within Facility 1.
Isolates representative of various sequence types were identified, with ST323 and ST661 shown to be the prevalent sequence types. In both Australia and worldwide, ST323 is associated with healthcare settings [55,84] and is recognized as an MDR sequence type [85]. This is consistent with our data, as all ST323 isolates herein displayed an MDR phenotype and were isolated from RACFs. Whilst our sampling was focused on asymptomatic carriage of K. pneumoniae, disease outbreaks caused by ST323 K. pneumoniae isolates have been identified in Africa and Australia [55,86]. Specifically, the ST323 outbreak in the Australian report was within a geriatric care ward [55]. With the ST323 clones identified here spreading across three residents and colonizing sinks, these data demonstrate that MDR K. pneumoniae can persist and disseminate within these environments, and that they represent an ongoing threat to health [114]. This is particularly troubling in an environment such as an RACF, housing vulnerable residents particularly susceptible to opportunistic pathogens. RACFs may, therefore, represent potential outbreak sites for AMR isolates warranting continued and increased surveillance. The presence of these ST323 isolates within an Australian RACF, all of which were ceftazidime-resistant, may therefore pose a real threat to the elderly residents.
ST661 isolates have been reported in healthcare settings in the UK and aquatic environments in South America, and on both occasions they were characterized by widespread dissemination throughout the localized environment [87,88]. The ST661 isolates identified here displayed MDR phenotypes; therefore, the observed persistence within the environment and outward spread between residents, coupled with the known ability of this ST to become disseminated widely over time, is therefore a clear risk factor for disease outbreak and would be an avenue for continued research of isolates within this sequence type. No reported instances of the other sequenced isolates for which a sequence type was identified, ST4726, ST997 or ST603, were found in the literature. However, given that these are STs which are now identified as correlating to isolation from RACFs and with AMR phenotypes, surveillance for these STs may be warranted. Here, clonal dissemination of K. pneumoniae was observed from intestinal carriage to the colonization of sinks, and other residents, consistent with other findings [49,115,116]. Crucially, the colonization of sinks by K. pneumoniae isolates has been repeatedly traced back as the cause of outbreaks and is therefore a risk factor for future outbreaks [49,115,116]. Sinks can act as reservoirs allowing for the persistence of pathogenic organisms like K. pneumoniae, with exposure causing repeated infections and potentiating outbreaks. The high potential risk of a K. pneumoniae outbreak within an RACF, as demonstrated by the persistence within environments and clonal spread across residents in this study, underscores the importance of continued AMR surveillance within environments housing vulnerable, immunocompromised persons such as RACFs.
5. Conclusions
This pilot study represents the first surveillance of AMR K. pneumoniae isolates within RACFs, and the first quantification of the resistome of these isolates. We found that 53% of the K. pneumoniae complex isolates from RACFs assessed in this study were multidrug resistant. Genomic analyses failed to conclusively identify genetic resistance determinants for the observed resistance to cefepime and colistin. Further, we identified cross-resident clonal spread and the persistence of these AMR isolates within bathroom sinks in the RACFs—representing a significant outbreak threat. These findings were consistent with a growing trend of AMR K. pneumoniae isolates with unclear mechanisms of resistance, and they highlight the need for further studies to elucidate these resistance determinants and for continued surveillance in this space.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12040751/s1, Supplementary Table S1: Mutations in genes known to confer antimicrobial resistance, with isolate A031 as reference, Supplementary Table S2: Matrix of SNP pair counts for ST323 isolates, with A409 used as reference, Supplementary Table S3: Matrix of SNP pair counts for ST661 isolates, with A922 used as a reference.
Author Contributions
Conceptualisation, J.M.B., S.A.S. and H.V.; Experimental, J.M.B., N.L.S., S.A.S. and A.A.; Analysis, J.M.B., S.A.S., B.J.H., N.L.S. and H.V.; Writing—Original Draft Preparation, J.M.B.; Writing—Review and Editing, J.M.B., S.A.S., N.L.S., M.S.W., L.E.X.L. and H.V.; Supervision, S.A.S. and H.V.; Project Administration, H.V.; Funding Acquisition; H.V. All authors have read and reviewed the manuscript prior to publication. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Medical Research Future Fund (MRFF, GN1152556) to H.V., J.M.B. and N.L.S. are the recipients of Australian government research training program for domestic students (RTP) scholarships. A.A. is the recipient of an Australian government research training program for international students (RTPi) scholarship.
Data Availability Statement
The collective WGSs of isolates used in this study were deposited to the NCBI database BioProject ‘PRJNA949397’ (Table 1).
Acknowledgments
The authors extend their gratitude to the aged care provider for participation in this project and to Barry Lowe for sampling the residents and facilities. The carbapenemase-positive control strains used in this study were kindly provided by Jan Bell from the Australian Group on Antimicrobial Resistance. We would also like to thank Gianluca Brunetti (FII, UniSA) for help with the wastewater collection and Michael Short (FII, UniSA) for advice on wastewater sampling. We thank Jon Whittall and the technical staff from Microbiology at the University of South Australia for technical assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- World Health Organisation (WHO). Antimicrobial Resistance Fact Sheet. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 31 May 2022).
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Wellcome Trust and HM Government. 2014. Available online: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed on 1 March 2022).
- Dong, N.; Yang, X.; Chan, E.W.-C.; Zhang, R.; Chen, S. Klebsiella species: Taxonomy, hypervirulence and multidrug resistance. eBioMedicine 2022, 79, 103998. [Google Scholar] [CrossRef] [PubMed]
- Australian Group on Antimicrobial Resistance (AGAR). Antimicrobial Use and Resistance in Human Health in Australia (AURA) 2023 Fifth Australian Report on Antimicrobial Use and Resistance in Human Health; Australian Commission on Safety and Quality in Health Care: Sydney, Australia, 2021. Available online: https://www.safetyandquality.gov.au/sites/default/files/2023-11/aura_2023_fifth_australian_report_on_antimicrobial_use_and_resistance_in_human_health.pdf (accessed on 1 March 2024).
- Kareem, S.M.; Al-Kadmy, I.M.; Kazaal, S.S.; Mohammed Ali, A.N.; Aziz, S.N.; Makharita, R.R.; Algammal, A.M.; Al-Rejaie, S.; Behl, T.; Batiha, G.E.-S. Detection of gyrA and parC mutations and prevalence of plasmid-mediated quinolone resistance genes in Klebsiella pneumoniae. Infect. Drug Resist. 2021, 14, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Y.; Nam, Y.S.; Lee, H.J. Prevalence of plasmid-mediated quinolone resistance genes among ciprofloxacin-nonsusceptible Escherichia coli and Klebsiella pneumoniae isolated from blood cultures in Korea. Can. J. Infect. Dis. Med. Microbiol. 2014, 25, 163–169. [Google Scholar] [CrossRef]
- Rocha, J.; Henriques, I.; Gomila, M.; Manaia, C.M. Common and distinctive genomic features of Klebsiella pneumoniae thriving in the natural environment or in clinical settings. Sci. Rep. 2022, 12, 10441. [Google Scholar] [CrossRef] [PubMed]
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J.; et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 2015, 112, E3574–E3581. [Google Scholar] [CrossRef] [PubMed]
- Bialek-Davenet, S.; Criscuolo, A.; Ailloud, F.; Passet, V.; Jones, L.; Delannoy-Vieillard, A.-S.; Garin, B.; Le Hello, S.; Arlet, G.; Nicolas-Chanoine, M.-H. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 2014, 20, 1812. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Chen, Q.; Shen, F.; Jiang, Y.; Wu, X.; Hua, X.; Fu, Y.; Yu, Y. Resistance Evolution of Hypervirulent Carbapenem-resistant Klebsiella pneumoniae ST11 during Treatment with Tigecycline and Polymyxin. Emerg. Microbes Infect. 2021, 10, 1129–1136. [Google Scholar] [CrossRef]
- Kaczmarek, F.M.; Dib-Hajj, F.; Shang, W.; Gootz, T.D. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla ACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob. Agents Chemother. 2006, 50, 3396–3406. [Google Scholar] [CrossRef]
- Yu, Q.; Hu, Z.; Tian, B.; Li, Y.; Li, J.; Xu, H. Drug-resistant genes and virulence factors of carbepenem-resistant Klebsiella peumoniae. Chin. J. Clin. Infect. Dis. 2016, 6, 52–58. [Google Scholar]
- Wong, J.L.; Romano, M.; Kerry, L.E.; Kwong, H.-S.; Low, W.-W.; Brett, S.J.; Clements, A.; Beis, K.; Frankel, G. OmpK36-mediated carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat. Commun. 2019, 10, 3957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, X.; Wang, Y.; Li, G.; Tian, Y.; Liu, H.; Ai, F.; Ma, Y.; Wang, B.; Ruan, F.; et al. Contribution of β-Lactamases and Porin Proteins OmpK35 and OmpK36 to Carbapenem Resistance in Clinical Isolates of KPC-2-Producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2014, 58, 1214–1217. [Google Scholar] [CrossRef] [PubMed]
- Stuart, R.L.; Wilson, J.; Bellaard-Smith, E.; Brown, R.; Wright, L.; Vandergraaf, S.; Gillespie, E.E. Antibiotic use and misuse in residential aged care facilities. Intern. Med. J. 2012, 42, 1145–1149. [Google Scholar] [CrossRef] [PubMed]
- Raban, M.Z.; Gates, P.J.; Gasparini, C.; Westbrook, J.I. Temporal and regional trends of antibiotic use in long-term aged care facilities across 39 countries, 1985–2019: Systematic review and meta-analysis. PLoS ONE 2021, 16, e0256501. [Google Scholar] [CrossRef] [PubMed]
- Surveillance Outcome Programs Bloodstream Infections 2021 Report; Australian Group on Antimicrobial Resistance: East Perth, Australia, 2021.
- Cowan, R.U.; Kishan, D.; Walton, A.L.; Sneath, E.; Cheah, T.; Butwilowsky, J.; Friedman, N.D. Cleaning, resistant bacteria, and antibiotic prescribing in residential aged care facilities. Am. J. Infect. Control 2016, 44, e19–e21. [Google Scholar] [CrossRef] [PubMed]
- Tronsmo, A.; Gjøen, T.; Sørum, H.; Godfroid, J.; Yazdankhah, S.P.; Jelmert, A.; Klein, J.; Okoli, A.S.; Ytrehus, B.; Skaar, I. Antimicrobial Resistance due to the Use of Biocides and Heavy Metals: A Literature Review; Norwegian Scientific Committee for Food Safety: Oslo, Norway, 2016. [Google Scholar]
- Mao, Y.-C.; Chang, C.-L.; Huang, Y.-C.; Su, L.-H.; Lee, C.-T. Laboratory investigation of a suspected outbreak caused by Providencia stuartii with intermediate resistance to imipenem at a long-term care facility. J. Microbiol. Immunol. Infect. 2018, 51, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Ingarfield, S.L.; Finn, J.C.; Jacobs, I.G.; Gibson, N.P.; Holman, C.D.; Jelinek, G.A.; Flicker, L. Use of emergency departments by older people from residential care: A population based study. Age Ageing 2009, 38, 314–318. [Google Scholar] [CrossRef] [PubMed]
- O’Fallon, E.; Kandel, R.; Schreiber, R.; D’Agata, E.M. Acquisition of multidrug-resistant gram-negative bacteria: Incidence and risk factors within a long-term care population. Infect. Control Hosp. Epidemiol. 2010, 31, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Dowson, L.; Rajkhowa, A.; Buising, K.; Kong, D.C.; Stuart, R.L.; Thursky, K.; Bennett, N. The 2018 Aged Care National Antimicrobial Prescribing Survey: Results show room for improvement. Aust. Prescr. 2019, 42, 200–203. [Google Scholar] [CrossRef]
- Stuart, R.L.; Marshall, C.; Orr, E.; Bennett, N.; Athan, E.; Friedman, D.; Reilly, M.; Racrig, M.O. Survey of infection control and antimicrobial stewardship practices in Australian residential aged-care facilities. Intern. Med. J. 2015, 45, 576–580. [Google Scholar] [CrossRef]
- Mody, L.; Foxman, B.; Bradley, S.; McNamara, S.; Lansing, B.; Gibson, K.; Cassone, M.; Armbruster, C.; Mantey, J.; Min, L. Longitudinal assessment of multidrug-resistant organisms in newly admitted nursing facility patients: Implications for an evolving population. Clin. Infect. Dis. 2018, 67, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Kahvecioglu, D.; Ramiah, K.; McMaughan, D.; Garfinkel, S.; McSorley, V.E.; Nguyen, Q.N.; Yang, M.; Pugliese, C.; Mehr, D.; Phillips, C.D. Multidrug-resistant organism infections in US nursing homes: A national study of prevalence, onset, and transmission across care settings, October 1, 2010–December 31, 2011. Infect. Control Hosp. Epidemiol. 2014, 35, S48–S55. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Carbapenem-resistant Klebsiella pneumoniae associated with a long-term-care facility—West Virginia, 2009–2011. MMWR. Morb. Mortal. Wkly. Rep. 2011, 60, 1418–1420. [Google Scholar]
- Munoz-Price, L.S.; Hayden, M.K.; Lolans, K.; Won, S.; Calvert, K.; Lin, M.; Sterner, A.; Weinstein, R.A. Successful control of an outbreak of Klebsiella pneumoniae carbapenemase—Producing K. pneumoniae at a long-term acute care hospital. Infect. Control Hosp. Epidemiol. 2010, 31, 341–347. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 13.0; EUCAST: Växjö, Sweden, 2023. [Google Scholar]
- Wand, M.E.; Bock, L.J.; Bonney, L.C.; Sutton, J.M. Mechanisms of Increased Resistance to Chlorhexidine and Cross-Resistance to Colistin following Exposure of Klebsiella pneumoniae Clinical Isolates to Chlorhexidine. Antimicrob. Agents Chemother. 2017, 61, e01162-16. [Google Scholar] [CrossRef] [PubMed]
- Amsalu, A.; Sapula, S.A.; De Barros Lopes, M.; Hart, B.J.; Nguyen, A.H.; Drigo, B.; Turnidge, J.; Leong, L.E.; Venter, H. Efflux Pump-Driven Antibiotic and Biocide Cross-Resistance in Pseudomonas aeruginosa Isolated from Different Ecological Niches: A Case Study in the Development of Multidrug Resistance in Environmental Hotspots. Microorganisms 2020, 8, 1647. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, I.; Oggioni, M.R.; Knight, D.; Curiao, T.; Coque, T.; Kalkanci, A.; Martinez, J.L.; Consortium, B. Evaluation of epidemiological cut-off values indicates that biocide resistant subpopulations are uncommon in natural isolates of clinically-relevant microorganisms. PLoS ONE 2014, 9, e86669. [Google Scholar] [CrossRef] [PubMed]
- Sundaramoorthy, N.S.; Suresh, P.; Selva Ganesan, S.; GaneshPrasad, A.; Nagarajan, S. Restoring colistin sensitivity in colistin-resistant E. coli: Combinatorial use of MarR inhibitor with efflux pump inhibitor. Sci. Rep. 2019, 9, 19845. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.-H.; Lo, C.-C.; Chou, M.-C.; Yeh, T.-H.; Chen, K.-L.; Liao, W.-Y.; Lo, H.-R. Synergistic Actions of Benzyl Isothiocyanate with Ethylenediaminetetraacetic Acid and Efflux Pump Inhibitor Phenylalanine-Arginine β-Naphthylamide against Multidrug-Resistant Escherichia coli. Microb. Drug Resist. 2020, 26, 468–474. [Google Scholar] [CrossRef]
- Venter, H.; Mowla, R.; Ohene-Agyei, T.; Ma, S. RND-type drug efflux pumps from Gram-negative bacteria: Molecular mechanism and inhibition. Front. Microbiol. 2015, 6, 377. [Google Scholar] [CrossRef]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M. TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Cantu, V.A.; Sadural, J.; Edwards, R. PRINSEQ++, a multi-threaded tool for fast and efficient quality control and preprocessing of sequencing datasets. PeerJ Prepr. 2019, 7, e27553v27551. [Google Scholar]
- Joshi, N.A.; Fass, J.N. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33) [Software]. Available online: https://github.com/najoshi/sickle (accessed on 1 January 2022).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.-R.; Florensa, A.F. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- CSIphylogeny. Available online: https://cge.food.dtu.dk/services/CSIPhylogeny/ (accessed on 1 January 2022).
- Berglund, B.; Hoang, N.T.B.; Lundberg, L.; Le, N.K.; Tärnberg, M.; Nilsson, M.; Bornefall, E.; Khu, D.T.K.; Welander, J.; Le, H.T.; et al. Clonal spread of carbapenem-resistant Klebsiella pneumoniae among patients at admission and discharge at a Vietnamese neonatal intensive care unit. Antimicrob. Resist. Infect. Control 2021, 10, 162. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, S.; Wei, L.; Feng, Y.; Cai, L.; Dunn, S.; McNally, A.; Zong, Z. Arm race among closely-related carbapenem-resistant Klebsiella pneumoniae clones. ISME Commun. 2022, 2, 76. [Google Scholar] [CrossRef]
- Feng, Y.; Wei, L.; Zhu, S.; Qiao, F.; Zhang, X.; Kang, Y.; Cai, L.; Kang, M.; McNally, A.; Zong, Z. Handwashing sinks as the source of transmission of ST16 carbapenem-resistant Klebsiella pneumoniae, an international high-risk clone, in an intensive care unit. J. Hosp. Infect. 2020, 104, 492–496. [Google Scholar] [CrossRef]
- PlasmidFinder 2.1.1. Available online: https://bioweb.pasteur.fr/packages/pack@PlasmidFinder@2.1.1 (accessed on 1 January 2023).
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Kampf, G. Adaptive bacterial response to low level chlorhexidine exposure and its implications for hand hygiene. Microb. Cell 2019, 6, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, Y.; Xu, C.; Zhang, X.; Li, J.; Dong, G.; Cao, J.; Zhou, T. Chlorhexidine exposure of clinical Klebsiella pneumoniae strains leads to acquired resistance to this disinfectant and to colistin. Int. J. Antimicrob. Agents 2019, 53, 864–867. [Google Scholar] [CrossRef] [PubMed]
- Gorrie, C.L.; Mirceta, M.; Wick, R.R.; Judd, L.M.; Wyres, K.L.; Thomson, N.R.; Strugnell, R.A.; Pratt, N.F.; Garlick, J.S.; Watson, K.M.; et al. Antimicrobial-Resistant Klebsiella pneumoniae Carriage and Infection in Specialized Geriatric Care Wards Linked to Acquisition in the Referring Hospital. Clin. Infect. Dis. 2018, 67, 161–170. [Google Scholar] [CrossRef]
- Stuart, R.L.; Kotsanas, D.; Webb, B.; Vandergraaf, S.; Gillespie, E.E.; Hogg, G.G.; Korman, T.M. Prevalence of antimicrobial-resistant organisms in residential aged care facilities. Med. J. Aust. 2011, 195, 530–533. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, A.; Sonbol, F.; Elbanna, T.; El-Ekhnawy, E. Exposure to sublethal concentrations of benzalkonium chloride induces antimicrobial resistance and cellular changes in Klebsiellae pneumoniae clinical isolates. Microb. Drug Resist. 2019, 25, 631–638. [Google Scholar] [CrossRef]
- Srinivasan, V.B.; Rajamohan, G. KpnEF, a New Member of the Klebsiella pneumoniae Cell Envelope Stress Response Regulon, Is an SMR-Type Efflux Pump Involved in Broad-Spectrum Antimicrobial Resistance. Antimicrob. Agents Chemother. 2013, 57, 4449–4462. [Google Scholar] [CrossRef]
- Satlin, M.J.; Lewis, J.S.; Weinstein, M.P.; Patel, J.; Humphries, R.M.; Kahlmeter, G.; Giske, C.G.; Turnidge, J. Clinical and Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility Testing Position Statements on Polymyxin B and Colistin Clinical Breakpoints. Clin. Infect. Dis. 2020, 71, e523–e529. [Google Scholar] [CrossRef]
- EUCAST. E.C.o.A.S.T. Colistin Guidance. 2022. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Guidance_documents/Colistin_guidance_2022.pdf (accessed on 11 February 2023).
- Hammond, D.S.; Schooneveldt, J.M.; Nimmo, G.R.; Huygens, F.; Giffard, P.M. blaSHV Genes in Klebsiella pneumoniae: Different Allele Distributions Are Associated with Different Promoters within Individual Isolates. Antimicrob. Agents Chemother. 2005, 49, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Drieux, L.; Brossier, F.; Sougakoff, W.; Jarlier, V. Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: Review and bench guide. Clin. Microbiol. Infect. 2008, 14, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. AmpC β-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Tian, L.; Li, G.; Qu, H.; Sun, J.; Liang, W.; Li, X.; Wang, X.; Deng, Z.; Liu, J.; et al. Emergence of the third-generation cephalosporin-resistant hypervirulent Klebsiella pneumoniae due to the acquisition of a self-transferable blaDHA-1-carrying plasmid by an ST23 strain. Virulence 2018, 9, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Pishtiwan, A.H.; Khadija, K.M. Prevalence of blaTEM, blaSHV, and blaCTX-M Genes among ESBL-Producing Klebsiella pneumoniae and Escherichia coli Isolated from Thalassemia Patients in Erbil, Iraq. Mediterr J Hematol Infect Dis 2019, 11, e2019041. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, I.; Chenouf, N.S.; Carvalho, J.A.; Castro, A.P.; Silva, V.; Capita, R.; Alonso-Calleja, C.; Enes Dapkevicius, M.L.N.; Igrejas, G.; Torres, C.; et al. Multidrug-resistant Klebsiella pneumoniae harboring extended spectrum β-lactamase encoding genes isolated from human septicemias. PLoS ONE 2021, 16, e0250525. [Google Scholar] [CrossRef] [PubMed]
- Damjanova, I.; Toth, A.; Paszti, J.; Hajbel-Vékony, G.; Jakab, M.; Berta, J.; Milch, H.; Füzi, M. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type β-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005—The new ‘MRSAs’? J. Antimicrob. Chemother. 2008, 62, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, A.A.; Abdelaziz, N.A.; Amin, M.A.; Aziz, R.K. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase-producing Escherichia coli. Sci. Rep. 2019, 9, 4224. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Gniadkowski, M.; Nordmann, P. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum β-lactamase CTX-M-15 and of its structurally related β-lactamase CTX-M-3. J. Antimicrob. Chemother. 2002, 50, 1031–1034. [Google Scholar] [CrossRef]
- Williamson, D.A.; Roberts, S.A.; Smith, M.; Heffernan, H.; Tiong, A.; Pope, C.; Freeman, J.T. High rates of susceptibility to ceftazidime among globally prevalent CTX-M-producing Escherichia coli: Potential clinical implications of the revised CLSI interpretive criteria. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 821–824. [Google Scholar] [CrossRef]
- Stoesser, N.; Batty, E.M.; Eyre, D.W.; Morgan, M.; Wyllie, D.H.; Del Ojo Elias, C.; Johnson, J.R.; Walker, A.S.; Peto, T.E.A.; Crook, D.W. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J. Antimicrob. Chemother. 2013, 68, 2234–2244. [Google Scholar] [CrossRef] [PubMed]
- White, P.A.; McIver, C.J.; Deng, Y.-M.; Rawlinson, W.D. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 2000, 182, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.W.; Lim, J.; Kim, S.; Kim, J.; Kwon, G.C.; Koo, S.H. Characterization of trimethoprim-sulfamethoxazole resistance genes and their relatedness to class 1 integron and insertion sequence common region in gram-negative bacilli. J. Microbiol. Biotechnol. 2015, 25, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin, Y.; Wang, Z.; Hu, N.; Liu, Q.; Zhou, W.; Li, X.; Hu, L.; Guo, J.; Huang, X. Molecular mechanisms of colistin resistance in Klebsiella pneumoniae in a tertiary care teaching hospital. Front. Cell Infect. Microbiol. 2021, 11, 678. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Chiou, J.; Zeng, Z.; Liu, L.; Chen, X.; Zeng, L.; Chan, E.W.C.; Liu, J.-H.; Chen, S. Residues Distal to the Active Site Contribute to Enhanced Catalytic Activity of Variant and Hybrid β-Lactamases Derived from CTX-M-14 and CTX-M-15. Antimicrob. Agents Chemother. 2015, 59, 5976–5983. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, L.; Pascual, A.; Hernández-Allés, S.; Alvarez-Díaz, D.; Suárez, A.I.; Tran, J.; Benedí, V.J.; Jacoby, G.A. Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 1999, 43, 1669–1673. [Google Scholar] [CrossRef] [PubMed]
- Doménech-Sánchez, A.; Martínez-Martínez, L.; Hernández-Allés, S.; del Carmen Conejo, M.; Pascual, A.; Tomás, J.M.; Albertí, S.; Benedí, V.J. Role of Klebsiella pneumoniae OmpK35 porin in antimicrobial resistance. Antimicrob. Agents Chemother. 2003, 47, 3332–3335. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Allés, S.; Albertí, S.; Álvarez, D.; Doménech-Sánchez, A.; Martínez-Martínez, L.; Gil, J.; Tomás, J.M.; Benedí, V.J. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 1999, 145, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Ardanuy, C.; Liñares, J.; Domínguez, M.A.; Hernández-Allés, S.; Benedí, V.J.; Martínez-Martínez, L. Outer membrane profiles of clonally related Klebsiella pneumoniae isolates from clinical samples and activities of cephalosporins and carbapenems. Antimicrob. Agents Chemother. 1998, 42, 1636–1640. [Google Scholar] [CrossRef]
- Castanheira, M.; Mendes, R.E.; Sader, H.S. Low Frequency of Ceftazidime-Avibactam Resistance among Enterobacteriaceae Isolates Carrying blaKPC Collected in U.S. Hospitals from 2012 to 2015. Antimicrob. Agents Chemother. 2017, 61, e02369-16. [Google Scholar] [CrossRef]
- Tsai, Y.-K.; Fung, C.-P.; Lin, J.-C.; Chen, J.-H.; Chang, F.-Y.; Chen, T.-L.; Siu, L.K. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 2011, 55, 1485–1493. [Google Scholar] [CrossRef] [PubMed]
- Ni, R.T.; Onishi, M.; Mizusawa, M.; Kitagawa, R.; Kishino, T.; Matsubara, F.; Tsuchiya, T.; Kuroda, T.; Ogawa, W. The role of RND-type efflux pumps in multidrug-resistant mutants of Klebsiella pneumoniae. Sci. Rep. 2020, 10, 10876. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, W.; Onishi, M.; Ni, R.; Tsuchiya, T.; Kuroda, T. Functional study of the novel multidrug efflux pump KexD from Klebsiella pneumoniae. Gene 2012, 498, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Geraci, D.; Bonura, C.; Giuffrè, M.; Saporito, L.; Graziano, G.; Aleo, A.; Fasciana, T.; Di Bernardo, F.; Stampone, T.; Palma, D. Is the monoclonal spread of the ST258, KPC-3-producing clone being replaced in southern Italy by the dissemination of multiple clones of carbapenem-nonsusceptible, KPC-3-producing Klebsiella pneumoniae? Clin. Microbiol. Infect. 2015, 21, e15–e17. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef] [PubMed]
- Henson, S.P.; Boinett, C.J.; Ellington, M.J.; Kagia, N.; Mwarumba, S.; Nyongesa, S.; Mturi, N.; Kariuki, S.; Scott, J.A.G.; Thomson, N.R. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int. J. Med. Microbiol. 2017, 307, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.; Phan, H.T.T.; Findlay, J.; Stoesser, N.; Pankhurst, L.; Navickaite, I.; De Maio, N.; Eyre, D.W.; Toogood, G.; Orsi, N.M.; et al. Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: Long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. J. Antimicrob. Chemother. 2017, 72, 3025–3034. [Google Scholar] [CrossRef] [PubMed]
- Toledano-Tableros, J.E.; Gayosso-Vázquez, C.; Jarillo-Quijada, M.D.; Fernández-Vázquez, J.L.; Morfin-Otero, R.; Rodríguez-Noriega, E.; Giono-Cerezo, S.; Gutkind, G.; Di Conza, J.; Santos-Preciado, J.I. Dissemination of bla NDM–1 Gene among Several Klebsiella pneumoniae Sequence Types in Mexico Associated with Horizontal Transfer Mediated by IncF-Like Plasmids. Front. Microbiol. 2021, 12, 611274. [Google Scholar] [CrossRef] [PubMed]
- Gorrie, C.L.; Mirčeta, M.; Wick, R.R.; Judd, L.M.; Lam, M.M.; Gomi, R.; Abbott, I.J.; Thomson, N.R.; Strugnell, R.A.; Pratt, N.F. Genomic dissection of Klebsiella pneumoniae infections in hospital patients reveals insights into an opportunistic pathogen. Nat. Commun. 2022, 13, 3017. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, X.; Torres, V.V.L.; Liu, H.; Rocker, A.; Zhang, Y.; Wang, J.; Chen, L.; Bi, W.; Lin, J. An outbreak of carbapenem-resistant and hypervirulent Klebsiella pneumoniae in an intensive care unit of a major teaching hospital in Wenzhou, China. Front. Public Health 2019, 7, 229. [Google Scholar] [CrossRef]
- Emeraud, C.; Figueiredo, S.; Bonnin, R.A.; Khecharem, M.; Ouzani, S.; Leblanc, P.-E.; Jousset, A.B.; Fortineau, N.; Duranteau, J.; Dortet, L. Outbreak of CTX-M-15 Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae ST394 in a French Intensive Care Unit Dedicated to COVID-19. Pathogens 2021, 10, 1426. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.W.; Mustapha, M.M.; Griffith, M.P.; Evans, D.R.; Ezeonwuka, C.; Pasculle, A.W.; Shutt, K.A.; Sundermann, A.; Ayres, A.M.; Shields, R.K. Evolution of outbreak-causing carbapenem-resistant Klebsiella pneumoniae ST258 at a tertiary care hospital over 8 years. mBio 2019, 10, e01945-19. [Google Scholar] [CrossRef] [PubMed]
- Arteaga-Livias, K.; Pinzas-Acosta, K.; Perez-Abad, L.; Panduro-Correa, V.; Rabaan, A.A.; Pecho-Silva, S.; Dámaso-Mata, B. A multidrug-resistant Klebsiella pneumoniae outbreak in a Peruvian hospital: Another threat from the COVID-19 pandemic. Infect. Control Hosp. Epidemiol. 2022, 43, 267–268. [Google Scholar] [CrossRef] [PubMed]
- Rosello, A.; Hayward, A.C.; Hopkins, S.; Horner, C.; Ironmonger, D.; Hawkey, P.M.; Deeny, S.R. Impact of long-term care facility residence on the antibiotic resistance of urinary tract Escherichia coli and Klebsiella. J. Antimicrob. Chemother. 2017, 72, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Ulstad, C.R.; Solheim, M.; Berg, S.; Lindbæk, M.; Dahle, U.R.; Wester, A.L. Carriage of ESBL/AmpC-producing or ciprofloxacin non-susceptible Escherichia coli and Klebsiella spp. in healthy people in Norway. Antimicrob. Resist. Infect. Control 2016, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-P.; Wang, J.-T.; Chang, S.-C.; Chang, F.-Y.; Fung, C.-P.; Chuang, Y.-C.; Chen, Y.-S.; Shiau, Y.-R.; Tan, M.-C.; Wang, H.-Y. The antimicrobial susceptibility of Klebsiella pneumoniae from community settings in Taiwan, a trend analysis. Sci. Rep. 2016, 6, 36280. [Google Scholar] [CrossRef] [PubMed]
- Andriatahina, T.; Randrianirina, F.; Hariniana, E.R.; Talarmin, A.; Raobijaona, H.; Buisson, Y.; Richard, V. High prevalence of fecal carriage of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a pediatric unit in Madagascar. BMC Infect. Dis. 2010, 10, 204. [Google Scholar] [CrossRef]
- Sapula, S.A.; Amsalu, A.; Whittall, J.J.; Hart, B.J.; Siderius, N.L.; Nguyen, L.; Gerber, C.; Turnidge, J.; Venter, H. The scope of antimicrobial resistance in Residential Aged Care Facilities determined through analysis of Escherichia coli and the total wastewater resistome. mSpectrum, 2023; 11, e00731-23. [Google Scholar]
- Mulder, M.; Kiefte-de Jong, J.C.; Goessens, W.H.F.; de Visser, H.; Hofman, A.; Stricker, B.H.; Verbon, A. Risk factors for resistance to ciprofloxacin in community-acquired urinary tract infections due to Escherichia coli in an elderly population. J. Antimicrob. Chemother. 2016, 72, 281–289. [Google Scholar] [CrossRef]
- Tran, J.H.; Jacoby, G.A. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 2002, 99, 5638–5642. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Strahilevitz, J.; Hooper, D.C. Plasmid-mediated quinolone resistance. Plasmids Biol. Impact Biotechnol. Discov. 2015, 475–503. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.M.; Cano, M.E.; Velasco, C.; Martínez-Martínez, L.; Pascual, A. Plasmid-mediated quinolone resistance: An update. J. Infect. Chemother. 2011, 17, 149–182. [Google Scholar] [CrossRef] [PubMed]
- Agyepong, N.; Govinden, U.; Owusu-Ofori, A.; Amoako, D.G.; Allam, M.; Janice, J.; Pedersen, T.; Sundsfjord, A.; Essack, S. Genomic characterization of multidrug-resistant ESBL-producing Klebsiella pneumoniae isolated from a Ghanaian teaching hospital. Int. J. Infect. Dis. 2019, 85, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Mbelle, N.M.; Feldman, C.; Sekyere, J.O.; Maningi, N.E.; Modipane, L.; Essack, S.Y. Pathogenomics and evolutionary epidemiology of multi-drug resistant clinical Klebsiella pneumoniae isolated from Pretoria, South Africa. Sci. Rep. 2020, 10, 1232. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Li, X.; Chen, Y.; Jiang, Y.; Zhou, Z.; Zhang, H.; Sun, L.; Ruan, Z.; Feng, Y.; Akova, M. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: A multicentre longitudinal study. Lancet Infect. Dis. 2017, 17, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Zhou, Y.; Li, J.; Yin, W.; Wang, S.; Zhang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-Q.; Li, Y.-X.; Lei, C.-W.; Zhang, A.-Y.; Wang, H.-N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1791–1795. [Google Scholar] [CrossRef]
- Ah, Y.-M.; Kim, A.-J.; Lee, J.-Y. Colistin resistance in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 2014, 44, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Elias, R.; Duarte, A.; Perdigão, J. A molecular perspective on colistin and Klebsiella pneumoniae: Mode of action, resistance genetics, and phenotypic susceptibility. Diagnostics 2021, 11, 1165. [Google Scholar] [CrossRef]
- Miro, E.; Rossen, J.W.; Chlebowicz, M.A.; Harmsen, D.; Brisse, S.; Passet, V.; Navarro, F.; Friedrich, A.W.; García-Cobos, S. Core/whole genome multilocus sequence typing and core genome SNP-based typing of OXA-48-producing Klebsiella pneumoniae clinical isolates from Spain. Front. Microbiol. 2020, 10, 2961. [Google Scholar] [CrossRef]
- Popa, L.I.; Gheorghe, I.; Barbu, I.C.; Surleac, M.; Paraschiv, S.; Măruţescu, L.; Popa, M.; Pîrcălăbioru, G.G.; Talapan, D.; Niţă, M. Multidrug resistant Klebsiella pneumoniae ST101 clone survival chain from inpatients to hospital effluent after chlorine treatment. Front. Microbiol. 2021, 11, 610296. [Google Scholar] [CrossRef]
- Pérez-Vázquez, M.; Oteo, J.; García-Cobos, S.; Aracil, B.; Harris, S.R.; Ortega, A.; Fontanals, D.; Hernández, J.M.; Solís, S.; Campos, J. Phylogeny, resistome and mobile genetic elements of emergent OXA-48 and OXA-245 Klebsiella pneumoniae clones circulating in Spain. J. Antimicrob. Chemother. 2016, 71, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Oteo, J.; Pérez-Vázquez, M.; Bautista, V.; Ortega, A.; Zamarrón, P.; Saez, D.; Fernández-Romero, S.; Lara, N.; Ramiro, R.; Aracil, B. The spread of KPC-producing Enterobacteriaceae in Spain: WGS analysis of the emerging high-risk clones of Klebsiella pneumoniae ST11/KPC-2, ST101/KPC-2 and ST512/KPC-3. J. Antimicrob. Chemother. 2016, 71, 3392–3399. [Google Scholar] [CrossRef] [PubMed]
- Kotsanas, D.; Wijesooriya, W.; Korman, T.M.; Gillespie, E.E.; Wright, L.; Snook, K.; Williams, N.; Bell, J.M.; Li, H.Y.; Stuart, R.L. “Down the drain”: Carbapenem-resistant bacteria in intensive care unit patients and handwashing sinks. Med. J. Aust. 2013, 198, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Starlander, G.; Melhus, Å. Minor outbreak of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in an intensive care unit due to a contaminated sink. J. Hosp. Infect. 2012, 82, 122–124. [Google Scholar] [CrossRef]
- Decraene, V.; Phan, H.; George, R.; Wyllie, D.; Akinremi, O.; Aiken, Z.; Cleary, P.; Dodgson, A.; Pankhurst, L.; Crook, D. A large, refractory nosocomial outbreak of Klebsiella pneumoniae carbapenemase-producing Escherichia coli demonstrates carbapenemase gene outbreaks involving sink sites require novel approaches to infection control. Antimicrob. Agents Chemother. 2018, 62, e01689-18. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).