Multidrug-Resistant Staphylococcus sp. and Enterococcus sp. in Municipal and Hospital Wastewater: A Longitudinal Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites and Sample Collection
2.2. Microbial Analysis and Identification
2.3. Antimicrobial Susceptibility Testing
3. Results
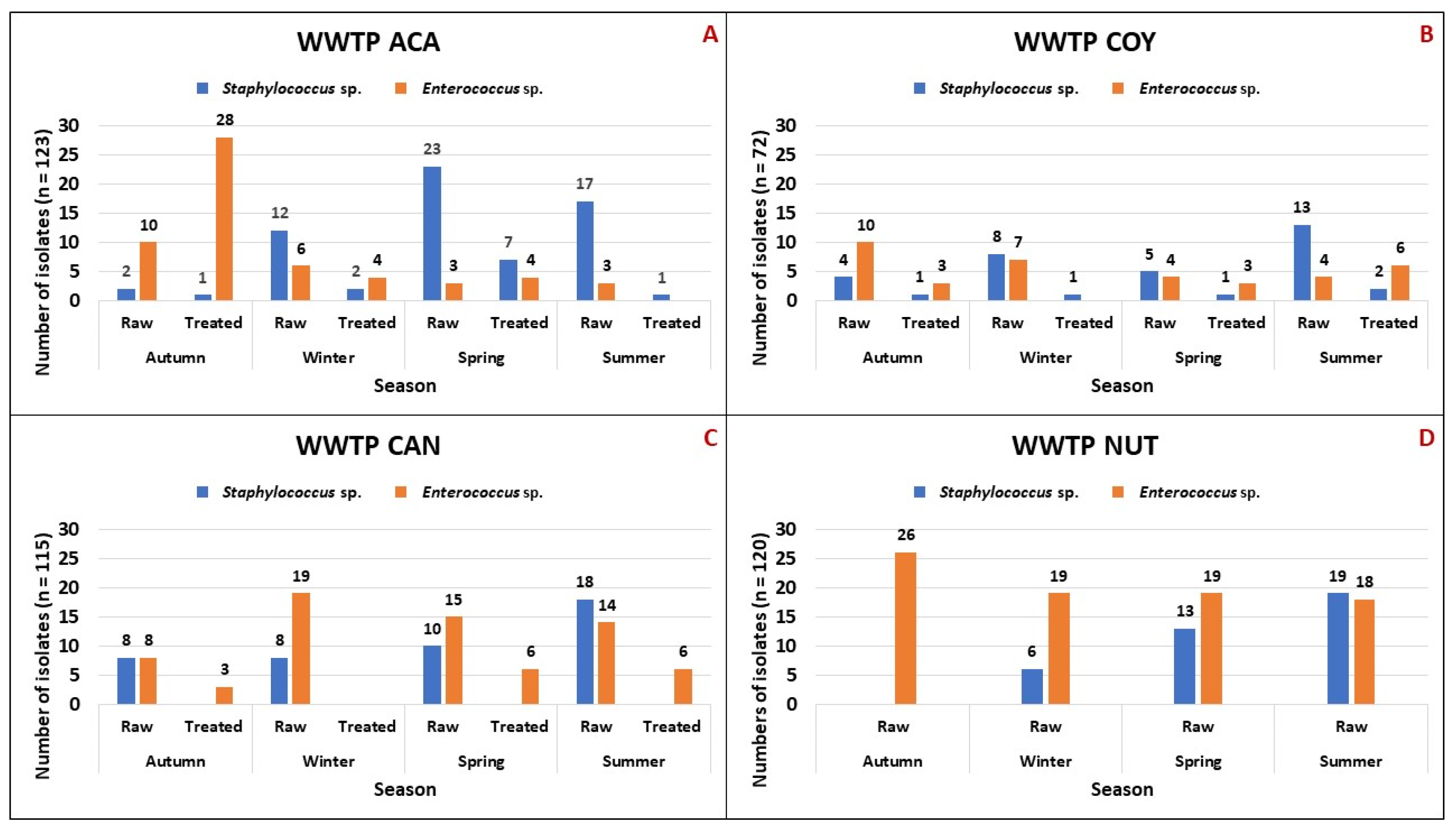
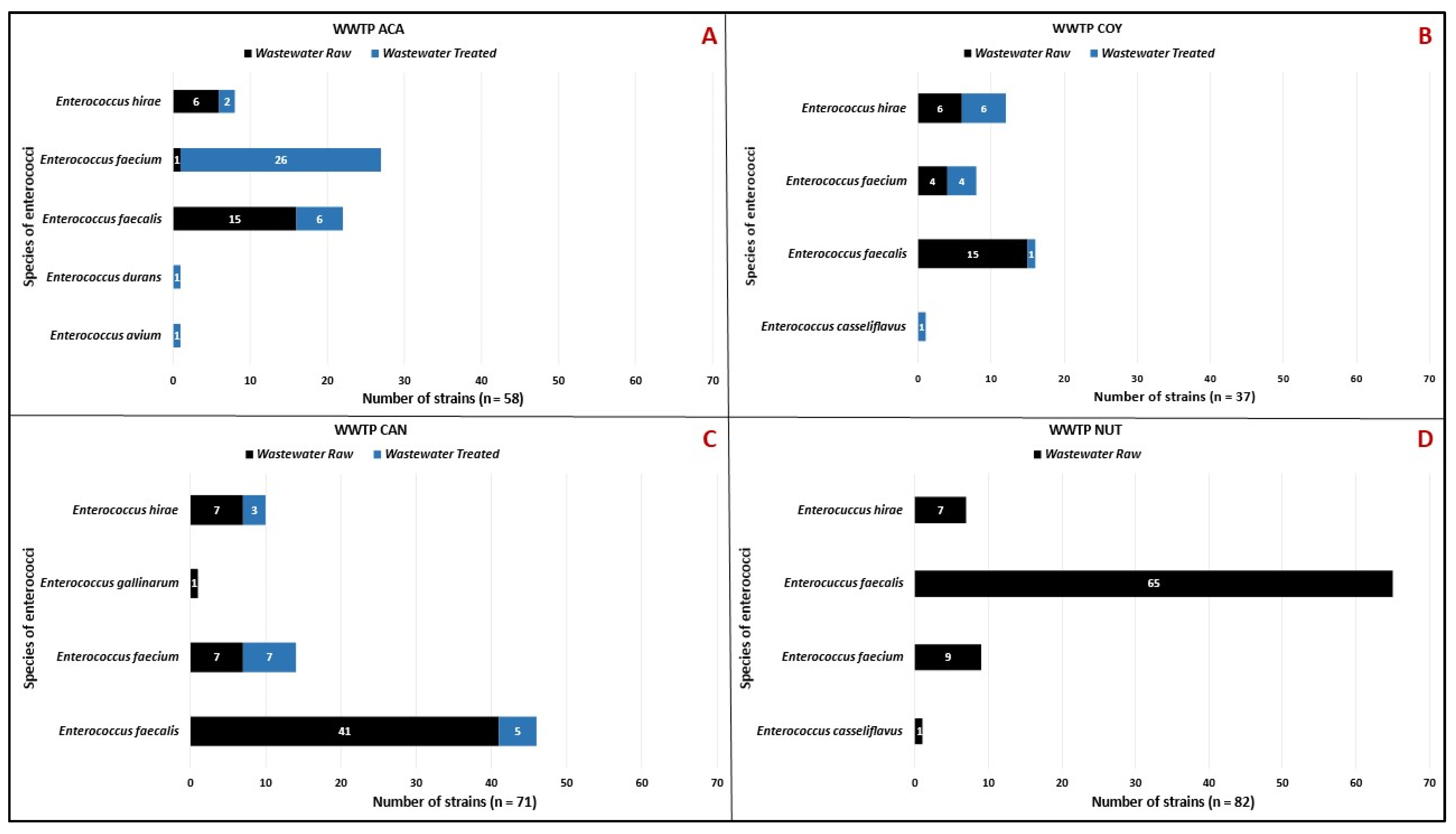
3.1. Staphylococcus sp. and Enterococcus sp. Isolates
3.2. Antimicrobial Susceptibility of Staphylococcus sp. and Enterococcus sp. Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, B.; Webster, T.J. Bacteria Antibiotic Resistance: New Challenges and Opportunities for Implant-Associated Orthopedic Infections. J. Orthop. Res. 2018, 36, 22–32. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. 2014. Available online: https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (accessed on 15 December 2023).
- Bhatia, R. Antimicrobial Resistance: Threat, Consequences and Options. Natl. Med. J. India 2018, 31, 133–135. [Google Scholar] [CrossRef]
- Mauldin, P.D.; Salgado, C.D.; Hansen, I.S.; Durup, D.T.; Bosso, J.A. Attributable Hospital Cost and Length of Stay Associated with Health Care-Associated Infections Caused by Antibiotic-Resistant Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2010, 54, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Hutinel, M.; Larsson, D.G.J.; Flach, C.F. Antibiotic Resistance Genes of Emerging Concern in Municipal and Hospital Wastewater from a Major Swedish City. Sci. Total Environ. 2022, 812, 151433. [Google Scholar] [CrossRef] [PubMed]
- Pärnänen, K.M.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic Resistance in European Wastewater Treatment Plants Mirrors the Pattern of Clinical Antibiotic Resistance Prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Woolhouse, M.E.J. Using Sewage for Surveillance of Antimicrobial Resistance. Science 2020, 367, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Sims, N.; Kasprzyk-Hordern, B. Future Perspectives of Wastewater-Based Epidemiology: Monitoring Infectious Disease Spread and Resistance to the Community Level. Environ. Int. 2020, 139, 105689. [Google Scholar] [CrossRef]
- Hitzenbichler, F.; Simon, M.; Salzberger, B.; Hanses, F. Clinical Significance of Coagulase-Negative Staphylococci Other than S. epidermidis Blood Stream Isolates at a Tertiary Care Hospital. Infection 2017, 45, 179–186. [Google Scholar] [CrossRef]
- Garza-González, E.; López, D.; Pezina, C.; Muruet, W.; Bocanegra-García, V.; Muñoz, I.; Ramírez, C.; LLaca-Díaz, J.M. Diversity of Staphylococcal Cassette Chromosome Mec Structures in Coagulase-Negative Staphylococci and Relationship to Drug Resistance. J. Med. Microbiol. 2010, 59, 323–329. [Google Scholar] [CrossRef]
- Werner, G.; Couto, N.; Feil, E.J.; Novais, A.; Hegstad, K.; Howden, B.P.; Friedrich, A.W.; Reuter, S. Taking Hospital Pathogen Surveillance to the next Level. Microb. Genom. 2023, 9, 001008. [Google Scholar] [CrossRef]
- Bin Kim, Y.; Seo, K.W.; Shim, J.B.; Son, S.H.; Noh, E.B.; Lee, Y.J. Molecular Characterization of Antimicrobial-Resistant Enterococcus faecalis and Enterococcus faecium Isolated from Layer Parent Stock. Poult. Sci. 2019, 98, 5892–5899. [Google Scholar] [CrossRef]
- Kalode, V.; Patil, P. Enterococcus Species: A Systemic Review. J. Pure Appl. Microbiol. 2023, 17, 761–767. [Google Scholar] [CrossRef]
- Rincón, S.; Panesso, D.; Díaz, L.; Carvajal, L.P.; Reyes, J.; Munita, J.M.; Arias, C.A. Resistance to “Last Resort” Antibiotics in Gram-Positive Cocci: The Post-Vancomycin Era. Biomedica 2014, 34 (Suppl. 1), 191–208. [Google Scholar] [CrossRef] [PubMed]
- Gouliouris, T.; Raven, K.E.; Moradigaravand, D.; Ludden, C.; Coll, F.; Blane, B.; Naydenova, P.; Horner, C.; Brown, N.M.; Corander, J.; et al. Detection of Vancomycin-Resistant Enterococcus faecium Hospital-Adapted Lineages in Municipal Wastewater Treatment Plants Indicates Widespread Distribution and Release into the Environment. Genome Res. 2019, 29, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. Reveals Distinct Species and Antimicrobial Resistance Diversity across a One-Health Continuum. Sci. Rep. 2020, 10, 3937. [Google Scholar] [CrossRef] [PubMed]
- Zieliński, W.; Korzeniewska, E.; Harnisz, M.; Hubeny, J.; Buta, M.; Rolbiecki, D. The Prevalence of Drug-Resistant and Virulent Staphylococcus spp. in a Municipal Wastewater Treatment Plant and Their Spread in the Environment. Environ. Int. 2020, 143, 105914. [Google Scholar] [CrossRef]
- Omid, M.R.; Jamali, H.; Kafilzadeh, F.; Borjian, A.; Arzanlou, M. Occurrence of Staphylococcus spp. in the wastewaters from Iran: Diversity, Antimicrobial Resistance, and Virulence Potential. J. Water Health 2023, 21, 178–191. [Google Scholar] [CrossRef]
- Duncan, D.; Harvey, F.; Walker, M. EPA Guidelines: Regulatory Monitoring and Testing. Water and Wastewater Sampling; 2007; Volume 3, ISBN 9781921125478. Available online: https://www.epa.sa.gov.au/files/8494_guide_wws.pdf (accessed on 15 December 2023).
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI Supplement M100; CLSI: Wayne, PA, USA, 2022; ISBN 978-1-68440-033-1. [Google Scholar]
- Kang, X.H.; Leng, Y.; O, M.M.; Zeng, X.Y.; Li, S.W. The Seasonal Changes of Core Bacterial Community Decide Sewage Purification in Sub-Plateau Municipal Sewage Treatment Plants. Bioprocess Biosyst. Eng. 2020, 43, 1609–1617. [Google Scholar] [CrossRef]
- He, Y.; Li, K.X.; Wang, J.W.; Wang, W.; Fan, P.C.; Chen, H.H.; Wang, J.J. Microbial Community Structure of Wastewater Treatment Plants in Different Seasons. Huanjing Kexue/Environ. Sci. 2021, 42, 1488–1495. [Google Scholar] [CrossRef]
- Regasa Dadi, B.; Girma, E.; Tesfaye, M.; Seid, M. Assessment of the Bacteriological Profile and Antibiotic Susceptibility Patterns of Wastewater in Health Facilities of Ethiopia. Int. J. Microbiol. 2021, 2021, 9969479. [Google Scholar] [CrossRef]
- Gómez, P.; Lozano, C.; Benito, D.; Estepa, V.; Tenorio, C.; Zarazaga, M.; Torres, C. Characterization of Staphylococci in Urban Wastewater Treatment Plants in Spain, with Detection of Methicillin Resistant Staphylococcus aureus ST398. Environ. Pollut. 2016, 212, 71–76. [Google Scholar] [CrossRef]
- Heß, S.; Gallert, C. Demonstration of Staphylococci with Inducible Macrolide-Lincosamide-Streptogramin B (MLSB) Resistance in Sewage and River Water and of the Capacity of Anhydroerythromycin to Induce MLSB. FEMS Microbiol. Ecol. 2014, 88, 48–59. [Google Scholar] [CrossRef][Green Version]
- Faria, C.; Vaz-Moreira, I.; Serapicos, E.; Nunes, O.C.; Manaia, C.M. Antibiotic Resistance in Coagulase Negative Staphylococci Isolated from Wastewater and Drinking Water. Sci. Total Environ. 2009, 407, 3876–3882. [Google Scholar] [CrossRef] [PubMed]
- Adesoji, T.O.; Egyir, B.; Shittu, A.O. Antibiotic-Resistant Staphylococci from the Wastewater Treatment Plant and Grey-Water Samples in Obafemi Awolowo University, Ile-Ife, Nigeria. J. Water Health 2020, 18, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Alduhaidhawi, A.H.M.; AlHuchaimi, S.N.; Al-Mayah, T.A.; Al-Ouqaili, M.T.S.; Alkafaas, S.S.; Muthupandian, S.; Saki, M. Prevalence of CRISPR-Cas Systems and Their Possible Association with Antibiotic Resistance in Enterococcus faecalis and Enterococcus faecium Collected from Hospital Wastewater. Infect. Drug Resist. 2022, 15, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Gotkowska-Płachta, A. The Prevalence of Virulent and Multidrug-Resistant Enterococci in River Water and in Treated and Untreated Municipal and Hospital Wastewater. Int. J. Environ. Res. Public Health 2021, 18, 563. [Google Scholar] [CrossRef] [PubMed]
- Molale-Tom, L.G.; Bezuidenhout, C.C. Prevalence, Antibiotic Resistance and Virulence of Enterococcus spp. from Wastewater Treatment Plant Effluent and Receiving Waters in South Africa. J. Water Health 2020, 18, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, H.; Ortega-Polo, R.; Zaheer, R.; Goji, N.; Amoako, K.K.; Brown, R.S.; Majury, A.; Liss, S.N.; McAllister, T.A. Comparative Genomics of Multidrug-Resistant Enterococcus spp. Isolated from Wastewater Treatment Plants. BMC Microbiol. 2020, 20, 20. [Google Scholar] [CrossRef] [PubMed]
- Patel, T.; Lewis, M.E.; Niesley, M.L.; Chowdhury, M. Postneurosurgical Central Nervous System Infection due to Enterococcus faecalis Successfully Treated with Intraventricular Vancomycin. Infect. Dis. Clin. Pract. 2016, 24, 174–176. [Google Scholar] [CrossRef][Green Version]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-Resistant Pathogens Associated with Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef]
- Tiwari, A.; Oliver, D.M.; Bivins, A.; Sherchan, S.P.; Pitkänen, T. Bathing Water Quality Monitoring Practices in Europe and the United States. Int. J. Environ. Res. Public Health 2021, 18, 5513. [Google Scholar] [CrossRef]
- Tiwari, A.; Hokajärvi, A.M.; Santo Domingo, J.W.; Kauppinen, A.; Elk, M.; Ryu, H.; Jayaprakash, B.; Pitkänen, T. Categorical Performance Characteristics of Method ISO 7899-2 and Indicator Value of Intestinal Enterococci for Bathing Water Quality Monitoring. J. Water Health 2018, 16, 711–723. [Google Scholar] [CrossRef]
- Tiwari, A.; Krolicka, A.; Tran, T.T.; Räisänen, K.; Ásmundsdóttir, Á.M.; Wikmark, O.G.; Lood, R.; Pitkänen, T. Antibiotic Resistance Monitoring in Wastewater in the Nordic Countries: A Systematic Review. Environ. Res. 2024, 246, 118052. [Google Scholar] [CrossRef]
- WHO. The 2019 WHO AWaRe Classification of Antibiotics for Evaluation and Monitoring of Use. Available online: https://www.who.int/publications/i/item/WHOEMPIAU2019.11 (accessed on 18 July 2022).
- Kumar, H.; Palaha, R.; Kaur, N.; Ratnakar, W.S.; Sodi, A.; Kaur, M.; Katiyar, R.; Sharma, M.; Kaur, C.; Kumar, V. Prevalence of Multidrug-Resistant, Coagulase-Positive Staphylococcus aureus in Nasal Carriage, Food, Wastewater and Paper Currency in Jalandhar City (North-Western), an Indian State of Punjab. Environ. Monit. Assess. 2015, 187, 4134. [Google Scholar] [CrossRef]
- Korzeniewska, E.; Harnisz, M. Relationship between Modification of Activated Sludge Wastewater Treatment and Changes in Antibiotic Resistance of Bacteria. Sci. Total Environ. 2018, 639, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Jendrzejewska, N.; Karwowska, E. Bacterial Resistance to β-Lactam Antibiotics in Municipal Wastewater: Insights from a Full-Scale Treatment Plant in Poland. Microorganisms 2022, 10, 2323. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg Goldstein, R.E.; Micallef, S.A.; Gibbs, S.G.; Davis, J.A.; He, X.; George, A.; Kleinfelter, L.M.; Schreiber, N.A.; Mukherjee, S.; Sapkota, A.; et al. Methicillin-Resistant Staphylococcus aureus (MRSA) Detected at Four U.S. Wastewater Treatment Plants. Environ. Health Perspect. 2012, 120, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Börjesson, S.; Melin, S.; Matussek, A.; Lindgren, P.E. A Seasonal Study of the MecA Gene and Staphylococcus aureus Including Methicillin-Resistant, S. aureus in a Municipal Wastewater Treatment Plant. Water Res. 2009, 43, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.; Chou, C. Class 1 Integrons and the Antiseptic Resistance Gene (QacEΔ1) in Municipal and Swine Slaughterhouse Wastewater Treatment Plants and Wastewater—Associated Methicillin-Resistant Staphylococcus aureus. Int. J. Environ. Res. Public Health 2015, 12, 6249–6260. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.M.; Gündoǧdu, A.; Stratton, H.M.; Katouli, M. Antibiotic Resistant Staphylococcus aureus in Hospital Wastewaters and Sewage Treatment Plants with Special Reference to Methicillin-Resistant Staphylococcus aureus (MRSA). J. Appl. Microbiol. 2013, 114, 44–54. [Google Scholar] [CrossRef]
- Nogueira, S.P.; Andrade, S.L.; Magalhães, P.F.C., Jr.; Procopio, R.E.L. Antibiotic Resistance of Staphylococcus spp. Isolated from Sewage in Manaus, Amazonas. Eur. J. Med. Health Sci. 2021, 3, 134–137. [Google Scholar] [CrossRef]
- Hanna, N.; Sun, P.; Sun, Q.; Li, X.; Yang, X.; Ji, X.; Zou, H.; Ottoson, J.; Nilsson, L.E.; Berglund, B.; et al. Presence of Antibiotic Residues in Various Environmental Compartments of Shandong Province in Eastern China: Its Potential for Resistance Development and Ecological and Human Risk. Environ. Int. 2018, 114, 131–142. [Google Scholar] [CrossRef]
- Limayem, A.; Wasson, S.; Mehta, M.; Pokhrel, A.R.; Patil, S.; Nguyen, M.; Chen, J.; Nayak, B. High-Throughput Detection of Bacterial Community and Its Drug-Resistance Profiling from Local Reclaimed Wastewater Plants. Front. Cell. Infect. Microbiol. 2019, 9, 303. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.H.; Alsallaq, R.; Parsons, J.B.; Ferrolino, J.; Hayden, R.T.; Rubnitz, J.E.; Rafiqullah, I.M.; Robinson, D.A.; Margolis, E.B.; Rosch, J.W.; et al. Vancomycin Heteroresistance and Clinical Outcomes in Bloodstream Infections Caused by Coagulase-Negative Staphylococci. Antimicrob. Agents Chemother. 2020, 64, e00944-20. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, P.B.; Massinhani, F.H.; dos Santos, K.R.N.; Chamon, R.C.; Silva, R.B.; Correa, F.E.L.; da Cunha Hueb Barata Oliveira, C.; Oliveira, A.G. Methicillin-Resistant Staphylococcus epidermidis Isolates with Reduced Vancomycin Susceptibility from Bloodstream Infections in a Neonatal Intensive Care Unit. J. Med. Microbiol. 2020, 69, 41–45. [Google Scholar] [CrossRef]
- Sakimura, T.; Kajiyama, S.; Adachi, S.; Chiba, K.; Yonekura, A.; Tomita, M.; Koseki, H.; Miyamoto, T.; Tsurumoto, T.; Osaki, M. Biofilm-Forming Staphylococcus epidermidis Expressing Vancomycin Resistance Early after Adhesion to a Metal Surface. Biomed Res. Int. 2015, 2015, 943056. [Google Scholar] [CrossRef]
- Crespo-Ortega, L.; Bonilla-Hernández, R.; Pedraza, A.; Lisker, A. Endocarditis Infecciosa Por Staphylococcus lentus. An. Médicos Asoc. Médica Cent. Médico ABC 2022, 67, 304–308. [Google Scholar] [CrossRef]
- Hay, C.Y.; Sherris, D.A. Staphylococcus lentus Sinusitis: A New Sinonasal Pathogen. Ear Nose Throat J. 2020, 99, NP62–NP63. [Google Scholar] [CrossRef]
- Hernández Sarmiento, R.; Álvarez Olmos, M.; Aguilera Martínez, S. Meningitis por Staphylococcus lentus Resistente a Meticilina Asociado a Derivación Ventriculoperitoneal en Lactante Menor. Pediatria 2019, 52, 118–121. [Google Scholar] [CrossRef]
- Rivera, M.; Dominguez, M.D.; Mendiola, N.R.; Roso, G.R.; Quereda, C. Staphylococcus lentus Peritonitis: A Case Report. Perit. Dial. Int. 2014, 34, 469–470. [Google Scholar] [CrossRef]
- Shaker, M.N.; Hmdan, T.A.; Issa, A.H. Isolation and Diagnosis of Staphylococcus lentus from Different Operation Theater Hospitals. Sci. J. Med. Res. 2018, 2, 177–181. [Google Scholar] [CrossRef]
- Nishiyama, M.; Praise, S.; Tsurumaki, K.; Baba, H.; Kanamori, H.; Watanabe, T. Prevalence of Antibiotic-Resistant Bacteria ESKAPE among Healthy People Estimated by Monitoring of Municipal wastewater. Antibiotics 2021, 10, 495. [Google Scholar] [CrossRef] [PubMed]
- Iweriebor, B.C.; Gaqavu, S.; Obi, L.C.; Nwodo, U.U.; Okoh, A.I. Antibiotic Susceptibilities of Enterococcus Species Isolated from Hospital and Domestic Wastewater Effluents in Alice, Eastern Cape Province of South Africa. Int. J. Environ. Res. Public Health 2015, 12, 4231–4246. [Google Scholar] [CrossRef] [PubMed]
- Hricová, K.; Röderová, M.; Fryčák, P.; Pauk, V.; Kurka, O.; Mezerová, K.; Štosová, T.; Bardoň, J.; Milde, D.; Kučová, P.; et al. Prevalence of Vancomycin-Resistant Enterococci and Antimicrobial Residues in Wastewater and Surface Water. Life 2021, 11, 1403. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, Z.; Hu, J.; Wang, B.; Rong, H.; Li, Z.; Sun, Y.; Wang, Y.; Zhang, X.; Wang, M.; et al. Evaluation of Culturable ‘Last-Resort’ Antibiotic Resistant Pathogens in Hospital Wastewater and Implications on the Risks of Nosocomial Antimicrobial Resistance Prevalence. J. Hazard. Mater. 2022, 438, 129477. [Google Scholar] [CrossRef] [PubMed]
- Bender, J.K.; Cattoir, V.; Hegstad, K.; Sadowy, E.; Coque, T.M.; Westh, H.; Hammerum, A.M.; Schaffer, K.; Burns, K.; Murchan, S.; et al. Update on Prevalence and Mechanisms of Resistance to Linezolid, Tigecycline and Daptomycin in Enterococci in Europe: Towards a Common Nomenclature. Drug Resist. Updat. 2018, 40, 25–39. [Google Scholar] [CrossRef]
- Jahne, M.A.; Rogers, S.W.; Ramler, I.P.; Holder, E.; Hayes, G. Hierarchal Clustering Yields Insight into Multidrug-Resistant Bacteria Isolated from a Cattle Feedlot Wastewater Treatment System. Environ. Monit. Assess. 2015, 187, 4168. [Google Scholar] [CrossRef]
- Freitas, A.R.; Elghaieb, H.; León-Sampedro, R.; Abbassi, M.S.; Novais, C.; Coque, T.M.; Hassen, A.; Peixe, L. Detection of OptrA in the African Continent (Tunisia) within a Mosaic Enterococcus faecalis Plasmid from Urban Wastewaters. J. Antimicrob. Chemother. 2017, 72, 3245–3251. [Google Scholar] [CrossRef]
- Szewczyk, E.M.; Nowak, T.; Cieślikowski, T.; Rózalska, M. Potential Role of Staphylococcus cohnii in a Hospital Environment. Microb. Ecol. Health Dis. 2003, 15, 51–56. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velazquez-Meza, M.E.; Galarde-López, M.; Cornejo-Juárez, P.; Carrillo-Quiroz, B.A.; Velázquez-Acosta, C.; Bobadilla-del-Valle, M.; Ponce-de-León, A.; Alpuche-Aranda, C.M. Multidrug-Resistant Staphylococcus sp. and Enterococcus sp. in Municipal and Hospital Wastewater: A Longitudinal Study. Microorganisms 2024, 12, 645. https://doi.org/10.3390/microorganisms12040645
Velazquez-Meza ME, Galarde-López M, Cornejo-Juárez P, Carrillo-Quiroz BA, Velázquez-Acosta C, Bobadilla-del-Valle M, Ponce-de-León A, Alpuche-Aranda CM. Multidrug-Resistant Staphylococcus sp. and Enterococcus sp. in Municipal and Hospital Wastewater: A Longitudinal Study. Microorganisms. 2024; 12(4):645. https://doi.org/10.3390/microorganisms12040645
Chicago/Turabian StyleVelazquez-Meza, Maria Elena, Miguel Galarde-López, Patricia Cornejo-Juárez, Berta Alicia Carrillo-Quiroz, Consuelo Velázquez-Acosta, Miriam Bobadilla-del-Valle, Alfredo Ponce-de-León, and Celia Mercedes Alpuche-Aranda. 2024. "Multidrug-Resistant Staphylococcus sp. and Enterococcus sp. in Municipal and Hospital Wastewater: A Longitudinal Study" Microorganisms 12, no. 4: 645. https://doi.org/10.3390/microorganisms12040645
APA StyleVelazquez-Meza, M. E., Galarde-López, M., Cornejo-Juárez, P., Carrillo-Quiroz, B. A., Velázquez-Acosta, C., Bobadilla-del-Valle, M., Ponce-de-León, A., & Alpuche-Aranda, C. M. (2024). Multidrug-Resistant Staphylococcus sp. and Enterococcus sp. in Municipal and Hospital Wastewater: A Longitudinal Study. Microorganisms, 12(4), 645. https://doi.org/10.3390/microorganisms12040645






