Abstract
The Pangong Tso lake is a high-altitude freshwater habitat wherein the resident microbes experience unique selective pressures, i.e., high radiation, low nutrient content, desiccation, and temperature extremes. Our study attempts to analyze the diversity of culturable bacteria by applying a high-throughput amplicon sequencing approach based on long read technology to determine the spectrum of bacterial diversity supported by axenic media. The phyla Pseudomonadota, Bacteriodetes, and Actinomycetota were retrieved as the predominant taxa in both water and sediment samples. The genera Hydrogenophaga and Rheinheimera, Pseudomonas, Loktanella, Marinomonas, and Flavobacterium were abundantly present in the sediment and water samples, respectively. Low nutrient conditions supported the growth of taxa within the phyla Bacteriodetes, Actinomycetota, and Cyanobacteria and were biased towards the selection of Pseudomonas, Hydrogenophaga, Bacillus, and Enterococcus spp. Our study recommends that media formulations can be finalized after analyzing culturable diversity through a high-throughput sequencing effort to retrieve maximum species diversity targeting novel/relevant taxa.
1. Introduction
Freshwater lakes are an ideal spot for understanding the critical role of the microbiome in ecosystem shaping [1]. Altitude and salinity are some of the major deterministic factors affecting patterns of microbial community of a lake ecosystem [2,3,4]. High-altitude lakes are comparatively pristine with a unique mix of microbial diversity selected by severe environmental pressures (i.e., radiation, temperatures, nutrient fluctuations, etc.) that reduce species richness and increase prokaryotic dominance [5,6,7,8,9]. Recently, psychrophilic and psychrotolerant microorganisms of cold habitats have drawn tremendous attention due to the complex secondary metabolisms assisting their survival, adaptation, and evolution. Perhaps these cold habitats could be an asset of new taxa, genes, and metabolites with remarkable biotechnological applications [10,11,12]. Thus, a detailed study is required to gain information about the relationship between lacustrine ecosystem function and their resident microbial diversity.
The Himalayan range lakes are one of the most fragile and unique ecosystems, with 4699 lakes being documented in the region according to an ISRO study [13]. Pangong Tso, the largest high altitude brackish water lake, possesses multiple freeze–thaw cycles, nutrient scarcity, salinity, and high UV radiation, which makes it the most stressed ecosystem in the Indian Himalayan region. Microbial diversity analysis of the lake has been previously undertaken using both culturable and molecular approaches. Yadav et al. [14] described the composition of psychrotrophic bacteria by using traditional cultivation-based approaches and identified the phylum Bacillota (genus Bacillus) to be predominant with other reports targeting psychrotrophic bacteria in an array of biotechnological applications and their role in biogeochemical cycles [15,16] Culture-independent analysis has identified Pseudomonadota (genera Loktanella, Rhizobium, Marinobacter, Pseudomonas, Methylophaga) as the dominant taxa followed by Bacteroidota (genus Gramella) and Bacillota (genus Zunogwangia) [16,17,18] and an analysis of a soil–water mix collected beneath frozen ice revealed the dominance of Bacteroidota followed by Bacillota and Pseudomonadota [19]. Further, in high-altitude lakes (Pangong Tso, Dashair, Gurudongmar, Chandratal, Dal) of the Indian trans-Himalayan region [20] and in other continents (Spain, Dry-Andes, Australian Alps, and California [21,22,23,24]), similar patterns of limited bacterial diversity have been reported. However, despite these insights, routine culturable strategies are still important for the isolation, taxonomic, and metabolic characterization of pure strains as they decipher the key roles in biogeochemical cycles, functional activities in plant growth promotion, and cold active enzymes. One of the biggest limitations of studying structural and functional microbial diversity is the inability to cultivate the majority of bacteria [25]. To address these knowledge gaps, we have combined high-throughput next-generation sequencing based on long read technology with conventional cultivation techniques to understand the bacterial diversity of the lake ecosystem and how media formulation affects the scale of retrieval.
2. Materials and Methods
2.1. Sample Collection
The Pangong Tso lake is located in the Eastern part of Leh, Ladakh (33°57.560′ N and 078°25.627′ E), at a height of about 4350 m A.S.L. (above mean sea level). It extends from India to China, with only 40% of its length in India. Samples (both sediment and water) were collected in the month of September 2017 in 50 mL sterile falcon tubes (Tarsons, Figure S1) and whirl pack bags (1 L, Hi-Media, Thane, MH, India). Samples were transported to the laboratory within 48 h after collection. No permit was required for sample collection and it was performed with support from the local DRDO laboratory.
2.2. Physico-Chemical Parameters
The pH of the samples (water and sediment) was measured on-site using pH strips [Hi-Media], which were further analyzed in the laboratory using a pH meter [(EUTECH instruments pH 700)]. The temperatures of the water and sediment samples were recorded using a thermometer during sample collection. TDS, SO42−, ammoniacal nitrogen, NO3, NO2, and Cl were analyzed as per standard protocols [26]. The analyses of Ca, Mg, Zn, Mn, Fe, B, Cu, Co, Mo, Cd, Cr, Al, B, Ni, Pb, Hg, Li, Va, Se, Si, and As were followed as per AOAC 990.08 standards [27]. TOC, TKN, conductivity, and salinity were recorded as per IS 2720 [28], AOAC 955.04 [29], APHA 2510B [30], and APHA 2520B [31] standards.
2.3. Sample Processing and Community DNA Isolation
About 1 g of sediment sample (PSD) was diluted in 10 mL of autoclaved distilled water and subjected to serial dilution by a factor of 10−1 to 10−7. Water samples (PW) were filtered through a 0.22 µm filter (Merck Millipore Ltd., Darmstadt, Germany) and the filter was suspended in 10 mL autoclaved distilled water in 50 mL falcon tubes (Tarsons, Kolkata, India), vortexed briefly, and serial dilutions were prepared (10−1 to 10−7). About 0.1 mL of the sediment and water dilutions was plated in duplicates on three types of media formulation prepared at two different pH levels (8.0 and 9.0), i.e., Reasoner’s 2A broth (R2A; Hi-Media), R2A broth, and tryptone soya broth (TSB, Hi-Media) diluted 100 times (R2A100) and 500 times (TSBA500) (Table S5), respectively, with distilled water, solidified with 1.5% (w/v) extra-pure agar (Hi-Media). The plates were incubated at 25 °C for up to 8 weeks considering slow growing bacteria, and the colony forming units (CFUs) were recorded. After incubation, colonies were flooded with sterile distilled water (2–5 mL) and bacterial suspensions were pooled together from all the dilutions (10−1 to 10−7) and pH (8.0–9.0), for all the media (Figure 1).
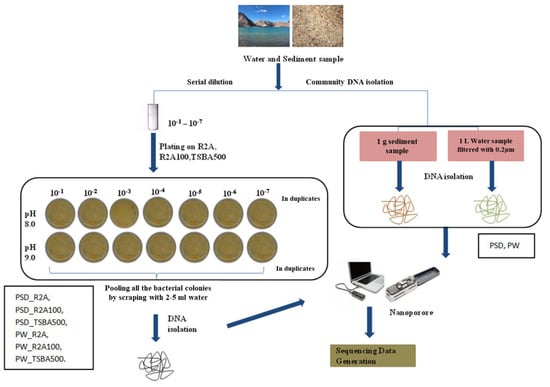
Figure 1.
Flowchart representing strategy of microbial diversity analysis of Pangong lake water and sediment samples.
Each pooled sample (PW_R2A, PW_R2A100, PW_TSBA500, PSD_R2A, PSD_R2A100 and PSD_TSBA500) and un-plated sediment (PSD)–water samples (PW), hereafter referred to as media and direct samples, respectively, were processed for DNA isolation using a FastDNA spin kit according to the manufacturer’s recommendations (MP Biomedical India Pvt. Ltd., Navi Mumbai, India).
2.4. Sequencing and Data Analysis
The total community DNA from all samples was subjected to 16S rRNA gene amplification with forward primer (5′-AGAGTTTGATCMTGGCTCAG-3′) and reverse primer (5′-CGGTTACCTTGTTACGACTT-3′) targeting the full-length gene (1.5 Kbp) amplification covering regions V1-V9 with LongAmp Taq 2× master mix (NEB). The thermocycling procedure involved an initial denaturation step at 95 °C for 1 min (1 cycle) followed by 25 cycles at 95 °C for 20 s, 25 cycles at 55 °C for 30 s (annealing), 25 cycles at 65 °C for 2 min (extension), and a final extension at 65 °C for 5 min (1 cycle). The PCR products were purified by using 1× Ampure XP beads (Beckmann Coulter, Brea, CA, USA). The purified PCR amplicons from each sample were pooled at an equimolar concentration. The pooled barcoded samples were then subjected to sequencing adapter ligation using the 16S Barcoding Kit (SQK-RAB204). Sequencing was performed on MinION Mk1b (Oxford Nanopore Technologies, Oxford, UK) using SpotON flow cell (FLO-MIN107) in a 48 h sequencing protocol on MinKNOW 1.10.11 with an error rate of 6–8%. Albacore v2.3.4 was used for basecalling with default parameters, which filters the reads having a Q score more than 7 that were considered as pass data and demultiplexed. The trimming of adapter sequences and chimeric sequences of raw data was performed using the Porechop tool and fastp reads were converted to fasta format using the seqtk tool. The reads were taxonomically assigned using Kraken2 (Galaxy Version 2.1.1) [32] using the RDP database (2020) with the default parameters. Data visualization was performed with the Pavian metagenomic tool [33]. In order to compare the bacterial diversity indices, PAST software version 3.1 [34] was used for the statistical analysis. The Shannon, Simpson, chao-1 diversity indices, and test of significant differences were performed using the Euclidean square root of the sum of the square distance clustering method. Principle component analysis (PCA) for abundant data normalization at the genus and species level was displayed by using the clustvis web tool [35]. For clustering analysis, the taxa with less than <0.1% were excluded from further analysis. For an analysis of shared and unique taxa between different samples, jvenn2 was used [36]. Differential abundance was plotted using GraphPad Prism 8 software to determine statistical significance using Mann–Whitney and p value with Bonferroni correction as the number of samples was uneven. Microbial genera with p value < 0.005 were considered significant. For the comparative phylogenetic analysis, the reads of all 8 samples analyzed in this study along with six samples of Chaudhari et al. (CH1–CH6) [18], one sample each of Bisht et al. (BI) [19], and Rathour et al. (RA) [16] were classified through Kraken2 using RDP (RDP database version 2020). The obtained classified reads output was concatenated using Pavian [33] and used as an input in MEGAN (MEGAN CE V6.17.0) to visualize the phylogenetic tree along with the quantitative data of each taxa.
3. Results and Discussion
3.1. Enumeration of Bacteria on Microbiological Media
Table 1 highlights CFU obtained on different microbiological media. Significant variations in the CFU were observed among the different media, pH, and sample types. The CFU/mL varied from 3 × 103–8.6 × 104 in the water sample to 2.3 × 104–3.1 × 105 CFU/g in the sediment sample. At pH 8.0, R2A agar supported the highest number of colonies for both water (8.6 × 104 CFU/mL) and sediment (3.1 × 105 CFU/g) samples, whereas at pH 9.0, R2A100 times diluted medium recovered the maximum number of colonies in the water (6.8 × 104 CFU/mL) and sediment (7.8 × 104 CFU/g) sample (Table 1).

Table 1.
Average CFU of water and sediment samples on different media at pH 8.0 and pH 9.0.
3.2. Physico-Chemical Analysis
Pangong lake is a brackish water, high-altitude lake, which remains frozen during winter for three months (December, January, and February, Bhat et al. [19]). The lake ecosystem faces extreme environmental conditions such as high salinity, alkalinity, electrical conductivity, and temperature in which microbes are able to survive with adaptive strategies [37]. At 0.12%, a negligible amount of TOC was detected, thus reflecting the oligotrophic conditions of the ecosystem. Both sediment and water samples were alkaline with respective pH values measured as 8.32 (1.565 mΩ/cm electrical conductivity) and pH 8.0–9.0 (Table S1), confirming the earlier findings of Bhat et al. [19], Rathour et al. [16], and Chaudhari et al. [18]. Metals such as calcium, magnesium, zinc, manganese, iron, boron, copper, cobalt, molybdenum, cadmium, chromium, aluminum, barium, nickel, lead, mercury, arsenic, chloride, lithium, vanadium, selenium, silicon, phosphorus, sulphate, ammoniacal nitrogen, nitrate nitrogen, and nitrite in the sediment sample were within permissible limits (Table S1) as per the WHO guidelines (https://www.omicsonline.org/articles-images/2161-0525-5-334-t011.html, accessed on 3 May 2023) and Alloway, B. J. [38].
3.3. Analysis of Bacterial Diversity by 16S rRNA Gene-Based Tag Sequencing
In traditional culturable techniques, only replicating cells that can form visually well-separated colonies can be sub-cultured, whereas abiotic parameters, i.e., temperature, pH, UV, nutrients, and absence of cell-to-cell communication may affect bacterial growth by forcing cells into dormancy, called the viable but non-culturable (VBNC) state [39,40]. These microbes are metabolically active but cannot form colonies on conventional media and grow only under favorable environmental conditions [39,41]. Further, it is a well-established fact that no bacteria types from an environmental sample can be cultured and identified [25,39,42], and conventional culturing processes are labor-intensive as well as logistically and economically intensive; therefore, in this study, the direct sequencing of colonies/cells through long read technology grown on media samples (i.e., total CFU on R2A, R2A100, and TSBA500) was compared with direct samples (PW, PSD), wherein total DNA was extracted from water and sediment samples directly to ascertain the effect of media formulations on the culturability pattern and to compare their effectiveness in retrieving the in situ diversity (Figure 1). Moreover, since the majority of the reads analyzed in all the samples were nearly full length (Table S2), the identification of classified reads at the genus level could be achieved with more confidence.
A total of 230,129 reads were obtained in the range of 11,795 to 45,633 (Table S2). Among the direct samples, i.e., PSD and PW, 100% of reads were classified, whereas for the media samples, these were in the range of 100% to 69.1% (Table S3). The classified reads belonging to the phyla Pseudomonadota, Bacteroidota, Cyanobacteria, Bacillota, Verrucomicrobiota, and Actinomycetota were shared by both PW and PSD samples, while the phyla Planctomycetota, Deinococcus-Thermus, Acidobacteriota, Gemmatimonadota, Hydrogenedentes, and Nitrospirota were only detected in the PSD sample, indicating that the sediment was most diverse (Figure 2; Table S3).
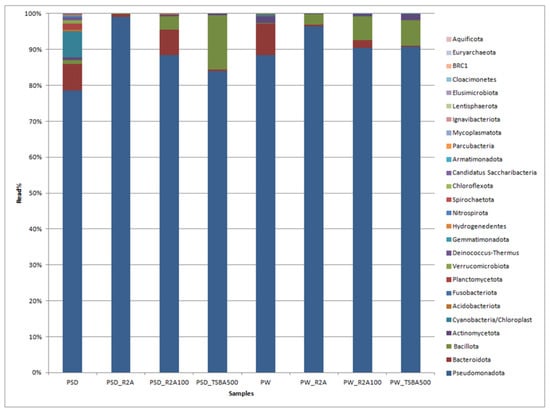
Figure 2.
Bar graph displaying the taxonomic distribution of bacterial diversity at the phyla level in direct and media samples.
It has been generally observed that sediments host a diverse bacterial composition due to the complex and heterogeneous gradients of substrate, pH, and redox potential, forming several microhabitats consisting of a vast matrix of organic and inorganic solid surfaces for bacterial growth [43,44,45]. The phyla Bacteroidota, Actinomycetota, and Cyanobacteria were relatively more abundant in PW and PSD compared to culture media samples (Figure 2; Table S3). Cyanobacteria were detected in direct samples (PSD, PW) and recovered only in diluted media (PW_R2A100, PW_TSBA500), indicating that low nutrient conditions were mandatory for their isolation. They are involved in key roles as autotrophic primary producers in carbon and nitrogen cycles through photosynthesis and nitrogen fixation, forming the base of the aquatic food chain and maintaining the ecology of the lake ecosystem [46,47]. Interestingly, for both the water and sediment samples, diluted media seemed to capture a higher diversity at the phyla level, which is evident by the detection of Actinomycetota in the sediment samples and Cyanobacteria in the water samples (Table S3). Some of the bacterial groups, i.e., Planctomycetota, Verrucomicrobiota, Acidobacteriota, and Gemmatimonadota, were recovered only in the direct samples (PSD and PW) and not in any of the culture media, with the latter three phyla detected only in the sediment sample (PSD; Table S3). It is a well-known fact that some of these taxa form part of the PVC superphylum and are extremely slow growing, very difficult to culture (require special media compositions), and several representatives of these lineages have only been recovered as metagenome assembled genomes (MAGs) [48]. It is also pertinent to note that Bacillota and Bacteriodetes were over- and under-represented, respectively, in the majority of the cultured media, thus undermining the biasness of the axenic media towards certain groups (Table S3). The Deinococcus-Thermus group detected only in the PSD sample withstands high temperatures and extreme UV radiation [49] as the Pangong Tso lake undergoes frequent temperature shifts and high UV radiation [7] due to its location at an extremely high altitude. Similar patterns of diversity with Pseudomonadota as the dominant phylum followed by Bacteroidota have been reported from sediment samples of the lake, although there is substantial variation in terms of the relative abundance of these taxa, which might be because of different sequencing and sampling methods [16,18].
At the class level, Gammaproteobacteria was most predominant in sample PW, whereas both Gamma- and Betaproteobacteria dominated the PSD sample, with the relative abundance of Alphaproteobacteria being much higher in PW compared to PSD, while Deltaproteobacteria was unique to the PSD sample (Table S3). It was interesting to observe this distribution pattern as previous reports of the lake have indicated a much higher abundance of either Gammaproteobacteria [16] or Gammaproteobacteria and Alphaproteobacteria [18], with Betaproteobacteria represented as a minor population. Abundant genera (≥1%) in PSD sample were Hydrogenophaga (~15.7%), Acinetobacter (4.6%), Pseudomonas (4.5%), Arenimonas (2.1%) followed by Comamonas, Stenotrophomonas, Rheinheimera, Methylotenera, Opitutus, Truepera, Rhodoferax, Variovorax, Burkholderia, Delftia, Acidovorax, Thiobacillus, and Lysobacter, while the top five genera in the PW sample were Rheinheimera (20.7%), Pseudomonas (15.3%), Loktanella (7.5%), Marinomonas (5.4%), and Flavobacterium (4.1%) with a moderate abundance of Escherichia/Shigella, Belliella, Providencia, Alishewanella, Sulfitobacter, Paracoccus, Serratia, Planktomarina, Enterobacter, Roseovarius, Owenweeksia, and Klebsiella (Figure 3 and Figure S3; Table S3).
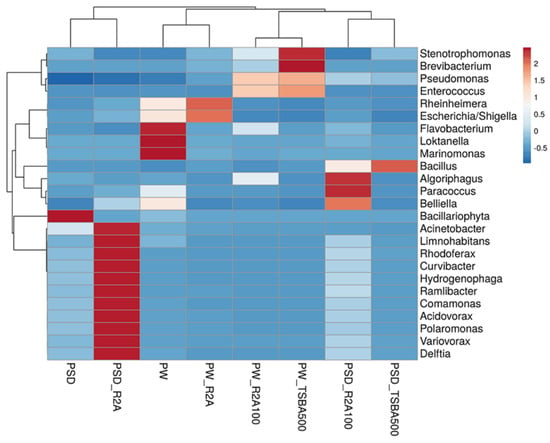
Figure 3.
Heatmap showing distribution of genera-level diversity in both direct samples and media samples.
The study of shared and unique taxa showed that PSD and PW samples contained 42 core genera, i.e., present in both water and sediment. The top five core genera were Rheinheimera, Pseudomonas, Hydrogenophaga, Loktanella, and Acinetobacter (Figure 4A). The analysis suggested that sediment samples contained a higher number of unique taxa (82), almost 50% more than the PW (40) sample (Figure 4B, Table S4).
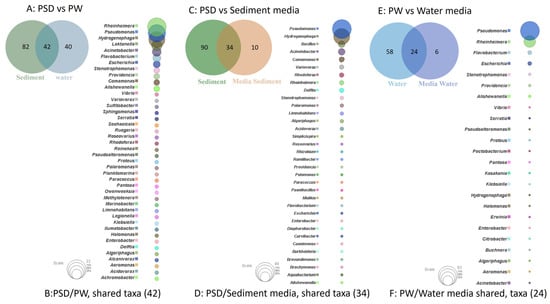
Figure 4.
Venn diagram representing details of total number of shared and unique taxa (at genus level) among sediment (PSD) and water (PW) samples (A); PSD and PSD media samples (C); and PW and PW media sample (E). Bubble plot depicts the identity of shared taxa among sediment (PSD) and water (PW) samples (B); PSD and PSD media samples (D); and PW and PW media sample (F).
The Hydrogenophaga spp. are facultative chemolithoautotrophs and majorly grow by the oxidation of organic compounds, but few species can oxidize H2/CO-as an energy source and utilize CO2 as a C source growing mixotrophically, and some have the capability to fix N2 and reduce NO3 under anaerobic conditions [50]. None of the previous reports, however, have identified this taxon from the sediment/water samples of the lake as an abundant taxa [14,15,16,17,18,19]; however, its versatile metabolic capabilities make it an interesting candidate for further investigation (Figure S3) [51,52]. It is important to note that in culture-based studies, several strains of the genus have been isolated in our lab and few seem to be novel species after preliminary characterization (unpublished data). The genus Loktanella of the Roseobacter group within the family Rhodobacteraceae in Alphaproteobacteria is a group of chemoorganoheterotrophic bacteria mostly isolated from marine environments with few species harboring bacteriochlorophyll a pigment and pufLM genes [53]. However, molecular studies targeting photosynthetic reaction centers (pufLM) and amplicon sequencing have revealed an abundance of Loktanella-like sequences in the saline lakes of the Tibetan plateau [54] and within the bacterial community of glacio-marine systems in the Arctic region [55]. Interestingly, Chaudhari et al. [18] have reported nearly half (49%) of the total bacterial community in the Pangong water sample as belonging to Loktanella (Figure S3). Although the authors described these sequences as belonging to aerobic anoxygenic phototrophic (AAP) bacteria, one needs to be cautious as none of the validly described species have an established role in photosynthesis and phylogenetic identity and the function of Loktanella-like sequences detected by Jiang et al. [54] have to be validated through more robust methods like whole-genome, metagenome, and culture-based approaches. The genera Pseudomonas, abundant in both lake sediment and water samples (~4.5% and 15% in PSD and PW, respectively), are strictly aerobic chemo-organotrophs that are ubiquitous in their distribution and have the ability to oxidatively degrade a variety of substrates including alkanes and aromatic compounds [56]. Many species are psychrotrophic, denitrifiers, and few have the ability to grow in low nutrient conditions [56,57]. Previous reports [16,18] have identified the group as an abundant part of the microbial community only in the sediment samples (Figure S3). Rheinheimera spp. highly abundant in the PW sample (~20%) and moderate in PSD (~2%) are chemoheterotrophic Gammaproteobacteria, growing optimally at salinity levels of 3% (w/v), rapidly degrade organic matter, and play a critical role in the biogeochemical cycling of carbon [58,59,60,61]. Importantly, previous culture-based [61,62,63] and functional metagenomic surveys [16] have identified this group as an important part of the microbial community of haloalkaline lakes (including Pangong) with the identification of genes related to ammonia, inorganic sulfur assimilation, and antimicrobial production [62]. The other two abundant taxa in the water sample, i.e., Marinomonas and Alishewanella within Gammaproteobacteria, belong to the families Oceanospirillaceae and Alteromonadaceae (recently transferred to the family Chromatiaceae [64]) that consist mainly of aerobic/facultatively anaerobic heterotrophs of mainly marine origin that require Na+ for growth with potential applications in the production of antimicrobials, melanin, and dye degradation, respectively [65,66]. Some of the Alishewanella spp., A. longhuensis [60], and A. alkalitolerans [64] have, in fact, been isolated from similar haloalkaline lakes, and it seems that soda lakes share similar compositions of the bacterial community with marine habitats in terms of the identity of few Pseudomonadota groups. Therefore, it may be prudent to use growth media like marine agar, etc., in future to target the isolation of native bacteria from such lake ecosystems since most of these microbes have an optimum Na+ requirement in the range of 3% (w/v) to maintain high osmotic pressure due to the high Cl− anion in the lake sediment samples (Table S1; [16,18]). This is a major contributor determining the microbial community composition [4,67,68]. Within the phylum Bacteroidota, Flavobacterium spp. were the most abundant (4%) in the water sample (PW, Table S3). Members are mostly psychrotolerant with optimum growth at 20–30 °C, abundant in cold aquatic ecosystems such as lakes, rivers, streams, etc. [69], and they degrade a variety of proteins and polysaccharides of algae, plants, fungi, insects, etc. [70,71,72,73,74,75]. Rathour et al. [16] and Chaudhari et al. [18], however, identified this taxon only in the sediment samples. The next abundant genus within the phylum was Belliella (Table S3), which are chemoheterotrophs isolated from alkaline, saline, and thermal habitats. In fact, three species, i.e., Belliella kenyensis [76], B. aquatica [77], and B. buryatensis [78] have been isolated from haloalkaline lakes located in Kenya, China, and the Republic of Buryatia (Russia), respectively. All the species described to date have optimum pH and salinity in the alkaline range and around 3% NaCl, respectively [78].
Among the moderately abundant taxa (~≤2%) in the sediment samples, Acidovorax, Burkholderia, Comamonas, Delftia, Rhodoferax, and Variovorax spp. of the order Burkholderiales within Betaproteobacteria are widely distributed in natural and man-made environments and are diverse in their metabolism. Rhodoferax spp. uses acetate, pyruvate, lactate, and succinate as the sole carbon sources [79]. The genera Variovorax, Delftia, and Comamonas spp. are capable of accumulating polyhydroxyalkanoates [80] and have a distinctive catabolic pattern involving protocatechuate meta- and ortho-cleavage pathways [81]. Some Variovorax spp. are chemoorganotrophs, capable of using a wide range of organic compounds for growth. Variovorax and Comamonas spp. were reported as minor populations in the Pangong sediment sample [18]. Burkholderia spp. display associative nitrogen fixation (nodule formation) in soil or sediment [79]. A Betaproteobacterium member, Methylotenera belonging to family Methylophilaceae, was observed only in the PSD sample (Figure S3). Methylotenera spp. are methylotrophs and use one carbon compound, methane or methanol, for their growth as a carbon source. Methylotrophs are mainly distributed in marine or brackish habitats [17], and previous studies of this ecosystem have reported the detection of Methylotenera in soil water mix collected from Pangong lake. Furthermore, Methylococcaceae, Methylothermaceae, Methylophilaceae, Methylobacteriaceae, and Methylocystaceae in a Pangong lake sediment sample were reported by Rathour et al. [16], wherein they identified Methylophaga as a major population (>10%, Gammaproteobacteria). Collectively, the facts suggest that methylotrophs are abundant in Pangong lake and play an important role in the methane cycle. Future functional metagenomic studies targeting methane metabolism can shed useful insights into the roles and mechanisms of methylotrophs in relation to C cycling. The detection of Escherichia/Shigella, Klebsilla, Serratia, Enterobacter, and Providencia spp. of the family Enterobacteriaceae in the water samples indicates the contamination of the lake by anthropogenic activities. Pangong lake is one of the tourist hot spots in Ladakh, India; thus human interference might affect its water quality.
With regard to our second objective to understand the effect of media formulation in terms of supporting the growth of a wide variety of species, analyses of diversity indices indicated that both R2A- and R2A100-times diluted media supported better species richness for the sediment sample with almost comparable values for Shannon, Simpson, chao-1, and total number of taxa, whereas for the water sample, the corresponding values of 2.306, 0.7624, 314.6, and 233 were highest for the R2A media (Table 2).

Table 2.
Diversity indices of bacterial diversity of direct and media samples.
Although R2A100 showed a slightly higher chao-1 index, one must take into account the fact that it is a non-parametric-based index and takes into consideration rare taxa, whereas the evenness-based Simpson index for the PW_R2A sample was much higher compared to PW_R2A100 (0.7624 vs. 0.2601, Table 2). The rarefaction curves revealed the requirement of more sequencing depth to cover the scale of diversity for PSD and PW samples, whereas all the media sample curves showed saturation, indicating good coverage (Figure S2). The phylum Pseudomonadota was the most abundant in direct and media samples. This ubiquitous predominance has been observed in different freshwater lakes of the Himalayan region as well as globally, especially in molecular surveys [82]. Bacteroidota was the second most abundant phylum in PSD, PW, PSD_R2A, and PSD_R2A100 samples, while Bacillota took this spot in media samples PSD_TSBA500, PW_TSBA500, PW_R2A, and PW_R2A100, highlighting the selective biasness of commonly used growth media in the over-representation of a few taxa in culturable diversity surveys. In fact, previous reports by Sahay et al. [15] and Yadav et al. [14,83] recovered bacterial groups belonging to Bacillota as the dominant population. However, none of the media could retrieve Planctomycetota, Verrucomicrobiota, Deinococcus-Thermus, Acidobacteriota, and Gemmatimonadota, indicating that the incubation time/culture media conditions were insufficient/inappropriate, respectively, for cultivation. A comparison of shared and unique taxa between total DNA samples (PSD and PW) and media samples was made in order to understand the number of taxa that can be retrieved in media. In the sediment sample, a total of 34 genera were detected in both PSD and PSD media samples, wherein Pseudomonas and Hydrogenophaga were the top two genera (Figure 4C). However, 90 genera were exclusively detected in the PSD sample, which could not be retrieved on any media samples, and only 10 genera were unique in media samples (Figure 4D, Table S4). In the case of water samples, 24 genera were core taxa in PW and PW media samples, whereas the top 2 genera were Pseudomonas and Rheinheimera (Figure 4E). A total of 58 genera were exclusively detected in PW samples that could not be retrieved on any media samples, whereas 6 genera were uniquely detected in PW media samples (Figure 4F, Table S4). The differential abundance analysis between sediment and water, considering all the samples, i.e., total DNA and media samples, revealed that the genera Hydrogenophaga and Pseudomonas were significantly associated with sediment and water samples, respectively. Although statistically non-significant, the genera Acinetobacter, Arenimonas, Rheinheimera, Escherichia, Loktanella, Marinomonas, and Flavobacterium were associated with sediment and water samples, respectively (Figure S5). Among Gammaproteobacteria, the genera Pseudomonas was observed in all the samples, while Rheinheimera and Flavobacterium were detected in all the samples except PW_TSBA500 and PSD_R2A, respectively. Some genera such as Methylotenera, Thiobacillus, Lysobacter, Opitutus, Truepera, Gemmatimonas, Bradymonas, Ilumatobacter, Marinobacter, Seohaeicola, and Reinekea were detected only in direct samples (PW and PSD; Figure 3, Table S3), whereas Roseovarius, Sulfitobacter, Alishewanella, Loktanella, Planktomarina, Ruegeria, Porphyrobacter, Novosphingobium, Erythrobacter, Staphylococcus, and Sphingobium were observed only in diluted media samples and direct samples (Figure 3, Table S3). The PCA plot represented a correlation of the genera Rheinheimera, Pseudomonas, Escherichia/Shigella, Providencia, etc., with water sample (PC3), whereas Hydrogenophaga, Bacillus, Thiobacillus, Algoriphagus, Methylotenera, etc., grouped with sediment samples (PC1) with cosmopolitan bacterial genera, i.e., Pseudomonas, and Bacillus, were over-represented especially in diluted media, indicating selection biasness in growth media and culture conditions (i.e., incubation times) (Figure S4). Interestingly, some of the abundant taxa, such as Hydrogenophaga, Pseudomonas, and Rheinheimera spp., were also recovered in our culture-dependent analysis, representing 3.5%, 25%, and 1.4% of the total isolates identified, respectively (Figure S6). In contrast, moderately abundant OTUs belonging to Acidovorax, Burkholderia, Delftia, Limnohabitans, and Rhodoferax were better represented in media samples, although none could be recovered in culture indicating the inherent procedural flaws in our conventional approach (Table S3). From the culturable work, we were able to isolate and identify a few facultative methylotrophs, i.e., Methylobacterium spp. (classified reads retrieved in PSD sample) and Methylorubrum spp., which are known to use C1 compounds like methanol and methylamines. This is not surprising given that the lake sediment contains a variety of trace elements, including Fe, Co, Mo, Mg, Mn, and Zn, as well as nitrogen, which is available as nitrate, nitrite, and ammonia, promoting the growth of such bacterial groups (Table S1). Interestingly, classified reads affiliated to genera Enterococcus, Jonesia, Streptophyta, Reyranella, Arthrobacter, Blastomonas, Gemmobacter, Mesorhizobium, Pseudorhodobacter, Ensifer, Haematobacter, Roseinatronobacter, Tabrizicola, Phyllobacterium, Aureimonas, Methylarculla, Ochrobactrum, Sphingorhabdus, and Nitrosospira were detected only in media samples, suggesting that these taxa might be less abundant in situ and may become enriched in growth media, as evident by the isolation of several strains of Tabrizicola spp., Blastomonas spp., and Jonesia spp. in our lab (Table S3, Figure 3). Previous reports on culturable diversity have identified Arthrobacter, Pseudomonas, Bacillus, Planococcus, Exiguobacterium, Paenibacillus, and Staphylococcus spp. as dominant groups involved in the production of several polymer degrading-enzymes [14,15,81]. It is pertinent here to mention that Planococcus antartcticus and Paenibacillus xylanexedens, reported previously, were successfully re-isolated and their functional activity was confirmed in our analyses, indicating niche specificity.
4. Conclusions
The present study attempted a comparison of the total and culturable bacterial diversity from high-altitude Pangong lake using long read sequencing technology, i.e., Oxford Nanopore, to understand the microbial composition within the saline high-altitude lake water and sediment samples. Our study indicated that the sediment hosted the most diverse bacterial community. Phyla Pseudomonadota, Bacteroidota, Cyanobacteria, Bacillota, Verrucomicrobiota, and Actinomycetota were observed both in sediment and water samples, while Planctomycetota, Deinococcus-Thermus, Acidobacteriota, Gemmatimonadota, Hydrogenedentes, and Nitrospirota were exclusive in the sediment. Many ecologically important genera like Hydrogenophaga, Loktanella, Pseudomonas, Rheinhemera, Flavobacterium, Marinomonas, Alishewanella, Acinetobacter, Arenimonas, and Comamonas were abundantly observed in both sediment and water samples of the ecosystem. The genera Hydrogenophaga, Rheinheimera, and Loktanella seemed to be unique and abundant taxa for the sediment and water samples, respectively. Loktanella appears to be an endemic microbial population as established in previous reports [18]. Some of the functionally important groups, i.e., Methylotenera, that play an important role in the C cycle through the utilization of C1 compounds were detected exclusively in the sediment. The presence of such phenotypic groups has been confirmed in previous metagenomic surveys of the habitat. The detection of Enterobacteriaceae in the water samples indicates contamination through anthropogenic activities.
In terms of culturability, R2A diluted 100 times and undiluted R2A seemed best suited for recovering bacterial diversity from sediment and water samples, respectively. Bacteria belonging to Planctomycetota, Verrucomicrobiota, Deinococcus-Thermus, Acidobacteriota, Gemmatimonadota, Hydrogenedentes, and Nitrospirota could not be detected in any of the culture media. One of the reasons for this was that culture conditions/timelines employed for isolation do not correspond with the above groups. Further, it might also be possible that the DNA extraction method could not lyse the cell walls of some groups (i.e., Deinococcus-Thermus) efficiently. It is pertinent to mention that future large scale culturing efforts could use marine agar as the majority of the abundant taxa detected have 3% salinity requirement for optimal growth. In recent years, through the advent of genome sequencing technologies, Actinomycetes and Cyanobacteria have been shown to encode the largest biosynthetic diversity [84], and diluted/low-strength media could be utilized for targeting the cultivation of novel lineages within these taxa.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/microorganisms12020397/s1, Figure S1: Sampling sites; Figure S2: Rarefaction curve showing comparative taxonomic richness in both direct and media samples; Figure S3: Comparative phylogenetic tree of direct and media samples along with six samples of Chaudhari et al. (CH1-CH6) [18], one sample each of Bisht et al. (BI) [19], and Rathour et al. (RA) [16]; Figure S4: Principal component analysis (PCA) plot showing the variation and clustering between the direct and media samples and genera; Figure S5: Median taxonomic abundance of common differentially abundant genera between sediment water (ns: non-significant); Figure S6: Top 30 genera among 16S rRNA targeted amplicon sequencing and isolates retrieved from culture-based studies; Table S1: Physico-chemical analysis of Pangong sediment sample; Table S2: Read statistics of all studied samples; Table S3: Percent read of all studied samples at different taxonomic level; Table S4: Shared and unique genera among PSD and PW; PSD and PSD media samples; PW and PW media samples; Table S5: Details of media composition used in this study.
Author Contributions
P.Y. performed the experiments, analyzed the initial data, and wrote the manuscript. J.D. and S.S.S. contributed towards the analyses of data and editing the manuscripts. S.K. designed and supervised the study, reviewed the data, and proofread the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by CSIR project OLP0149 “To decipher the prokaryotic diversity of hot springs and other interesting habitats of the region Ladakh” and P.Y., J.D. and S.S.S. are the recipients of the Department of Biotechnology (DBT), University Grands Commission (UGC), Council of Scientific & Industrial Research (CSIR) fellowships, respectively.
Data Availability Statement
All data were submitted to the NCBI Sequence Read Archive (SRA) under the following accession numbers: SRR11354751 (PW)-SRR11354747 (PSD); SRR11354746 (PSD_R2A); SRR11354745 (PSD_R2A100); SRR11354744 (PSD_TSBA500); SRR11354750 (PW_R2A) SRR11354749 (PW_R2A100); and SRR11354748 (PW_TSBA500).
Acknowledgments
We are thankful to Genotypic Technologies Pvt Ltd., Bengaluru, for sequencing and preliminary data analysis. We are thankful to Punjab Biotechnology Incubator (PBTI), Mohali, India for physio-chemical analysis of Pangong sediment sample. This is IMTECH communication number 14/2021. We thank Tsering Stobdan of Defence Institute of High Altitude Research (DIHAR) for arranging logistics for sampling. Thanks also to Anil Kaul (Former Director, CSIR-IMTECH) and Deepak Sharma for help in sampling.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Dong, H.; Jiang, H.; Yu, B.; Liu, X.; Zhang, C.; Chan, M.A. Impacts of Environmental Change and Human Activity on Microbial Ecosystems on the Tibetan Plateau, NW China. Gsa Today 2010, 20, 4–10. [Google Scholar] [CrossRef]
- Hayden, C.J.; Beman, J.M. Microbial Diversity and Community Structure along a Lake Elevation Gradient in Y Osemite N Ational P Ark, C Alifornia, USA. Environ. Microbiol. 2016, 18, 1782–1791. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ma, L.; Jiang, H.; Wu, G.; Dong, H. Salinity Shapes Microbial Diversity and Community Structure in Surface Sediments of the Qinghai-Tibetan Lakes. Sci. Rep. 2016, 6, 25078. [Google Scholar] [CrossRef]
- Zhao, D.; Zhang, S.; Xue, Q.; Chen, J.; Zhou, J.; Cheng, F.; Li, M.; Zhu, Y.; Yu, H.; Hu, S. Abundant Taxa and Favorable Pathways in the Microbiome of Soda-Saline Lakes in Inner Mongolia. Front. Microbiol. 2020, 11, 1740. [Google Scholar] [CrossRef] [PubMed]
- Heiri, O.; Lotter, A.F. 9000 Years of Chironomid Assemblage Dynamics in an Alpine Lake: Long-Term Trends, Sensitivity to Disturbance, and Resilience of the Fauna. J. Paleolimnol. 2003, 30, 273–289. [Google Scholar] [CrossRef]
- Lindström, E.S.; Kamst-Van Agterveld, M.P.; Zwart, G. Distribution of Typical Freshwater Bacterial Groups Is Associated with PH, Temperature, and Lake Water Retention Time. Appl. Environ. Microbiol. 2005, 71, 8201–8206. [Google Scholar] [CrossRef]
- Bhat, F.A.; Yousuf, A.R.; Aftab, A.; Arshid, J.; Mahdi, M.D.; Balkhi, M.H. Ecology and Biodiversity in Pangong Tso (Lake) and Its Inlet Stream in Ladakh, India. Int. J. Biodivers. Conserv. 2011, 3, 501–511. [Google Scholar]
- Margesin, R.; Miteva, V. Diversity and Ecology of Psychrophilic Microorganisms. Res. Microbiol. 2011, 162, 346–361. [Google Scholar] [CrossRef]
- Goordial, J.; Davila, A.; Lacelle, D.; Pollard, W.; Marinova, M.M.; Greer, C.W.; DiRuggiero, J.; McKay, C.P.; Whyte, L.G. Nearing the Cold-Arid Limits of Microbial Life in Permafrost of an Upper Dry Valley, Antarctica. ISME J. 2016, 10, 1613–1624. [Google Scholar] [CrossRef]
- Nedwell, D.B. Effect of Low Temperature on Microbial Growth: Lowered Affinity for Substrates Limits Growth at Low Temperature. FEMS Microbiol. Ecol. 1999, 30, 101–111. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Bruni, V.; Michaud, L. Characterization of Antarctic Psychrotrophic Bacteria with Antibacterial Activities against Terrestrial Microorganisms. J. Basic Microbiol. 2007, 47, 496–505. [Google Scholar] [CrossRef]
- Qin, Y.; Huang, Z.; Liu, Z. A Novel Cold-Active and Salt-Tolerant α-Amylase from Marine Bacterium Zunongwangia Profunda: Molecular Cloning, Heterologous Expression and Biochemical Characterization. Extremophiles 2014, 18, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Romshoo, S.; Panigrahy, S.; Patel, J.; Parihar, J. High Altitude Lakes of India; Space Applications Centre, ISRO: Ahmedabad, India, 2012. [Google Scholar]
- Yadav, A.N.; Sachan, S.G.; Verma, P.; Tyagi, S.P.; Kaushik, R.; Saxena, A.K. Culturable Diversity and Functional Annotation of Psychrotrophic Bacteria from Cold Desert of Leh Ladakh (India). World J. Microbiol. Biotechnol. 2015, 31, 95–108. [Google Scholar] [CrossRef]
- Sahay, H.; Babu, B.K.; Singh, S.; Kaushik, R.; Saxena, A.K.; Arora, D.K. Cold-active Hydrolases Producing Bacteria from Two Different Sub-glacial Himalayan Lakes. J. Basic Microbiol. 2013, 53, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Rathour, R.; Gupta, J.; Mishra, A.; Rajeev, A.C.; Dupont, C.L.; Thakur, I.S. A Comparative Metagenomic Study Reveals Microbial Diversity and Their Role in the Biogeochemical Cycling of Pangong Lake. Sci. Total Environ. 2020, 731, 139074. [Google Scholar] [CrossRef]
- Rathour, R.; Gupta, J.; Kumar, M.; Hiloidhari, M.; Mehrotra, A.K.; Thakur, I.S. Metagenomic Sequencing of Microbial Communities from Brackish Water of Pangong Lake of the Northwest Indian Himalayas. Genome Announc. 2017, 5, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, D.S.; Dhotre, D.P.; Jani, K.; Sharma, A.; Singh, Y.; Shouche, Y.S.; Rahi, P. Bacterial Communities Associated with the Biofilms Formed in High-Altitude Brackish Water Pangong Tso Located in the Himalayan Plateau. Curr. Microbiol. 2020, 77, 4072–4084. [Google Scholar] [CrossRef]
- Bisht, G.; Sourirajan, A.; Baumler, D.J.; Dev, K. 16S RRNA Gene Amplicon Data Set-Based Bacterial Diversity in a Water-Soil Sample from Pangong Tso Lake, a High-Altitude Grassland Lake of the Northwest Himalayas. Microbiol. Resour. Announc. 2018, 7, e01192-18. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.N.; Verma, P.; Sachan, S.G.; Kaushik, R.; Saxena, A.K. Biodiversity and Biotechnological Applications of Psychrotrophic Microbes Isolated from Indian Himalayan Regions. EC Microbiol. 2017, ECO.01, 48–54. [Google Scholar]
- Humayoun, S.B.; Bano, N.; Hollibaugh, J.T. Depth Distribution of Microbial Diversity in Mono Lake, a Meromictic Soda Lake in California. Appl. Environ. Microbiol. 2003, 69, 1030–1042. [Google Scholar] [CrossRef]
- Catalan, J.; Camarero, L.; Felip, M.; Pla, S.; Ventura, M.; Buchaca, T.; Bartumeus, F.; de Mendoza, G.; Miró, A.; Casamayor, E.O. High Mountain Lakes: Extreme Habitats and Witnesses of Environmental Changes. Limnetica 2006, 25, 551–584. [Google Scholar] [CrossRef]
- Hörtnagl, P.; Perez, M.T.; Zeder, M.; Sommaruga, R. The Bacterial Community Composition of the Surface Microlayer in a High Mountain Lake. FEMS Microbiol. Ecol. 2010, 73, 458–467. [Google Scholar] [CrossRef]
- Aszalós, J.M.; Krett, G.; Anda, D.; Márialigeti, K.; Nagy, B.; Borsodi, A.K. Diversity of Extremophilic Bacteria in the Sediment of High-Altitude Lakes Located in the Mountain Desert of Ojos Del Salado Volcano, Dry-Andes. Extremophiles 2016, 20, 603–620. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertilsson, S. A Guide to the Natural History of Freshwater Lake Bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 14–49. [Google Scholar] [CrossRef] [PubMed]
- Baruah, T.C.; Barthakur, H.P. A Text Book of Soil Analysis; Vikas Publishing House: Noida, India, 1997. [Google Scholar]
- AOAC 990.08; Metals in Solid Wastes by Inductively Coupled Plasma Atomic Emission Method. AOAC International: Rockville, MD, USA, 1998.
- IS 27720; Methods of Test for Soils. Indian standard Institution: New Delhi, India, 1983.
- AOAC 955.04; Nitrogen (Total) in Fertilizers. Kjeldahl Method. AOAC International: Rockville, MD, USA, 1997.
- APHA 2510B; Standard Methods for the. Examination of Water and Wastewater—Laboratory Method. American Public Health Organization: Washington, DC, USA, 1992.
- APHA 2520B; Standard Methods for the Examination of Water and Wastewater—Electrical Conductivity Method. American Public Health Organization: Washington, DC, USA, 1999.
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast Metagenomic Sequence Classification Using Exact Alignments. Genome Biol. 2014, 15, 1–12. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Salzberg, S.L. Pavian: Interactive Analysis of Metagenomics Data for Microbiome Studies and Pathogen Identification. Bioinformatics 2020, 36, 1303–1304. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T. Past: Paleontological Statistics Software Package for Educaton and Data Anlysis. Palaeontol. Electron. 2001, 4, 1. [Google Scholar]
- Metsalu, T.; Vilo, J. ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data Using Principal Component Analysis and Heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An Interactive Venn Diagram Viewer. BMC Bioinform. 2014, 15, 1–7. [Google Scholar] [CrossRef]
- Kohli, I.; Joshi, N.C.; Mohapatra, S.; Varma, A. Extremophile–an Adaptive Strategy for Extreme Conditions and Applications. Curr. Genom. 2020, 21, 96–110. [Google Scholar] [CrossRef]
- Alloway, B.J. Heavy Metals in Soil and Plants; John Wiley and Sons: New York, NY, USA, 1990. [Google Scholar]
- Amann, R.I.; Ludwig, W.; Schleifer, K.-H. Phylogenetic Identification and in Situ Detection of Individual Microbial Cells without Cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Bin Mannan, K.S.; Andrews, S. Viable but Nonculturable Bacteria: Food Safety and Public Health Perspective. ISRN Microbiol. 2013, 2013, 703813. [Google Scholar] [CrossRef]
- Roszak, D.B.; Colwell, R. Survival Strategies of Bacteria in the Natural Environment. Microbiol. Rev. 1987, 51, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Staley, J.T.; Konopka, A. Measurement of in Situ Activities of Nonphotosynthetic Microorganisms in Aquatic and Terrestrial Habitats. Annu. Rev. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, V.; Øvreås, L.; Thingstad, T.F. Prokaryotic Diversity—Magnitude, Dynamics, and Controlling Factors. Science 2002, 296, 1064–1066. [Google Scholar] [CrossRef]
- Inagaki, F.; Kuypers, M.M.M.; Tsunogai, U.; Ishibashi, J.; Nakamura, K.; Treude, T.; Ohkubo, S.; Nakaseama, M.; Gena, K.; Chiba, H. Microbial Community in a Sediment-Hosted CO2 Lake of the Southern Okinawa Trough Hydrothermal System. Proc. Natl. Acad. Sci. USA 2006, 103, 14164–14169. [Google Scholar] [CrossRef]
- Jørgensen, B.B.; Boetius, A. Feast and Famine—Microbial Life in the Deep-Sea Bed. Nat. Rev. Microbiol. 2007, 5, 770–781. [Google Scholar] [CrossRef]
- Nweze, N.O. Ecological Implications and Roles of Cyanobacteria (Cyanophyta) in Food Security–A Review. Plant Prod. Res. J. 2009, 13, 8–14. [Google Scholar] [CrossRef]
- Wong, H.L.; Smith, D.-L.; Visscher, P.T.; Burns, B.P. Niche Differentiation of Bacterial Communities at a Millimeter Scale in Shark Bay Microbial Mats. Sci. Rep. 2015, 5, 15607. [Google Scholar] [CrossRef]
- Vollmers, J.; Frentrup, M.; Rast, P.; Jogler, C.; Kaster, A.-K. Untangling Genomes of Novel Planctomycetal and Verrucomicrobial Species from Monterey Bay Kelp Forest Metagenomes by Refined Binning. Front. Microbiol. 2017, 8, 472. [Google Scholar] [CrossRef]
- Theodorakopoulos, N.; Bachar, D.; Christen, R.; Alain, K.; Chapon, V. Exploration of Deinococcus-Thermus Molecular Diversity by Novel Group-specific PCR Primers. Microbiologyopen 2013, 2, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Willems, A.; Gillis, M. Hydrogenophaga Willems, Busse, Goor, Pot, Falsen, Jantzen, Hoste, Gillis, Kersters, Auling and De Ley 1989, 329VP. In Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA, 2005; Volume 2, pp. 710–716. [Google Scholar]
- Banerjee, S.; Tancsics, A.; Toth, E.; Revesz, F.; Boka, K.; Kriszt, B. Hydrogenophaga aromaticivorans sp. Nov., Isolated from a Para-Xylene-Degrading Enrichment Culture, Capable of Degrading Benzene, Meta-and Para-Xylene. Int. J. Syst. Evol. Microbiol. 2021, 71, 004743. [Google Scholar] [CrossRef] [PubMed]
- Contzen, M.; Moore, E.R.B.; Blümel, S.; Stolz, A.; Kämpfer, P. Hydrogenophaga intermedia sp. Nov., a 4-Aminobenzene-Sulfonate Degrading Organism. Syst. Appl. Microbiol. 2000, 23, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Pujalte, M.J.; Lucena, T.; Ruvira, M.A.; Arahal, D.R.; Macián, M.C. The Family Rhodobacteraceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Springer: Berlin/Heidelberg, Germany, 2014; pp. 439–512. [Google Scholar]
- Jiang, H.; Dong, H.; Yu, B.; Lv, G.; Deng, S.; Wu, Y.; Dai, M.; Jiao, N. Abundance and Diversity of Aerobic Anoxygenic Phototrophic Bacteria in Saline Lakes on the Tibetan Plateau. FEMS Microbiol. Ecol. 2009, 67, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.A.; Sinha, R.K.; Krishnan, K.P. Bacterial Community Structure of a Glacio-Marine System in the Arctic (Ny-Ålesund, Svalbard). Sci. Total Environ. 2020, 718, 135264. [Google Scholar] [CrossRef]
- Garrity, G.M.; Bell, J.A.; Lilburn, T. Phylum XIV. Proteobacteria phyl. nov. In Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA, 2005; Volume 2, p. 1. [Google Scholar]
- Favero, M.S.; Carson, L.A.; Bond, W.W.; Petersen, N.J. Pseudomonas Aeruginosa: Growth in Distilled Water from Hospitals. Science 1971, 173, 836–838. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, A.; Bianchi, M. Bacterial Diversity and Ecosystem Maintenance: An Overview. In Microbial Diversity and Ecosystem Function; CAB International (UNEP): Wallingford, CT, USA, 1995; pp. 185–198. [Google Scholar]
- Pinhassi, J.; Berman, T. Differential Growth Response of Colony-Forming α-and γ-Proteobacteria in Dilution Culture and Nutrient Addition Experiments from Lake Kinneret (Israel), the Eastern Mediterranean Sea, and the Gulf of Eilat. Appl. Environ. Microbiol. 2003, 69, 199–211. [Google Scholar] [CrossRef]
- Poretsky, R.S.; Bano, N.; Buchan, A.; LeCleir, G.; Kleikemper, J.; Pickering, M.; Pate, W.M.; Moran, M.A.; Hollibaugh, J.T. Analysis of Microbial Gene Transcripts in Environmental Samples. Appl. Environ. Microbiol. 2005, 71, 4121–4126. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, J.-T.; Xu, C.-J.; Liu, Y.-H.; Song, X.-F.; Li, H.; Liu, Z.-P. Rheinheimera longhuensis sp. Nov., Isolated from a Slightly Alkaline Lake, and Emended Description of Genus Rheinheimera Brettar et al. 2002. Int. J. Syst. Evol. Microbiol. 2012, 62, 2927–2933. [Google Scholar] [CrossRef]
- Gupta, H.K.; Gupta, R.D.; Singh, A.; Chauhan, N.S.; Sharma, R. Genome Sequence of Rheinheimera sp. Strain A13L, Isolated from Pangong Lake, India. J. Bacteriol. 2011, 193, 5873–5874. [Google Scholar] [CrossRef]
- Zhong, Z.-P.; Liu, Y.; Liu, L.-Z.; Wang, F.; Zhou, Y.-G.; Liu, Z.-P. Rheinheimera tuosuensis sp. Nov., Isolated from a Saline Lake. Int. J. Syst. Evol. Microbiol. 2014, 64, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Sisinthy, S.; Chakraborty, D.; Adicherla, H.; Gundlapally, S.R. Emended Description of the Family Chromatiaceae, Phylogenetic Analyses of the Genera Alishewanella, Rheinheimera and Arsukibacterium, Transfer of Rheinheimera longhuensis LH2-2 T to the Genus Alishewanella and Description of Alishewanella alkalitolerans sp. nov. from Lonar Lake, India. Antonie Van Leeuwenhoek 2017, 110, 1227–1241. [Google Scholar] [CrossRef] [PubMed]
- Solano, F.; Garcia, E.; Perez, D.E.; Sanchez-Amat, A. Isolation and Characterization of Strain MMB-1 (CECT 4803), a Novel Melanogenic Marine Bacterium. Appl. Environ. Microbiol. 1997, 63, 3499–3506. [Google Scholar] [CrossRef]
- Kolekar, Y.M.; Nemade, H.N.; Markad, V.L.; Adav, S.S.; Patole, M.S.; Kodam, K.M. Decolorization and Biodegradation of Azo Dye, Reactive Blue 59 by Aerobic Granules. Bioresour. Technol. 2012, 104, 818–822. [Google Scholar] [CrossRef]
- Banciu, H.L.; Muntyan, M.S. Adaptive Strategies in the Double-Extremophilic Prokaryotes Inhabiting Soda Lakes. Curr. Opin. Microbiol. 2015, 25, 73–79. [Google Scholar] [CrossRef]
- Zorz, J.K.; Sharp, C.; Kleiner, M.; Gordon, P.M.K.; Pon, R.T.; Dong, X.; Strous, M. A Shared Core Microbiome in Soda Lakes Separated by Large Distances. Nat. Commun. 2019, 10, 4230. [Google Scholar] [CrossRef]
- Van Trappen, S.; Vandecandelaere, I.; Mergaert, J.; Swings, J. Flavobacterium fryxellicola sp. Nov. and Flavobacterium psychrolimnae Sp. Nov., Novel Psychrophilic Bacteria Isolated from Microbial Mats in Antarctic Lakes. Int. J. Syst. Evol. Microbiol. 2005, 55, 769–772. [Google Scholar] [CrossRef] [PubMed]
- Stanier, R.Y. Studies on Nonfruiting Myxobacteria: I. Cytophaga johnsonae, n. sp., a Chitin-Decomposing Myxobacterium. J. Bacteriol. 1947, 53, 297–315. [Google Scholar] [CrossRef]
- Van der Meulen, H.J.; Harder, W.; Veldkamp, H. Isolation and Characterization of Cytophaga flevensis sp. Nov., a New Agarolytic Flexibacterium. Antonie Van Leeuwenhoek 1974, 40, 329–346. [Google Scholar] [CrossRef]
- Liao, C.-H.; Wells, J.M. Properties of Cytophaga johnsonae Strains Causing Spoilage of Fresh Produce at Food Markets. Appl. Environ. Microbiol. 1986, 52, 1261–1265. [Google Scholar] [CrossRef]
- Reichenbach, H. Genus I Cytophaga Winogradsky 1929. In Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA, 1989; Volume 3, pp. 2015–2050. [Google Scholar]
- Humphry, D.R.; George, A.; Black, G.W.; Cummings, S.P. Flavobacterium frigidarium sp. nov., an Aerobic, Psychrophilic, Xylanolytic and Laminarinolytic Bacterium from Antarctica. Int. J. Syst. Evol. Microbiol. 2001, 51, 1235–1243. [Google Scholar] [CrossRef]
- Bernardet, J.-F.; Bowman, J.P. The Genus Flavobacterium. Prokaryotes 2006, 7, 481–531. [Google Scholar]
- Akhwale, J.K.; Göker, M.; Rohde, M.; Schumann, P.; Klenk, H.-P.; Boga, H.I. Belliella kenyensis sp. Nov., Isolated from an Alkaline Lake. Int. J. Syst. Evol. Microbiol. 2015, 65 Pt 2, 457–462. [Google Scholar] [CrossRef]
- Zhong, Z.-P.; Liu, Y.; Hou, T.-T.; Zhou, Y.-G.; Liu, H.-C.; Liu, Z.-P. Belliella aquatica sp. Nov., Isolated from a Saline Lake. Int. J. Syst. Evol. Microbiol. 2015, 65, 1622–1627. [Google Scholar] [CrossRef] [PubMed]
- Kozyreva, L.; Egorova, D.; Anan’ina, L.; Plotnikova, E.; Ariskina, E.; Prisyazhnaya, N.; Radnaeva, L.; Namsaraev, B. Belliella Buryatensis sp. Nov., Isolated from Alkaline Lake Water. Int. J. Syst. Evol. Microbiol. 2016, 66, 137–143. [Google Scholar] [CrossRef]
- Coenye, T. The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Delafield, F.P.; Doudoroff, M.; Palleroni, N.J.; Lusty, C.J.; Contopoulos, R. Decomposition of Poly-β-Hydroxybutyrate by Pseudomonads. J. Bacteriol. 1965, 90, 1455–1466. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pantoja, D.; Donoso, R.; Agulló, L.; Córdova, M.; Seeger, M.; Pieper, D.H.; González, B. Genomic Analysis of the Potential for Aromatic Compounds Biodegradation in Burkholderiales. Environ. Microbiol. 2012, 14, 1091–1117. [Google Scholar] [CrossRef]
- Yadav, A.N.; Yadav, N.; Kour, D.; Kumar, A.; Yadav, K.; Kumar, A.; Rastegari, A.A.; Sachan, S.G.; Singh, B.; Chauhan, V.S. Bacterial Community Composition in Lakes. In Freshwater Microbiology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–71. [Google Scholar]
- Yadav, A.N.; Verma, P.; Kumar, M.; Pal, K.K.; Dey, R.; Gupta, A.; Padaria, J.C.; Gujar, G.T.; Kumar, S.; Suman, A. Diversity and Phylogenetic Profiling of Niche-Specific Bacilli from Extreme Environments of India. Ann. Microbiol. 2015, 65, 611–629. [Google Scholar] [CrossRef]
- Gavriilidou, A.; Kautsar, S.A.; Zaburannyi, N.; Krug, D.; Müller, R.; Medema, M.H.; Ziemert, N. Compendium of Specialized Metabolite Biosynthetic Diversity Encoded in Bacterial Genomes. Nat. Microbiol. 2022, 7, 726–735. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).