The Great Gobi A Strictly Protected Area: Characterization of Soil Bacterial Communities from Four Oases
Abstract
1. Introduction
2. Materials and Methods
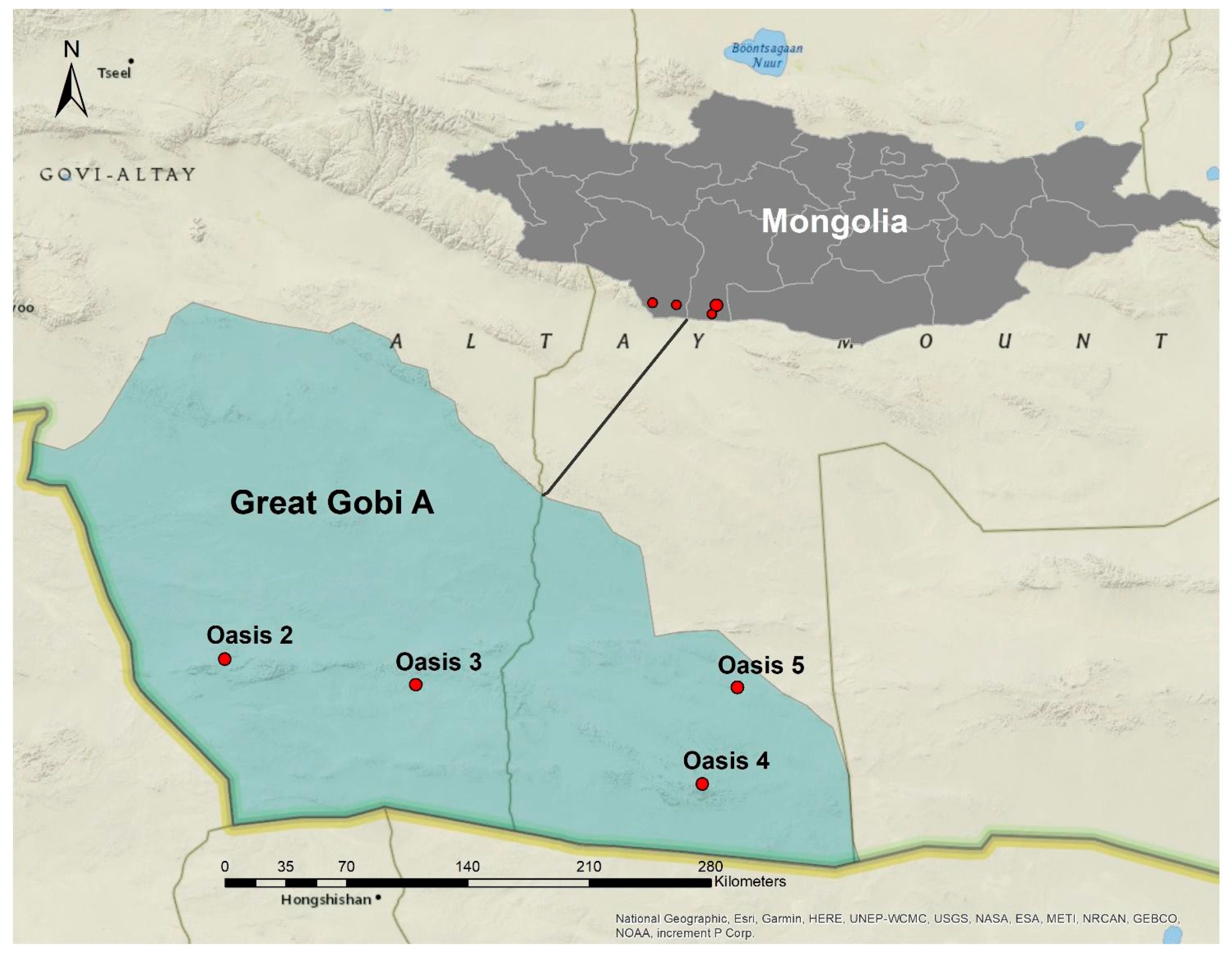
2.1. Study Area and Oases Soil Sampling
2.2. Chemical and Physical Characterisation of the Soil Samples
2.3. Extraction of Total DNA and Next-Generation Sequencing
2.4. Sequence Analysis
2.5. Statistical Analysis
3. Results
3.1. Characterization of Soil Physicochemical Features
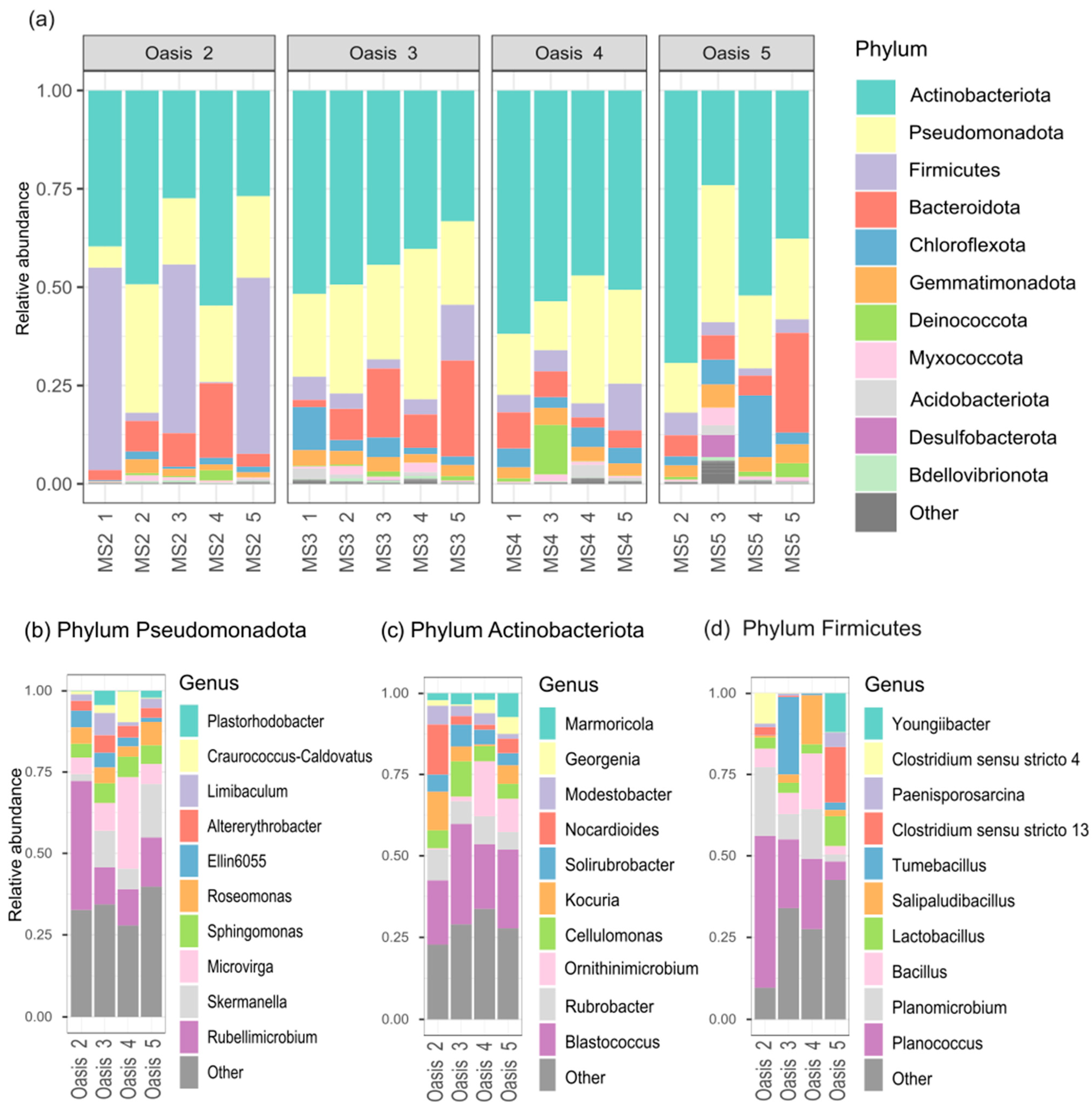
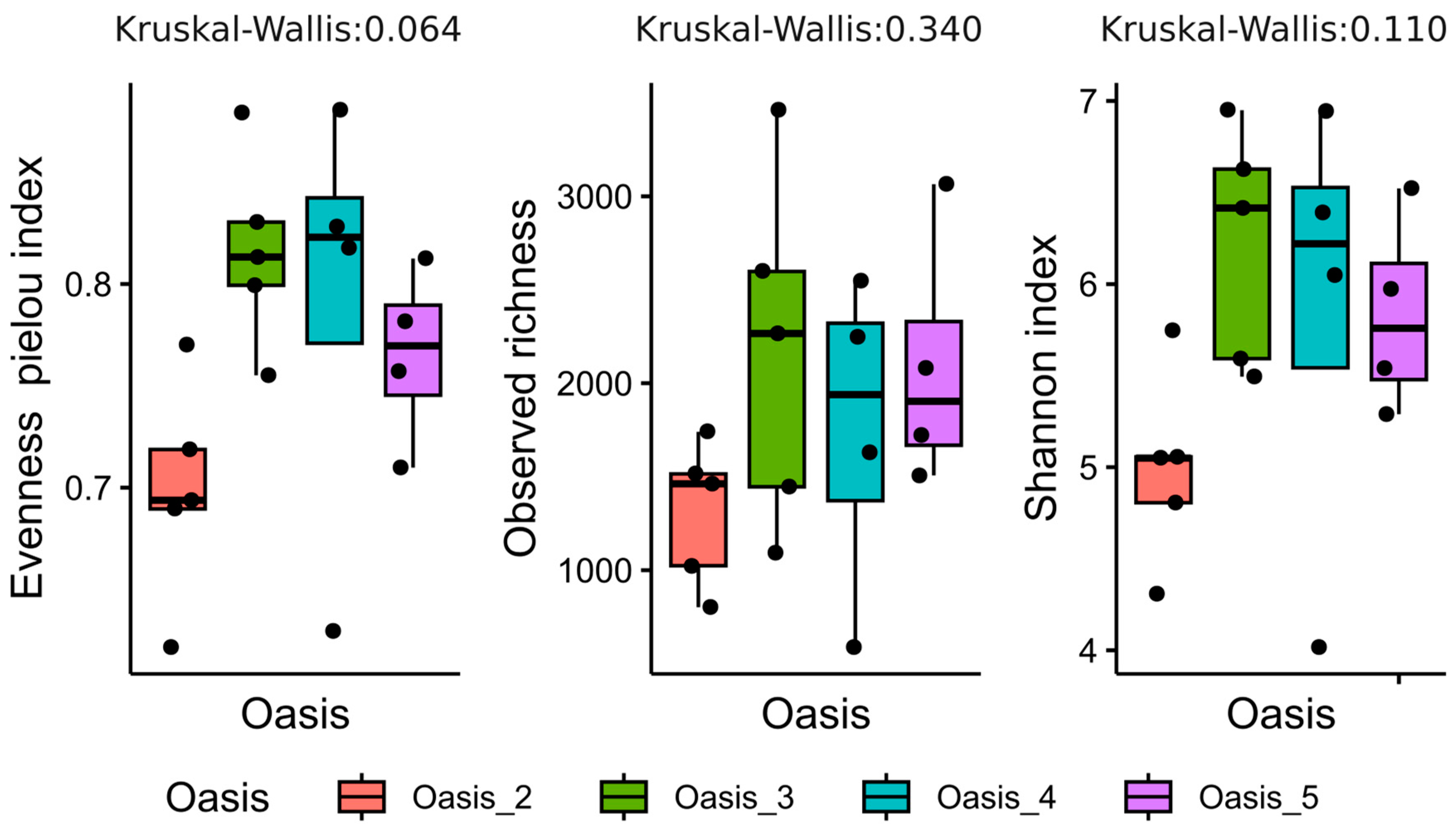
3.2. Characterization of Soil Bacterial Communities
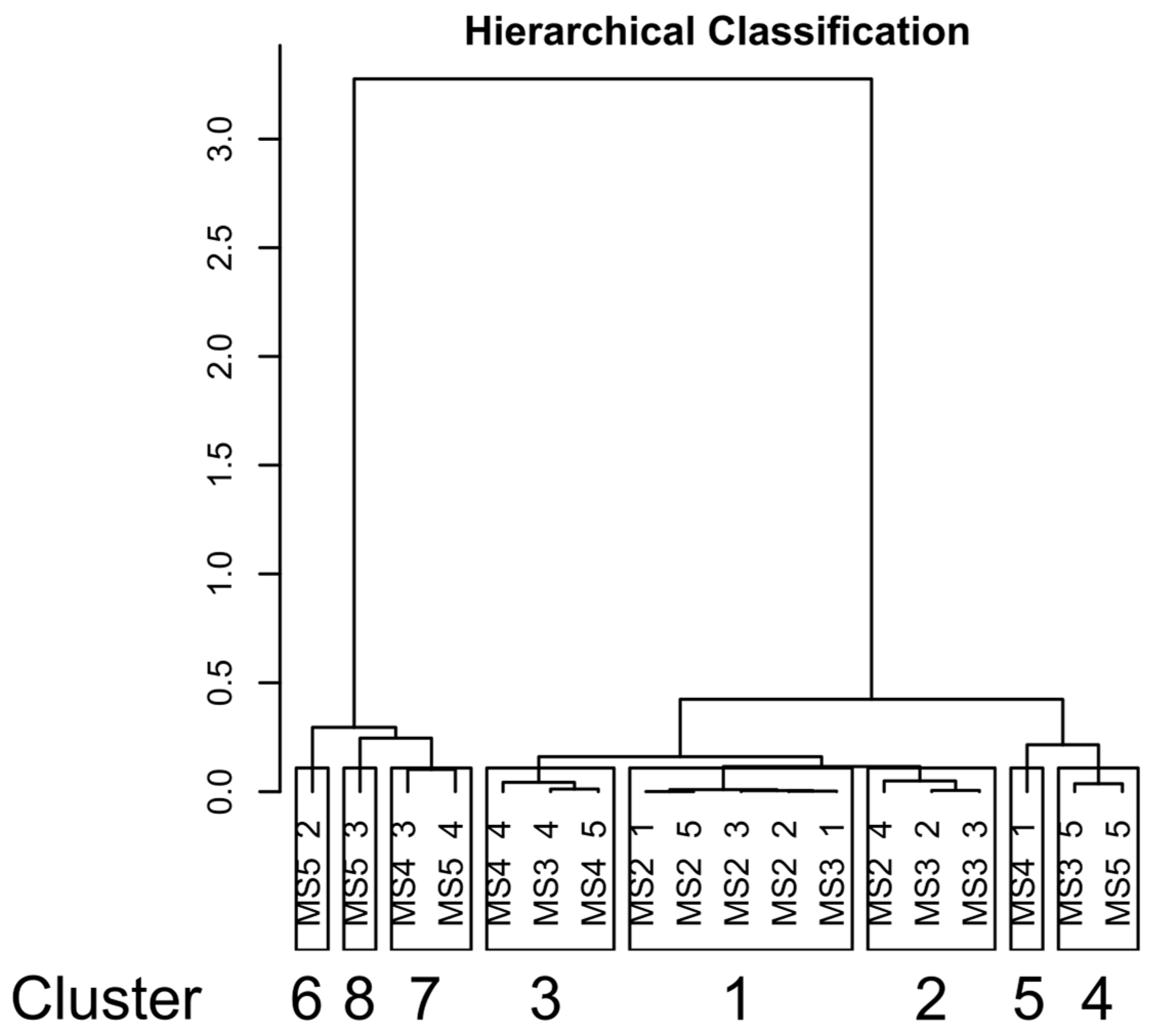
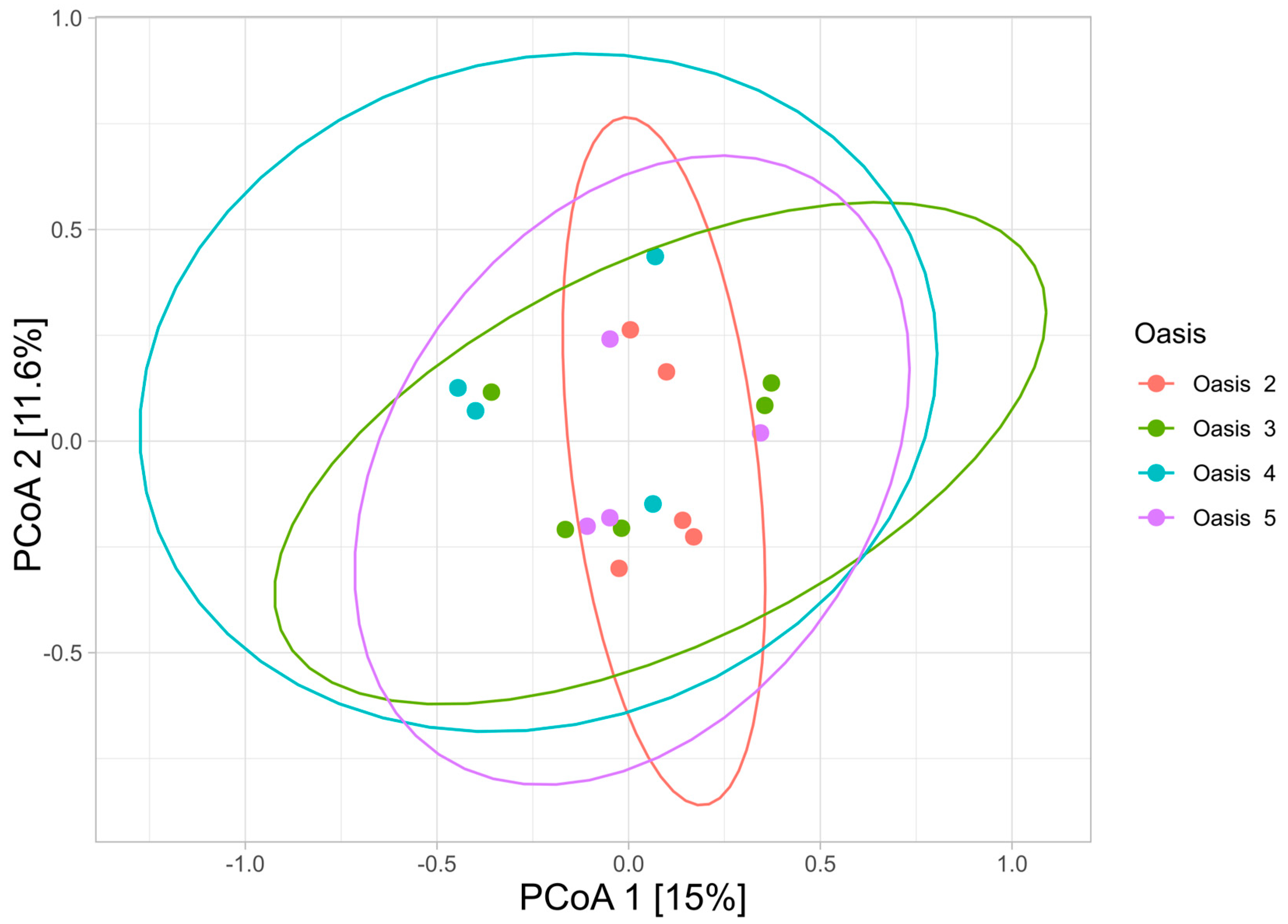
3.3. Beta Diversity Analysis
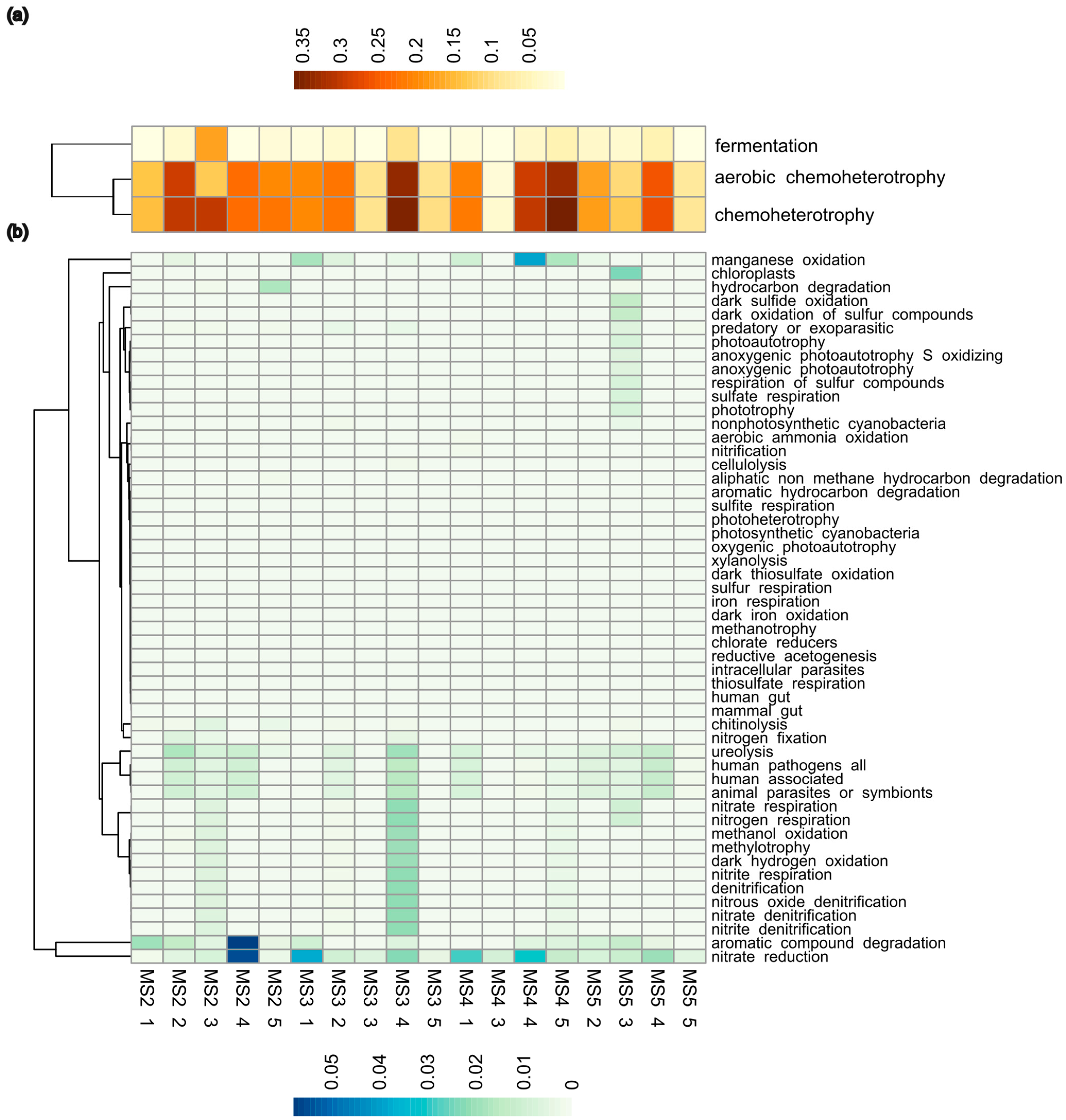
3.4. Functional Potential of Soil Microbiomes
4. Discussion and Conclusions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gobi Desert—An overview. Available online: https://www.sciencedirect.com/topics/earth-and-planetary-sciences/gobi-desert (accessed on 23 December 2022).
- Sternberg, T.; Rueff, H.; Middleton, N. Contraction of the Gobi Desert, 2000–2012. Remote Sens. 2015, 7, 1346–1358. [Google Scholar] [CrossRef]
- McCarthy, C.; Sternberg, T.; Hoshino, B.; Banfill, J.; Enkhjargal, E.; Konagaya, Y.; Phillips, S. Preserving the Gobi: Identifying potential UNESCO world heritage in Mongolia’s Gobi Desert. J. Asia-Pac. Biodivers. 2022, 15, 500–517. [Google Scholar] [CrossRef]
- Jargal, J. Protected area status in Mongolia. Mong. J. Biol. Sci. 2003, 1, 49–54. [Google Scholar]
- Great Gobi Biosphere Reserve, Mongolia. Available online: https://en.unesco.org/biosphere/aspac/great-gobi (accessed on 23 December 2022).
- Shu, W.S.; Huang, L.N. Microbial diversity in extreme environments. Nat. Rev. Microbiol. 2022, 20, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Noy-Meir, I. Desert ecosystems: Environment and producers. Annu. Rev. Ecol. Syst. 1973, 4, 25–51. [Google Scholar] [CrossRef]
- Roesch, L.F.W.; Fulthorpe, R.R.; Riva, A.; Casella, G.; Hadwin, A.K.M.; Kent, A.D.; Daroub, S.H.; Camargo, F.A.O.; Farmerie, W.G.; Triplett, E.W. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007, 1, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Castronovo, L.M.; Del Duca, S.; Chioccioli, S.; Vassallo, A.; Fibbi, D.; Coppini, E.; Fani, R. Biodiversity of Soil Bacterial Communities from the Sasso Fratino Integral Nature Reserve. Microbiol. Res. 2021, 12, 862–877. [Google Scholar] [CrossRef]
- Zhou, J.; Davey, M.E.; Figueras, J.B.; Rivkina, E.; Gilichinsky, D.; Tiedje, J.M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 1997, 143, 3913–3919. [Google Scholar] [CrossRef]
- Perera, I.; Subashchandrabose, S.R.; Venkateswarlu, K.; Naidu, R.; Megharaj, M. Consortia of cyanobacteria/microalgae and bacteria in desert soils: An underexplored microbiota. Appl. Microbiol. Biotechnol. 2018, 102, 7351–7363. [Google Scholar] [CrossRef]
- Amond, J.B.; Baxter, J.; Maggs-Kölling, G.; Martínez-Alvarez, L.; Read, D.A.; León-Sobrino, C.; Cowan, D.A. Microbial ecology of the Namib Desert. In Model Ecosystems in Extreme Environments; Academic Press: Cambridge, MA, USA, 2019; pp. 113–143. [Google Scholar]
- Orlando, J.; Alfaro, M.; Bravo, L.; Guevara, R.; Carú, M. Bacterial diversity and occurrence of ammonia-oxidizing bacteria in the Atacama Desert soil during a “desert bloom” event. Soil Biol. Biochem. 2010, 42, 1183–1188. [Google Scholar] [CrossRef]
- Chen, L.; Li, C.; Zhang, Z.; Feng, Q.; Xi, H.; Guo, R.; Wei, Y. Landscape differentiation of soil bacteria and bacteria-soil-vegetation interactions in desert-oasis ecosystems. Glob. Planet. Change 2024, 232, 104323. [Google Scholar] [CrossRef]
- Baubin, C.; Farrell, A.M.; Šťovíček, A.; Ghazaryan, L.; Giladi, I.; Gillor, O. Seasonal and spatial variability in total and active bacterial communities from desert soil. Pedobiologia 2019, 74, 7–14. [Google Scholar] [CrossRef]
- Jordaan, K.; Lappan, R.; Dong, X.; Aitkenhead, I.J.; Bay, S.K.; Chiri, E.; Greening, C. Hydrogen-oxidizing bacteria are abundant in desert soils and strongly stimulated by hydration. Msystems 2020, 5, e01131-20. [Google Scholar] [CrossRef] [PubMed]
- Ameen, F.; AlYahya, S.A.; AlNadhari, S.; Alasmari, H.; Alhoshani, F.; Wainwright, M. Phosphate solubilizing bacteria and fungi in desert soils: Species, limitations and mechanisms. Arch. Agron. Soil Sci. 2019, 65, 1446–1459. [Google Scholar] [CrossRef]
- Aanniz, T.; Ouadghiri, M.; Melloul, M.; Swings, J.; Elfahime, E.; Ibijbijen, J.; Amar, M. Thermophilic bacteria in Moroccan hot springs, salt marshes and desert soils. Braz. J. Microbiol. 2015, 46, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Steinberger, Y. Immediate effect of wetting event on microbial biomass and carbohydrate production-mediated aggregation in desert soil. In Soil Structure/Soil Biota Interrelationships; Elsevier: Amsterdam, The Netherlands, 1993; pp. 599–607. [Google Scholar]
- Wen, Y.; Zhang, G.; Zhang, W.; Liu, G. Distribution patterns and functional characteristics of soil bacterial communities in desert ecosystems of northern China. Sci. Total Environ. 2023, 905, 167081. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Shi, Q.; Li, H.; Zhou, X.; Dai, Y.; Kahaer, Y.; Peng, L. The combined effect of surface water and groundwater on environmental heterogeneity reveals the basis of beta diversity pattern in desert oasis communities. PLoS ONE 2022, 17, e0279704. [Google Scholar] [CrossRef] [PubMed]
- de Grenade, R.; Nabhan, G.P. Baja California Peninsula oases: An agro-biodiversity of isolation and integration. Appl. Geogr. 2013, 41, 24–35. [Google Scholar] [CrossRef]
- Liang, Y.; Liu, L. Simulating land-use change and its effect on biodiversity conservation in a watershed in northwest China. Ecosyst. Health Sustain. 2017, 3, 1335933. [Google Scholar] [CrossRef]
- Gunin, P.D.; Zolotokrylin, A.N. General characteristics of climate. In Deserts of the Trans-Altai Gobi: Natural Conditions, Ecosystems and Regioning; Nauka Publishing House: Moscow, Russia, 1986; pp. 27–29. [Google Scholar]
- Takhtajan, A.L. Higher Taxa of Vascular Plants, Except for Flowering Plants. In Problems of Paleobotany; Nauka Publishing House: Moscow, Russia, 1986; pp. 135–143. [Google Scholar]
- Grubov, V.I. Key to the Vascular Plants of Mongolia; Nauka: Leningrad, Russia, 1982; p. 442. [Google Scholar]
- Baasanmunkh, S.; Urgamal, M.; Oyuntsetseg, B.; Sukhorukov, A.P.; Tsegmed, Z.; Son, D.C.; Choi, H.J. Flora of Mongolia: Annotated checklist of native vascular plants. PhytoKeys 2022, 192, 63. [Google Scholar] [CrossRef]
- von Wehrden, H.; Hilbig, W.; Wesche, K. Plant communities of the Mongolian Transaltay Gobi. Feddes Reper. 2006, 117, 526–570. [Google Scholar] [CrossRef]
- Valboa, G.; Lagomarsino, A.; Brandi, G.; Agnelli, A.E.; Simoncini, S.; Papini, R.; Pellegrini, S. Long-term variations in soil organic matter under different tillage intensities. Soil Tillage Res. 2015, 154, 126–135. [Google Scholar] [CrossRef]
- Sequi, P.; De Nobili, M. Carbonio organico. Angeli F Minist. Politiche Agric. E For. Oss. Naz. Pedol. Qual. Suolo 2000, 7, 1–13. [Google Scholar]
- Andrenelli, M.C.; Fiori, V.; Pellegrini, S. Soil particle-size analysis up to 250 μm by X-ray granulometer: Device set-up and regressions for data conversion into pipette-equivalent values. Geoderma 2013, 192, 380–393. [Google Scholar] [CrossRef]
- Chakravorty, S.; Helb, D.; Burday, M.; Connell, N.; Alland, D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 2007, 69, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Mori, H.I.R.O.S.H.I.; Maruyama, F.U.M.I.T.O.; Kato, H.I.R.O.M.I.; Toyoda, A.T.S.U.S.H.I.; Dozono, A.Y.U.M.I.; Ohtsubo, Y.O.S.H.I.Y.U.K.I.; Kurokawa, K.E.N. Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 2014, 21, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Jones, E.; Harden, S.; Crawley, M.J. The R Book; John Wiley & Sons: Hoboken, NJ, USA, 2022. [Google Scholar]
- Wright, E.S. Using DECIPHER v2. 0 to analyze big biological sequence data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Maestre, F.T.; Gallardo, A.; Bowker, M.A.; Wallenstein, M.D.; Quero, J.L.; Zaady, E. Decoupling of soil nutrient cycles as a function of aridity in global drylands. Nature 2013, 502, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xu, X.; Ding, J.; Bao, F.; De Costa, Y.G.; Zhuang, W.; Wu, B. The responses to long-term water addition of soil bacterial, archaeal, and fungal communities in a desert ecosystem. Microorganisms 2021, 9, 981. [Google Scholar] [CrossRef] [PubMed]
- Ronca, S.; Ramond, J.B.; Jones, B.E.; Seely, M.; Cowan, D.A. Namib Desert dune/interdune transects exhibit habitat-specific edaphic bacterial communities. Front. Microbiol. 2015, 6, 845. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Couteau, C.; Luo, F.; Neveu, J.; DuBow, M.S. Bacterial diversity of surface sand samples from the Gobi and Taklamaken deserts. Microb. Ecol. 2013, 66, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, T.; Li, J.; Wu, M.; Liu, G.; Zhang, W.; Zhang, G. High proportions of radiation-resistant strains in Culturable bacteria from the Taklimakan Desert. Biology 2022, 11, 501. [Google Scholar] [CrossRef]
- Heulin, T.; De Luca, G.; Barakat, M.; Gommeaux, M.; de Groot, A.; Blanchard, L.; Achouak, W. Bacterial Adaptation to Hot and Dry Deserts; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 75–98. [Google Scholar]
- Xia, Q.; Rufty, T.; Shi, W. Soil microbial diversity and composition: Links to soil texture and associated properties. Soil Biol. Biochem. 2020, 149, 107953. [Google Scholar] [CrossRef]
- Reith, F.; Brugger, J.; Zammit, C.M.; Gregg, A.L.; Goldfarb, K.C.; Andersen, G.L.; Wakelin, S.A. Influence of geogenic factors on microbial communities in metallogenic Australian soils. ISME J. 2012, 6, 2107–2118. [Google Scholar] [CrossRef]
- Ardley, J.K.; Parker, M.A.; De Meyer, S.E.; Trengove, R.D.; O’Hara, G.W.; Reeve, W.G.; Howieson, J.G. Microvirga lupini sp. nov., Microvirga lotononidis sp. nov. and Microvirga zambiensis sp. nov. are alphaproteobacterial root-nodule bacteria that specifically nodulate and fix nitrogen with geographically and taxonomically separate legume hosts. Int. J. Syst. Evol. Microbiol. 2012, 62 Pt 11, 2579–2588. [Google Scholar] [CrossRef] [PubMed]
- Radl, V.; Simoes-Araujo, J.L.; Leite, J.; Passos, S.R.; Martins LM, V.; Xavier, G.R.; Zilli, J.E. Microvirga vignae sp. nov., a root nodule symbiotic bacterium isolated from cowpea grown in semi-arid Brazil. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 3, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Subhash, Y.; Lee, S.S. Skermanella rosea sp. nov., isolated from hydrocarbon-contaminated desert sands. Int. J. Syst. Evol. Microbiol. 2016, 66, 3951–3956. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, B.X.; Ren, H.; Zhang, J. Composition and functional diversity of microbial community across a mangrove-inhabited mudflat as revealed by 16S rDNA gene sequences. Sci. Total Environ. 2018, 633, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Leung, P.M.; Bay, S.K.; Meier, D.V.; Chiri, E.; Cowan, D.A.; Gillor, O.; Greening, C. Energetic basis of microbial growth and persistence in desert ecosystems. Msystems 2020, 5, 10-1128. [Google Scholar] [CrossRef]
- Wu, D.; Senbayram, M.; Moradi, G.; Mörchen, R.; Knief, C.; Klumpp, E.; Bol, R. Microbial potential for denitrification in the hyperarid Atacama Desert soils. Soil Biol. Biochem. 2021, 157, 108248. [Google Scholar] [CrossRef]
- Neveu, J.; Regeard, C.; DuBow, M.S. Isolation and characterization of two serine proteases from metagenomic libraries of the Gobi and Death Valley deserts. Appl. Microbiol. Biotechnol. 2011, 91, 635–644. [Google Scholar] [CrossRef]
| Oasis | pH | TC % | TOC % | TN % | C/N | Tot CaCO3 % | Sand | Clay | Silt | Texture Profile | Temperature (°C) | Precipitation (mm) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MS2_1 | Oasis_2 | NA | 0.46 | 0.06 | 0.00 | NA | 3.30 | 91.21 | 2.97 | 5.83 | sand | 23.64 | 1.40 |
| MS2_2 | Oasis_2 | 7.85 | 0.47 | 0.24 | 0.00 | NA | 1.90 | 91.61 | 1.00 | 7.39 | sand | 23.64 | 1.40 |
| MS2_3 | Oasis_2 | 8.2 | 0.37 | 0.08 | 0.00 | NA | 2.40 | 86.22 | 3.03 | 10.75 | loamy_sand | 23.64 | 1.40 |
| MS2_4 | Oasis_2 | NA | 0.44 | 0.23 | 0.00 | NA | 1.80 | 71.37 | 1.00 | 27.63 | loamy_sand | 23.64 | 1.40 |
| MS2_5 | Oasis_2 | 7.8 | 0.52 | 0.15 | 0.00 | NA | 3.10 | 90.48 | 2.70 | 6.83 | sand | 23.64 | 1.40 |
| MS3_1 | Oasis_3 | 8.6 | 0.46 | 0.13 | 0.00 | NA | 2.70 | 89.12 | 1.00 | 9.88 | sand | 25.59 | 0.50 |
| MS3_2 | Oasis_3 | 7.95 | 1.17 | 1.04 | 0.04 | 23.30 | 1.10 | 82.67 | 1.00 | 16.33 | loamy_sand | 25.59 | 0.50 |
| MS3_3 | Oasis_3 | 8.29 | 1.33 | 1.03 | 0.07 | 15.40 | 2.50 | 82.96 | 1.54 | 15.50 | loamy_sand | 25.59 | 0.50 |
| MS3_4 | Oasis_3 | 7.8 | 1.44 | 0.98 | 0.06 | 17.30 | 3.80 | 90.52 | 1.00 | 8.48 | sand | 25.59 | 0.50 |
| MS3_5 | Oasis_3 | 8.52 | 0.95 | 0.36 | 0.15 | 2.40 | 4.90 | 72.61 | 12.85 | 14.54 | sandy_loam | 25.59 | 0.50 |
| MS4_1 | Oasis_4 | 8.49 | 2.88 | 1.91 | 0.13 | 14.90 | 8.20 | 67.22 | 4.19 | 28.59 | sandy_loam | 23.07 | 2.10 |
| MS4_2 | Oasis_4 | 8.12 | 14.92 | 9.15 | 0.76 | 12.10 | 48.10 | 11.42 | 19.14 | 69.44 | silt_loam | 23.07 | 2.10 |
| MS4_3 | Oasis_4 | 9.82 | 4.70 | 3.61 | 0.24 | 15.30 | 9.00 | 46.30 | 18.35 | 35.35 | loam | 23.07 | 2.10 |
| MS4_4 | Oasis_4 | 8.3 | 1.35 | 0.45 | 0.03 | 14.40 | 7.50 | 80.25 | 1.00 | 18.75 | loamy_sand | 23.07 | 2.10 |
| MS4_5 | Oasis_4 | 8.38 | 1.23 | 0.59 | 0.02 | 33.70 | 5.40 | 85.33 | 0.98 | 13.69 | loamy_sand | 23.07 | 2.10 |
| MS5_1 | Oasis_5 | NA | 16.70 | 15.73 | 1.48 | 10.60 | 8.20 | NA | NA | NA | NA | 27.20 | 0.60 |
| MS5_2 | Oasis_5 | 7.75 | 1.21 | 0.60 | 0.02 | 35.90 | 5.10 | 27.14 | 23.42 | 49.45 | loam | 27.20 | 0.60 |
| MS5_3 | Oasis_5 | NA | 2.52 | 0.75 | 0.04 | 18.60 | 14.70 | 30.76 | 18.69 | 50.55 | silt_loam | 27.20 | 0.60 |
| MS5_4 | Oasis_5 | 8.26 | 3.51 | 2.73 | 0.26 | 10.50 | 6.50 | 29.80 | 16.44 | 53.75 | silt_loam | 27.20 | 0.60 |
| MS5_5 | Oasis_5 | 8.82 | 0.83 | 0.43 | 0.01 | 43.50 | 3.40 | 60.27 | 11.79 | 27.94 | sandy_loam | 27.20 | 0.60 |
| TC | Tot CaCO3 | Sand | Clay | Silt | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | CV (%) | Sig. | Mean | SD | CV (%) | Sig. | Mean | SD | CV (%) | Sig. | Mean | SD | CV (%) | Sig. | Mean | SD | CV (%) | Sig. | |
| Oasis_2 | 0.45 | 0.05 | 11.82 | b | 2.50 | 0.70 | 28.22 | b | 86.18 | 8.55 | 9.92 | a | 2.14 | 1.04 | 48.96 | b | 11.68 | 9.10 | 77.92 | b |
| Oasis_3 | 1.07 | 0.39 | 36.20 | ab | 3.02 | 1.43 | 47.67 | ab | 83.57 | 7.08 | 8.47 | a | 3.48 | 5.24 | 150.79 | b | 12.94 | 3.53 | 27.27 | b |
| Oasis_4 | 2.54 | 1.62 | 63.96 | a | 7.51 | 1.57 | 20.85 | a | 69.77 | 17.40 | 24.95 | a | 6.13 | 8.29 | 135.18 | b | 24.09 | 9.72 | 40.35 | b |
| Oasis_5 | 2.02 | 1.23 | 60.82 | ab | 7.42 | 5.01 | 67.57 | ab | 36.99 | 15.59 | 42.15 | b | 17.58 | 4.83 | 27.49 | a | 45.42 | 11.79 | 25.97 | a |
| Cluster | Variables | V-Test | p-Value |
|---|---|---|---|
| 1 | sand | 2.19 | 0.028 |
| TC | −2.14 | 0.033 | |
| silt | −2.36 | 0.018 | |
| 6 | clay | 2.20 | 0.028 |
| sand | −1.99 | 0.047 | |
| 7 | TC | 3.31 | 0.00092 |
| silt | 2.11 | 0.034 | |
| clay | 2.05 | 0.040 | |
| sand | −2.17 | 0.030 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esposito, A.; Del Duca, S.; Vitali, F.; Bigiotti, G.; Mocali, S.; Semenzato, G.; Papini, A.; Santini, G.; Mucci, N.; Padula, A.; et al. The Great Gobi A Strictly Protected Area: Characterization of Soil Bacterial Communities from Four Oases. Microorganisms 2024, 12, 320. https://doi.org/10.3390/microorganisms12020320
Esposito A, Del Duca S, Vitali F, Bigiotti G, Mocali S, Semenzato G, Papini A, Santini G, Mucci N, Padula A, et al. The Great Gobi A Strictly Protected Area: Characterization of Soil Bacterial Communities from Four Oases. Microorganisms. 2024; 12(2):320. https://doi.org/10.3390/microorganisms12020320
Chicago/Turabian StyleEsposito, Antonia, Sara Del Duca, Francesco Vitali, Gaia Bigiotti, Stefano Mocali, Giulia Semenzato, Alessio Papini, Giacomo Santini, Nadia Mucci, Anna Padula, and et al. 2024. "The Great Gobi A Strictly Protected Area: Characterization of Soil Bacterial Communities from Four Oases" Microorganisms 12, no. 2: 320. https://doi.org/10.3390/microorganisms12020320
APA StyleEsposito, A., Del Duca, S., Vitali, F., Bigiotti, G., Mocali, S., Semenzato, G., Papini, A., Santini, G., Mucci, N., Padula, A., Greco, C., Nasanbat, B., Davaakhuu, G., Bazarragchaa, M., Riga, F., Augugliaro, C., Cecchi, L., Fani, R., & Zaccaroni, M. (2024). The Great Gobi A Strictly Protected Area: Characterization of Soil Bacterial Communities from Four Oases. Microorganisms, 12(2), 320. https://doi.org/10.3390/microorganisms12020320














