Abstract
Antimicrobial resistance poses one of the greatest threats to global health and there is an urgent need for new therapeutic options. Phages are viruses that infect and kill bacteria and phage therapy could provide a valuable tool for the treatment of multidrug-resistant infections. In this study, water samples collected by citizen scientists as part of the Citizen Phage Library (CPL) project, and wastewater samples from the Environment Agency yielded phages with activity against clinical strains Klebsiella pneumoniae BPRG1484 and Enterobacter cloacae BPRG1482. A total of 169 and 163 phages were found for K. pneumoniae and E. cloacae, respectively, within four days of receiving the strains. A third strain (Escherichia coli BPRG1486) demonstrated cross-reactivity with 42 E. coli phages already held in the CPL collection. Seed lots were prepared for four K. pneumoniae phages and a cocktail combining these phages was found to reduce melanisation in a Galleria mellonella infection model. The resources and protocols utilised by the Citizen Phage Library enabled the rapid isolation and characterisation of phages targeted against multiple strains. In the future, within a clearly defined regulatory framework, phage therapy could be made available on a named-patient basis within the UK.
1. Introduction
Antimicrobial resistance (AMR) has been declared by the World Health Organisation (WHO) as one of the top ten global public health threats facing humanity [1]. In 2019 there were an estimated 1.27 million deaths directly attributable to bacterial AMR, greater than the combined number of deaths from malaria and HIV/AIDS for that year [2]. Whilst there are innovative approaches to finding novel antibiotics [3,4] and progressive economic models to incentivise antibiotic development [5,6], developing new antibiotics remains scientifically and financially challenging. With the rapid rise of AMR there is an urgent need for new therapeutic options.
Bacteriophage (phage) therapy employs viruses that infect and kill bacterial pathogens [7]. Phage–bacteria interactions are highly specific and phage therapy has the advantage that, unlike antibiotics, there is no collateral damage to the host microbiome [8]. In Georgia, Russia and Poland, phage therapy has been widely used for decades [9,10] and has a proven track record for safety [11]. In western countries, the use of phages to treat bacterial infections fell by the wayside as the first antibiotics came into widespread use in the 1940s [12]. However, the rapid emergence of AMR has given rise to a resurgence in the use of phage therapy [13,14,15,16,17] and currently 15 phage therapy clinical trials listed on https://clinicaltrials.gov (accessed on 1 August 2023) [18] are actively recruiting.
One hurdle to achieving successful phage therapy is the development of bacterial resistance to the phage. Resistance to a particular phage can occur through a variety of mechanisms including altering surface phage receptors, degrading phage nucleic acids and triggering abortive infection systems [19,20]. An ideal broad-spectrum phage medicine would therefore take the form of a phage cocktail containing multiple different phages that not only collectively infect a large proportion of the circulating strains but also target different receptor binding proteins on host strains to minimise viable routes to evolved resistance [21]. To bring a new medicine to market in the UK it must have been prepared according to good manufacturing practice (GMP) standards and have undergone randomised controlled trials to assess safety and efficacy before an application can be made to license the product [22]. To achieve GMP production of phage therapy medicines in the UK will take time and require considerable investment. There is a strong case for an alternative, rapid turnaround, relatively low-cost pathway for phage therapy where patients with multidrug-resistant (MDR) infections have exhausted all other treatments.
Several countries are able to administer phage therapy via non-GMP pathways [14,15,23,24]. In the US for example, patients with life-threatening MDR infections can access phage therapy through the expanded access pathway [25] and in Belgium, a framework for phage therapy exists whereby magistral phage preparations prepared according to a standard monograph can be administered on a named-patient basis [22,26]. The STAMP study in Australia (standardised treatment and monitoring protocol to assess safety and tolerability of bacteriophage therapy for adult and paediatric patients) enables patients who have exhausted all other therapeutic options for control of their infection to access phage therapy [27]. Since 2019, there have been twelve cases of phage therapy in the UK using non-GMP prepared phage. Two cases have involved the treatment of Mycobacterium infections [14,28] (using phage discovered and prepared at the University of Pittsburgh, PA, USA) and in a more recent study a magistral phage preparation (phage discovered at the Eliava Institute, Georgia, phage prepared at the Queen Anne Military Hospital, Brussels, Belgium) was used to treat 10 patients with limb-threatening diabetic foot infections [29].
Phage therapy depends on the availability of a large and diverse bank of isolated phages, or the capacity to rapidly isolate suitable phages from the environment against a clinical pathogen. Numerous phage banks have been established globally and have provided phage for research and phage therapy [30]. One approach to building phage banks is through citizen scientist projects. In addition to educational benefits, engaging individuals in scientific research can increase scientific output. The SEA-PHAGES (Science Education Alliance–Phage Hunters Advancing Genomics and Evolutionary Science) program [31] has to date enrolled 1400 students and has banked 4471 fully sequenced Actinobacteriophages (https://phagesdb.org (accessed on 1 August 2023)). Phages from this program have been used to treat drug-resistant nontuberculous Mycobacterium infections, including infections in two patients in the UK as described above [14,28].
In 2020 the Citizen Phage Library (CPL, https://citizenphage.com (accessed on 1 December 2023)) was established in the UK to provide therapeutic phages to combat AMR through citizen science. Phage Hunters are invited to collect water samples from their local environment which are then sent to the CPL laboratory for the isolation of phages against WHO priority pathogens. The CPL currently holds over 1000 phages against pathogens such as Acinetobacter baumannii, Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus and Burkholderia spp. In addition, the CPL holds nearly 600 water samples sent in by citizen scientists and several thousand wastewater samples provided by the Environment Agency from wastewater monitoring. These resources mean the CPL is well placed to screen clinical isolates for cross-reactivity with the banked phage collection and if no phages are available, the screening of water samples can be initiated immediately. To date, the CPL has responded successfully to four urgent need requests for phages made through the PhageDirectory (https://phage.directory/alerts (accessed on 1 December 2023)).
The study described here outlines the CPL response to a request from a UK hospital (via PhageDirectory) for phages against three MDR pathogens. The study highlights how protocols for high-throughput screening established by the CPL could be replicated to provide rapid access to phages for named-patient clinical use, supported by a clearly defined regulatory framework. The timeline for the discovery of phages against these strains is presented and the selection criteria for deciding which phages to take forward for the preparation of high titre purified seed lots are described.
2. Materials and Methods
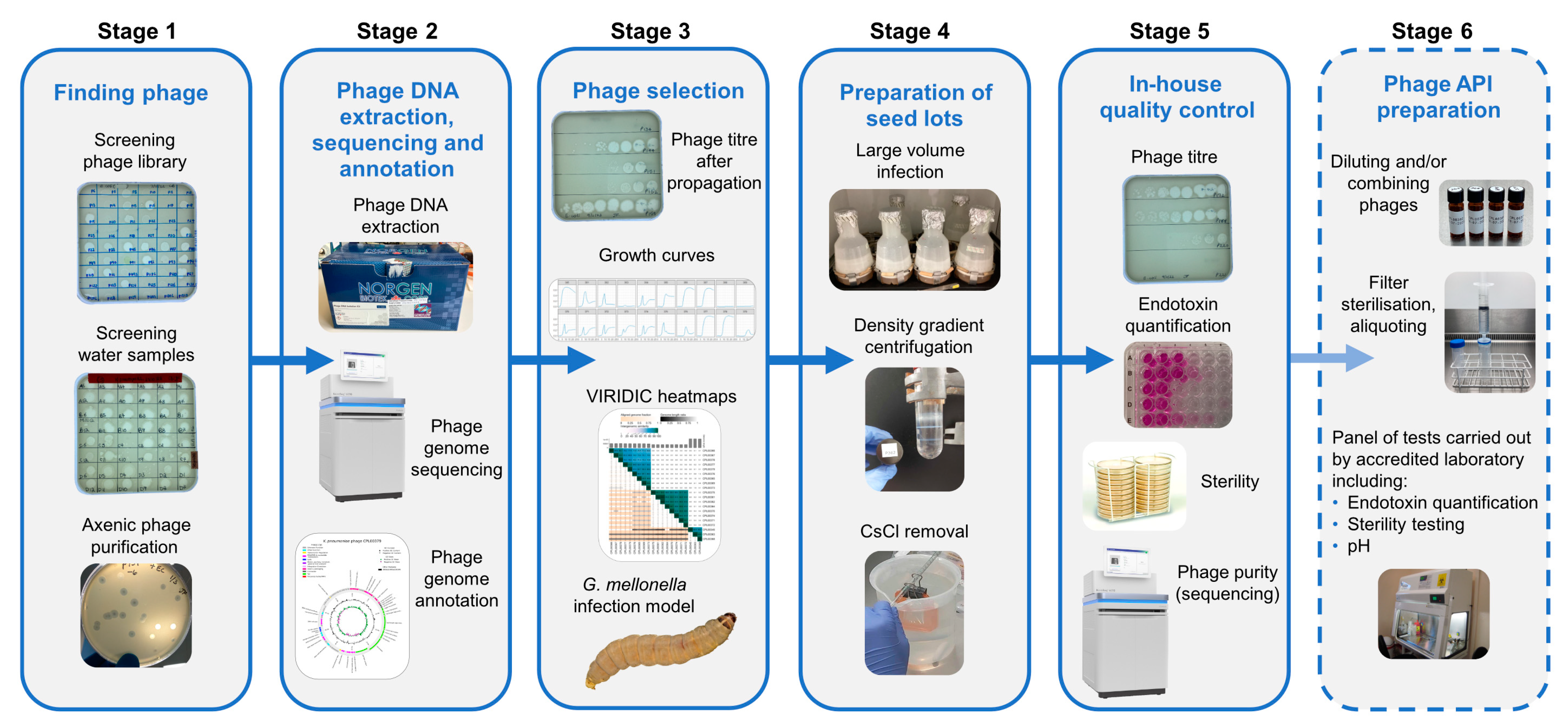
Figure 1 represents an overview of the workflow for finding and selecting phages against incoming strains through to quality control checks of phage seed lots.
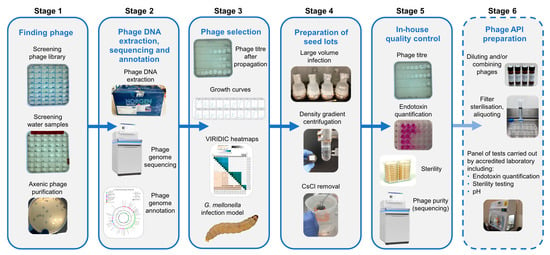
Figure 1.
Overview of bacteriophage isolation, selection and preparation of seed lots. Stage1. Phages active against the target strain held within the currently available library are identified. New phages are isolated from citizen scientist water and Environment Agency wastewater samples and are purified to give axenic preparations. Stage 2. DNA is extracted from overnight infections, sequenced and annotated. Stage 3. Phages are selected based on a number of criteria: phage titre on propagation, ability to suppress growth in liquid culture and genetic dissimilarity with other phages in the selection panel. Stage 4. Phages deemed safe for human use are cultivated in 2 L volumes. Following centrifugation, the supernatant is 0.22 µm filtered to remove bacterial debris. Phages are precipitated from the filtrate using PEG8000 and NaCl. Following centrifugation, the precipitated phage is resuspended in 20 mL of sterile saline and applied to a CsCl gradient. The phage band obtained via ultracentrifugation is removed and dialysed extensively against sterile saline. Stage 5. In-house quality control on the seed lot to determine phage titre, endotoxin content, sterility and phage purity. Stage 6. For the proposed preparation of an active pharmaceutical ingredient (API), working in an environment with specified air quality and cleanliness, seed lots would be diluted (and combined if required), passed through a medical grade 0.22 µm filter and aliquoted into pharmaceutical grade vials. Quality control tests including endotoxin quantification, pH and sterility would be undertaken in MHRA accredited laboratories.
2.1. Maintenance and Sequencing of Bacterial Strains
Bacterial strains received from the hospital’s clinical microbiology laboratory were streaked onto LB agar and cryostocks were subsequently prepared using Pro-Lab Microbank vials (Fisher Scientific, Loughborough, UK). Three strains were received, a Klebsiella pneumoniae strain (assigned as BPRG 1484), an Enterobacter cloacae strain (assigned as BPRG 1482) and an Escherichia coli strain (assigned as BPRG 1486). All three strains were positive for IMP (active-on-imipenem) carbapenemase (Table S1). Genomic DNA was extracted from the bacterial strains using the Nanobind CBB kit RT (PacBio, Menlo Park, CA, USA) according to the manufacturer’s instructions. Long-read sequencing of the bacterial genomes was carried out using a R9.4.1 (FLO-MIN 106) MinION flow cell (Oxford Nanopore Technologies (ONT), Oxford, UK). Library preparation was conducted using the Rapid Sequencing Kit (SQK-RAD004, ONT). Following priming of the flow cell, using the Flow Cell Priming Kit (EXP-FLP002, ONT), the library was loaded onto the flow cell. Data acquisition was achieved in real time using MinKNOW and basecalling was carried out using the Oxford Nanopore Guppy tool (v.6.0.1) using super-high accuracy models. Genomes were assembled using Trycycler (v0.5.3) and annotated with Prokka (v1.14.6).
2.2. Screening the Phage Collection against Bacterial Strains
Spot tests [32,33] were carried out as follows. LB broth supplemented with MgCl2 and CaCl2 (each at a final concentration of 10 mM, hereafter referred to as sLB) was inoculated with growth from the agar slope provided by the hospital laboratory and incubated at 37 °C with shaking (200 rpm) until the mid-logarithmic phase was reached (OD600 = 0.6). A 1 mL volume of the culture was added to 3 mL of cooled (55 °C) molten top agar, gently mixed and poured over the surface of pre-warmed bottom agar plates. Once set, phage lysates from the CPL collection were spotted (5 µL) onto the surface of the inoculated top agar and following overnight incubation at 37 °C, plates were examined for the presence of zones of lysis (ZOL). Top and bottom agar consisted of LB broth containing 0.65% w/v and 1% w/v bacteriological agar respectively. After cooling, MgCl2 and CaCl2 (each at a final concentration of 10 mM) were added to the top and bottom agar prior to use.
2.3. Screening Water Samples for New Phage against Bacterial Strains
Water samples sent in by citizen scientists and wastewater samples provided by the Environment Agency were screened for the presence of phage against the target K. pneumoniae and E. cloacae strains using a modified version of the high throughput method developed by Olsen et al. [33]. Citizen scientist water samples had been previously centrifuged at 10,000× g for 20 min (if required) to remove particulate matter, filtered through a 0.22 µm syringe filter (Sartorius, Goettingen, Germany) into sterile amber glass bottles and stored at 4 °C. Wastewater samples had been provided as either 1 mL aliquots in micro-centrifuge tubes stored at 4 °C or in frozen (−20 °C) 100 µL aliquots in microtitre trays. Wastewater samples were pooled (5–10 of the 1 mL aliquots or 30–40 of the 100 µL frozen aliquots), filtered (0.22 µm filter) into amber glass bottles and stored at 4 °C. For the screening assays, a 1 mL volume of each filtered water sample was added to a sterile 96 deep well plate (square well, V bottom, VWR, Radnor, PA, USA). To this was added 335 µL of 4.4 × concentrated LB broth, 60 µL of a 0.25 M MgCl2 plus 0.25 M CaCl2 stock solution and 70 µL of an overnight culture of the target strain. Plates were sealed with a SealPlate film (Merck, Darmstadt, Germany) and incubated overnight at 37 °C with shaking at 200 rpm (1st round enrichment). Following incubation, 200 µL from each well were transferred to a 96-well 0.45 µm MultiScreenHTS HV sterile filter plate (Millipore, Merck, Darmstadt, Germany) placed on top of a 96-well microtitre plate (Grenier Bio-One, Kremsmünster, Austria). The double plate stack was centrifuged at 900× g for 4 min. A 5 µL volume of the filtrate was transferred to a new 96-well microtitre plate containing 95 µL sLB and 5 µL of an overnight culture of the target strain in each well. The plate was sealed as before and incubated overnight at 37 °C with shaking at 200 rpm (2nd round enrichment). Following incubation, 200 µL from each well were filtered as described previously. Filtrates from the 2nd round enrichment were spotted (5 µL) onto inoculated top agar overlays and following overnight incubation at 37 °C, plates were examined for the presence of ZOL. To purify the phage contained within the ZOL (which may contain more than one phage type), a core was taken from each clear zone using a sterile tip and transferred to 100 µL of SM buffer (50 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 8 mM MgSO4). For axenic phage purification two methods were used and these are described in the supplementary material. For the wastewater samples, the volume of sample was limited and therefore for wastewater screening, the two target strains, K. pneumoniae and E. cloacae, were combined in the 1st round enrichment and 37.5 µL of an overnight culture of each strain were added to the deep well plate. The 2nd round enrichment was carried out on individual bacterial strains.
2.4. Phage DNA Extraction, Sequencing and Annotation
Phage infections were set up by adding 50 µL of purified phage and 500 µL of overnight culture to 20 mL of sLB (two tubes for each phage). Following overnight incubation at 37 °C, the infected cultures were centrifuged at 10,000× g for 20 min and the supernatant was filtered through a 0.22 µm syringe filter. A 30 mL volume of the resulting phage lysate was treated with 30 µL of DNase I and RNase A solution (10 mg/mL DNase I (Roche, Merck, Darmstadt, Germany) mixed with an equal volume of 20 mg/mL RNase A (Invitrogen, ThermoFisher Scientific, Waltham, MA, USA)) for 30 min at 37 °C to remove any remaining bacterial nucleic acid. Polyethylene glycol 8000 (PEG8000) and NaCl were added to the lysate to give final concentrations of 20% w/v and 2.5 M, respectively, and mixed gently by inversion until dissolved. The lysate was incubated at 4 °C overnight. Precipitated phages were concentrated via centrifugation at 10,000× g for 30 min and the phage pellet was re-suspended in 1 mL of SM buffer [34]. DNA was extracted from the phage using the Norgen phage DNA extraction kit (Norgen Biotek, Thorold, ON, Canada) following the manufacturer’s instructions. Phage DNA was quantified using the Qubit dsDNA broad range kit (Invitrogen, Waltham, MA, USA) and quality checked using the Genomic DNA ScreenTape assay on the Agilent 4200 TapeStation system. DNA libraries were prepared for short-read sequencing using NEBNext Ultra II FS Library Preparation and run on the Illumina Novaseq by the Exeter Sequencing Facility, to generate 2 × 150 bp paired-end reads. BBTools (v38.93) (https://sourceforge.net/projects/bbmap/ (accessed on 4 November 2022)) was used to remove optical duplicates, adapters and artefacts from the reads and error correction was carried out using Tadpole. Reads were mapped against host bacterial genomes to remove remaining residual host DNA and any DNA from prophages activated by lytic infection. Unmapped, corrected reads were assembled using Shovill (v1.1.0) using the following settings ‘--minlen 10,000 –mincov 20’. Assembled contigs were screened with CheckV (v1.01). Redundant overlapping ends were removed with apc (https://github.com/jfass/apc/ (accessed on 4 November 2022)). To evaluate whether new phages represented novel taxa, their closest relative at the nucleotide level was identified using the Mash distance as part of the INPHARED [35] pipeline. Reciprocal average nucleotide identity was then calculated between all closest relative genomes and all phage genomes isolated in this campaign using VIRIDIC [36], with novel species and genera assigned at 95% and 70% average nucleotide identity, as per the ICTV recommendations [37]. A phage was considered to represent a novel strain if its genome was less than 99% identical at the nucleotide level. Citizen scientists were invited to suggest names for their phage(s), which were then assigned as species names where appropriate. For further confirmation of the taxonomy of novel phages, nucleotide sequences from isolated phages were uploaded to the ViPTree Server (https://www.genome.jp/viptree (accessed on 9 January 2024), v 4.0) and aligned against dsDNA prokaryotic viruses in the Virus-Host DB to create a phylogenetic tree based on predicted proteome similarity [38]. The closest related phages in the database were assessed by alignment of phage genomes from the nearest shared inner branch of the generated tree. Full taxonomy information for each closest related phage was extracted using the NCBI Entrez Fetch command line tool and compared to nearest-neighbours identified by using INPHARED.
2.5. Phage Selection
Phages were selected for the preparation of seed lots from a panel of 20 K. pneumoniae phages and 22 E. coli phages according to the following criteria:
- Absence of temperate markers, antimicrobial resistance (AMR) genes and virulence factors. Phage genomes were annotated using Pharokka [39]. PhageAI (https://phage.ai/ (accessed on 8 November 2022)) [40] was used to predict the life cycle for each phage (virulent or temperate) and screening of phage genomes for AMR genes and virulence factors was carried out using Resistance Gene Identifier (RGI) [41] against the CARD database (https://card.mcmaster.ca/ (accessed on 8 November 2022)) and PhageLeads [42].
- Titre (PFU/mL) achieved on propagation on the hospital strains. Phage titres were determined using the small-drop plating assay [43]. A 1 mL volume of a mid-logarithmic culture (OD600 = 0.6) was added to 3 mL of cooled (55 °C) molten top agar, gently mixed and poured over the surface of a pre-warmed bottom agar plate. Ten-fold serial dilutions of the phage preparation were then spotted (5 µL) onto the inoculated agar overlay. Once the drops had dried, the plates were incubated at 37 °C and after overnight incubation, plaques were counted.
- Ability to suppress growth in liquid culture over a 24 h growth period. A mid-logarithmic culture of the bacterial host was diluted to an OD600 of 0.1 and 5 µL of this suspension was added to 95 µL of sLB broth in a microtitre plate. Phage lysates from the CPL collection (5 µL) were then added to each well as appropriate and the plate was incubated at 37 °C with periodic shaking in an Infinite® 200 Pro plate reader (Tecan, Männedorf, Switzerland). Measurements of OD600 were recorded every 30 min for 24 h. For the control wells, 5 µL of sLB broth were added in -place of phage lysates.
- Genetic clustering of phage genomes. Heatmaps for the K. pneumoniae and E. coli phage genomes were generated using VIRIDIC [36] to determine the intergenomic similarities of phages in the selection panel. Phages were downselected to maximise diversity by selecting phages from different genomic clusters in order to increase the likelihood of generating a cocktail that targeted multiple receptor binding sites.
2.6. Preparation of Seed Lots
Phages were propagated in sequentially larger volumes as follows [44]. On the first day a 20 mL volume of sLB was inoculated with 50 µL of phage lysate and 500 µL of a mid-logarithmic (OD600 = 0.6) culture of the host organism. In this case, the K. pneumoniae BPRG 1484 strain was used as the propagation host. Following overnight incubation at 37 °C with shaking at 200 rpm, the infected culture was centrifuged and filtered using a 0.22 µm syringe filter to give a mini phage lysate. A larger volume (100 mL) of sLB was then inoculated with 250 µL of the mini phage lysate and 2.5 mL of an overnight culture and incubated and filtered as previously to give a midi phage lysate. Next, three flasks each containing 667 mL of sLB were inoculated with 1.67 mL of the midi phage lysate and 16.7 mL of an overnight culture. Following overnight incubation, the infected cultures were pooled and vacuum filtered through a 0.22 µm bottle-top filter unit (Fisher Scientific, Loughborough, UK) into a sterile glass bottle. The combined 2 L of filtered maxi phage lysate were transferred to a sterile 5 L flask and PEG8000 and NaCl were added to give final concentrations of 20% w/v and 2.5 M, respectively. A sterile stirrer bar was added and the lysate was stirred at 4 °C overnight. The lysate was then centrifuged in 200 mL volumes at 10,000× g for 30 min at 4 °C and the phage pellets were resuspended in 0.9% w/v sterile saline to give a total volume of 20 mL of precipitated phage suspension. The CsCl (UltraPure, optical grade, ThermoFisher Scientific, Waltham, MA, USA) gradient was prepared by layering 3 mL of successively dense solutions (1.2, 1.4, 1.5 and 1.6 g/mL prepared in autoclaved Milli-Q water) of CsCl into a 26.3 mL ultracentrifuge tube (Beckman Coulter, Brea, CA, USA) starting with the most dense solution at the bottom of the tube. Half of the precipitated phage suspension was then applied to the gradient. Following ultracentrifugation at 120,000× g for 3 h at 4 °C, the phage band was removed in a volume of ~2 mL by careful pipetting and dialysed extensively against sterile saline over 24 h. A Slide-A-Lyser dialysis cassette (Fisher Scientific, Loughborough, UK) with a 10 kDa MWCO was pre-wetted with sterile saline and ~2 mL of air were injected into the cassette via the injection port. A 1.5 mL volume of the phage suspension was injected into the dialysis cassette, which was then suspended in a sterile beaker containing 1 L of sterile saline and placed at 4 °C with stirring. The saline was replaced with fresh sterile saline at 18, 21 and 24 h. PFU/mL counts were carried out at each step of the seed lot preparation.
2.7. The In-House Quality Control of Seed Lots
Phage titre (PFU/mL) of the seed lots was determined as described above. The endotoxin concentration of the phage preparation was determined using the ToxinSensor chromogenic lyophilised amoebocyte lysate (LAL) endotoxin assay kit (GenScript, Pistcataway, NJ, USA). The assay was carried out according to the manufacturer’s instructions, using one-tenth volumes, in a pyrogen-free round-bottomed 96 well plate (Grenier Bio-One, Kremsmünster, Austria). Endotoxin standards over the range 0.0–1.0 EU/mL were prepared in the LAL reagent water provided. Tenfold serial dilutions of the seed lots were prepared in cell culture grade sodium chloride solution (0.9% w/v, Sigma-Aldrich, St. Louis, MO, USA). The standards and dilutions of the phage preparation were assayed for endotoxin concentration in triplicate. Short-read DNA sequencing of each seed lot was carried out as described previously to ensure that there had been no cross contamination with other phages and that there was no carry over of bacterial DNA from the propagating strain. The sterility of the phage seed lots was determined by spotting multiple 5 µL volumes of the seed lot onto LB agar, incubating overnight at 37 °C and then checking for the growth of microorganisms.
2.8. In Vivo Galleria Mellonella Infection Assay
The efficacy of K. pneumoniae phage cocktails (prepared from seed lots) was assessed using an in vivo G. mellonella infection model based on that described by Champion et al. [45]. Full experimental details can be found in the supplementary material. In order to determine the dose of K. pneumoniae bacterial cells required to induce signs of infection (death or melanisation), larvae were inoculated with increasing doses of K. pneumoniae cells (from 4.6 × 103 to 4.6 × 106 CFU) delivered in a 10 µL volume. Survival and melanisation were evaluated every 2 h for the first 8 h, then again at 20, 22 and 24 h. To assess if K. pneumoniae phage cocktails had any effect on the survival and melanisation of larvae infected with K. pneumoniae, larvae were inoculated with K. pneumoniae only (4.0 × 106 CFU), phage cocktail only (2 × 107 PFU) or K. pneumoniae and phage cocktail combined, as detailed in Table S2. For larvae receiving both bacterial and phage injections, the bacterial inoculum was injected first, followed by a 30 min rest period prior to the phage cocktail injection. All larvae received two injections with control larvae receiving two saline injections. Once injections were complete, the larvae were transferred to 3D printed plastic trays with wells to separate and contain individual larvae (Figure S1). Trays were covered with fitted lids and incubated at 37 °C. To quantify melanisation, brightfield images of individual larvae were analysed using the software IMPACT2AMR (https://github.com/ashsmith88/IMPACT2AMR_galleria_imaging (accessed on 17 November 2022)), which identifies larvae within a boundary box and quantifies pixel brightness (inversely proportional to melanisation) within the boundary. Treatment groups (1–8) were distributed evenly across the plates to account for differences in light exposure across the plate that could affect melanisation scores (Table S3a,b). Survival and melanisation were recorded at 0, 14, 17, 21 and 24 h. Kaplan–Meier survival curves for the G. mellonella infection assays were plotted with the Survival Analysis Package v. 3.5–5 [46] using the survfit function and the survdiff function to test the difference between curves with a log rank test. To determine differences in the melanisation of larvae between treatment groups, the lme4 package v.1.1-35.1 [47] was used to run linear models, modelling melanisation as dependent on treatment, time (as a factor variable) and the interaction between treatment and time (melanisation~treatment*time).
3. Results
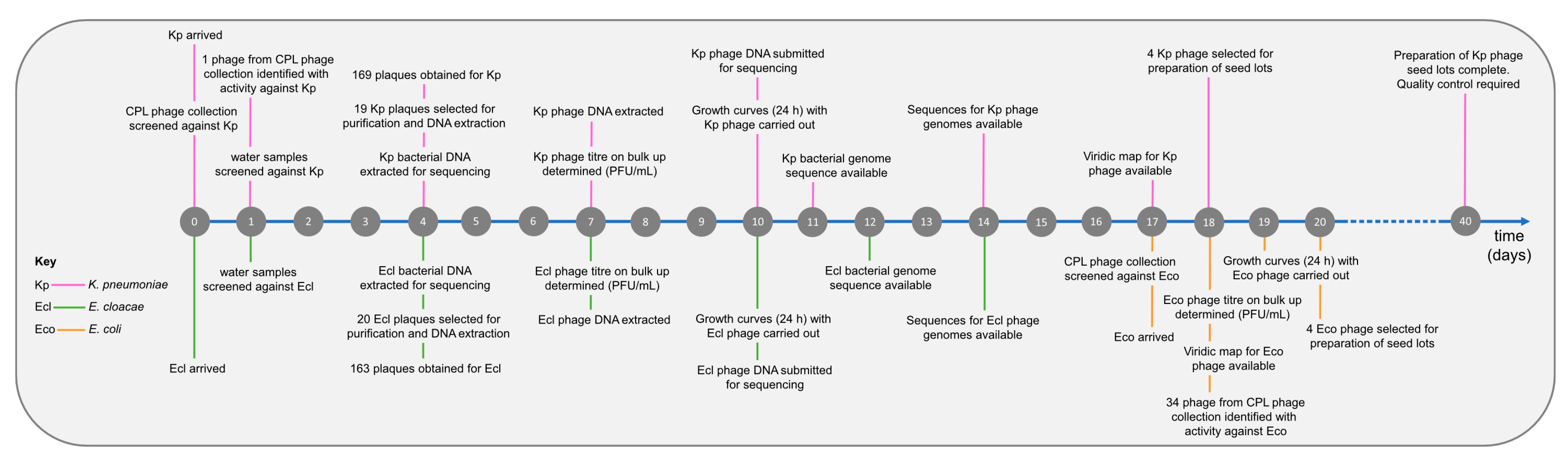
Initially, two strains were received from the clinical microbiology laboratory, a K. pneumoniae strain (BPRG 1484) and an E. cloacae strain (BPRG 1482), and work commenced on finding phages active against these two strains. Approximately two weeks later, an additional E. coli strain (BPRG 1486) was received and this was added to the schedule (Figure 2). Long-read whole genome sequencing was carried out for these strains and bacterial genome data, and associated reads are available on NCBI under BioProject PRJNA993854.
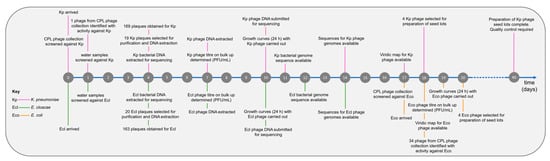
Figure 2.
Timeline for phage search and preparation of seed lots against the hospital strains.
3.1. Cross-Reactivity of Target Strains with Phages Held in the CPL Collection Identified within One Day
Cross-reactions between K. pneumoniae and E. coli and phages already held in the CPL collection were identified within one day of receiving the bacterial strains. For the K. pneumoniae strain, 23 phages were screened yielding two ZOL. Sequencing data were only available for one of the phages (CPL00345). This phage was exclusively lytic and contained no virulence factors, toxins or AMR genes and hence was included as a potential candidate suitable for human use. In the case of the E. coli strain, 161 phages were screened and 42 ZOL were obtained (Figure 3). Full genome data were available for 23 of the 42 E. coli phages, with one phage (CPL00091) eliminated from being taken forward due to the presence of an integrase gene and two phages (CPL00134L and CPL00134S) found to have identical genomes.

Figure 3.
Screening of the CPL collection against E. coli BPRG 1486 determined by placing 5 µL spots of banked phage lysates on soft agar overlays of the strain. ● Phage lysates selected for further characterisation.
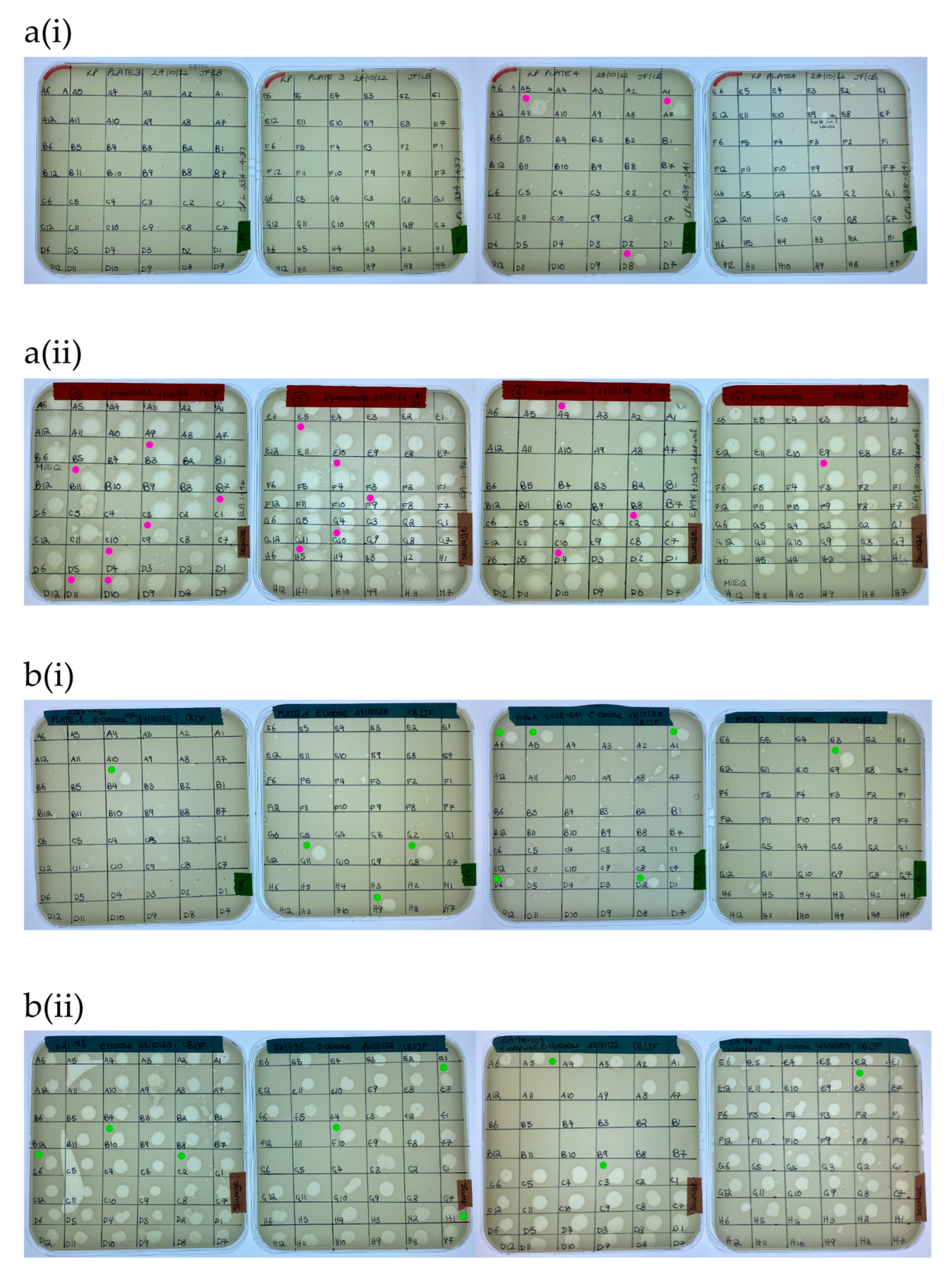
3.2. New Phage Isolated against Target Strains within Four Days
A total of 169 ZOL were obtained from the water sample screening with K. pneumoniae (Figure 4a). Of these, nineteen clear ZOL were selected for purification and sequencing, with three of these originating from citizen scientist water samples and sixteen from wastewater samples. In the case of E. cloacae, 163 ZOL were obtained from the water sample screening (Figure 4b). Twenty clear ZOL were selected for purification and sequencing, with ten isolated from citizen scientist water samples and ten from wastewater samples.
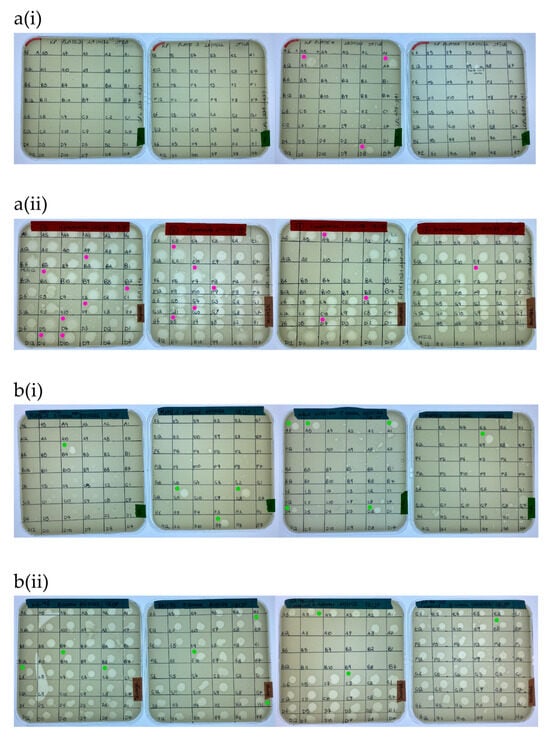
Figure 4.
Screening of (a) K. pneumoniae BPRG 1484 with (i) citizen scientist water samples and (ii) sewage water samples. Cores taken for purification and sequencing from K. pneumoniae ZOL (●). Screening of (b) E. cloacae BPRG 1482 with (i) citizen scientist water samples and (ii) sewage water samples. Cores taken for purification and sequencing from E. cloacae ZOL (●). Activity was determined by placing 5 µL spots of 2nd round enrichment filtrates on inoculated soft agar overlays of the strains.
3.3. Twenty-Nine Putatively Novel Phage Species Were Isolated
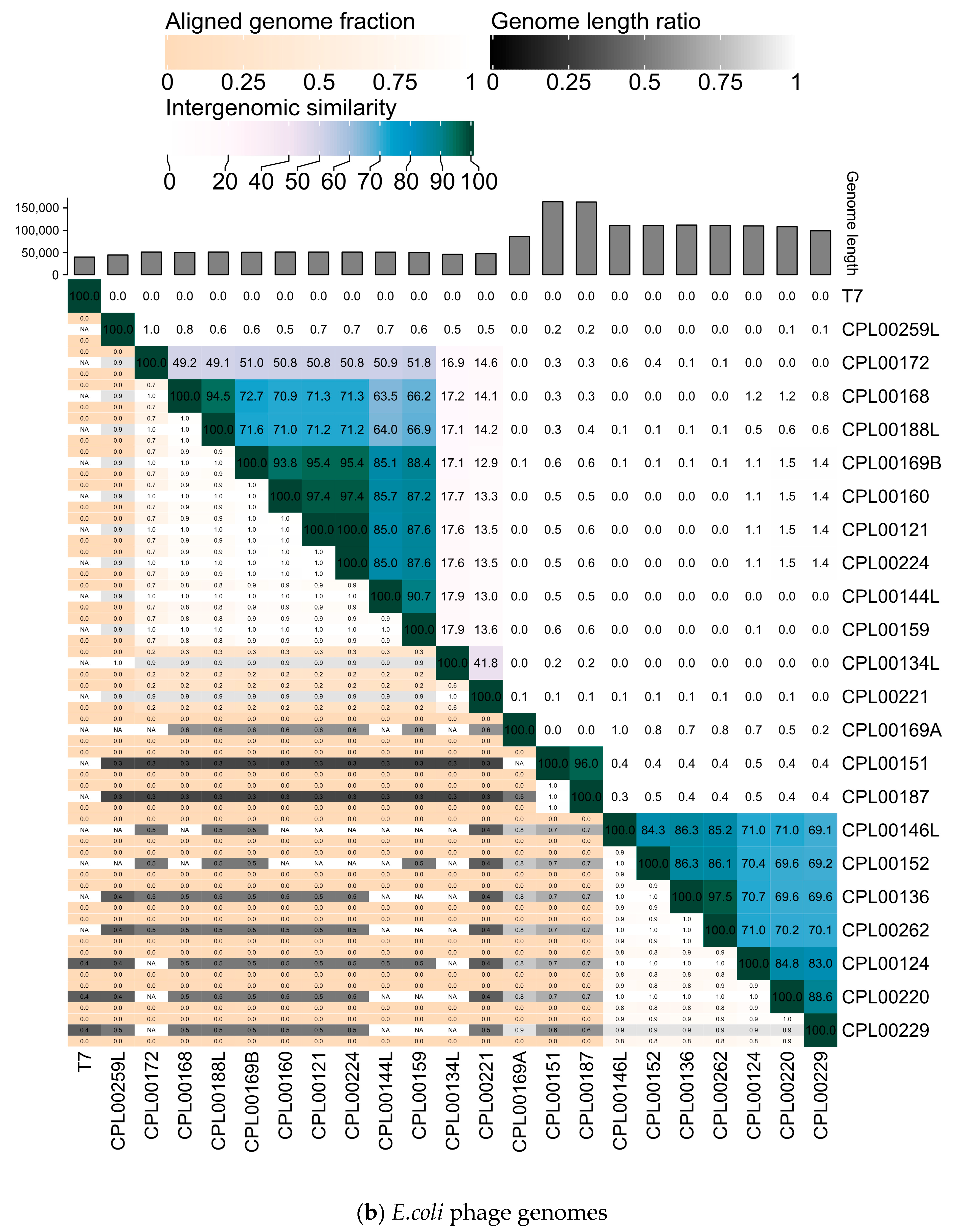
The newly isolated K. pneumoniae phage genomes were sequenced and characteristics of the phages together with the previously sequenced E. coli phages are presented in Table 1. Phylogenetic analysis revealed K. pneumoniae phages belonging to three different genera and E. coli phages belonging to nine different genera. Phages were considered to be novel species if they were <95% identical at the nucleotide level to previously known phages [37] and novel strains if they were <99% identical to previously known phages. In some cases, more than one isolated phage matched with the same closest known phage (Table S4a,b). Using these criteria, 16 novel species of K. pneumoniae phages were identified and 13 novel species of E. coli phages were identified, congruent with the greater number of phage genomes described for E. coli and hence greater phylogenetic coverage of E. coli phage genomes compared to K. pneumoniae phages, with 7075 and 1461 phage genomes currently registered with NCBI GenBank for Escherichia and Klebsiella, respectively. The taxonomic assignment of new phages to existing genera was consistent between ViPTree and INPHARED for all but two phages: CPL00221 and CPL00134L. Both of these phages were placed on their own branch in ViPTree (Figure S6) and shared <70% ANI from their nearest neighbour: E. coli phages CPL00221 (69.3% ANI with Escherichia phage AugustPiccard, MZ501051) and CPL00134L (47% ANI with Escherichia phage vB_EcoS_MM0, MK373793). These phages shared 41% ANI with each other, and <18% ANI with any other phage in this study. It is likely that these phages are the first representatives of a novel phage subfamily. Future sequencing efforts of phages from citizen samples will hopefully assist in finding additional members to support phylogenetic placement. The pooling of wastewater samples (16/20 K. pneumoniae phages were isolated from wastewater samples) may also lead to reduced diversity as the fittest phage in each pool will dominate. In contrast, 21/21 E. coli phages were isolated from individual environmental water samples from citizen scientists.

Table 1.
(a) Characteristics of the 20 phages active against the K. pneumoniae BPRG 1484 strain and their citizen scientist given names. (b) Characteristics of the 21 phages active against the E. coli BPRG 1486 strain and their citizen scientist given names.
3.4. Titre Achieved on Propagation
The titre achieved on propagation in the hospital strains of K. pneumoniae (BPRG 1484) and E. coli (BPRG 1486) is given in Table 2. The K. pneumoniae phages produced titres ranging from 4.0 × 103 to 4.4 × 1011 PFU/mL. These phages were isolated using the same K. pneumoniae strain (BPRG 1484), with the exception of CPL00345 which had been isolated using a strain of K. pneumoniae from a diabetic foot infection (DFI 20.19). CPL00345 gave the lowest titre of all of the K. pneumoniae phages when propagated in the K. pneumoniae BPRG 1484 strain (4.0 × 103 PFU/mL). Of the 13 E. coli phages tested (which had been isolated initially on E. coli BW25113), 10 produced titres ranging from 1.5 × 106 to 1.0 × 1011 PFU/mL when propagated in the hospital E. coli strain (BPRG 1486). Three of the phages (CPL00134L, CPL00221 and CPL00229) did not produce any plaques on the lowest dilution tested (×10−2). Thus, whilst the lysates of these three phages were able to produce a clear ZOL in the library collection screening assay (Figure 3) with E. coli BPRG 1486, they did not propagate well in this strain in liquid culture.

Table 2.
Titre (PFU/mL) achieved for the K. pneumoniae phages and E. coli phages when propagated in the hospital strains.
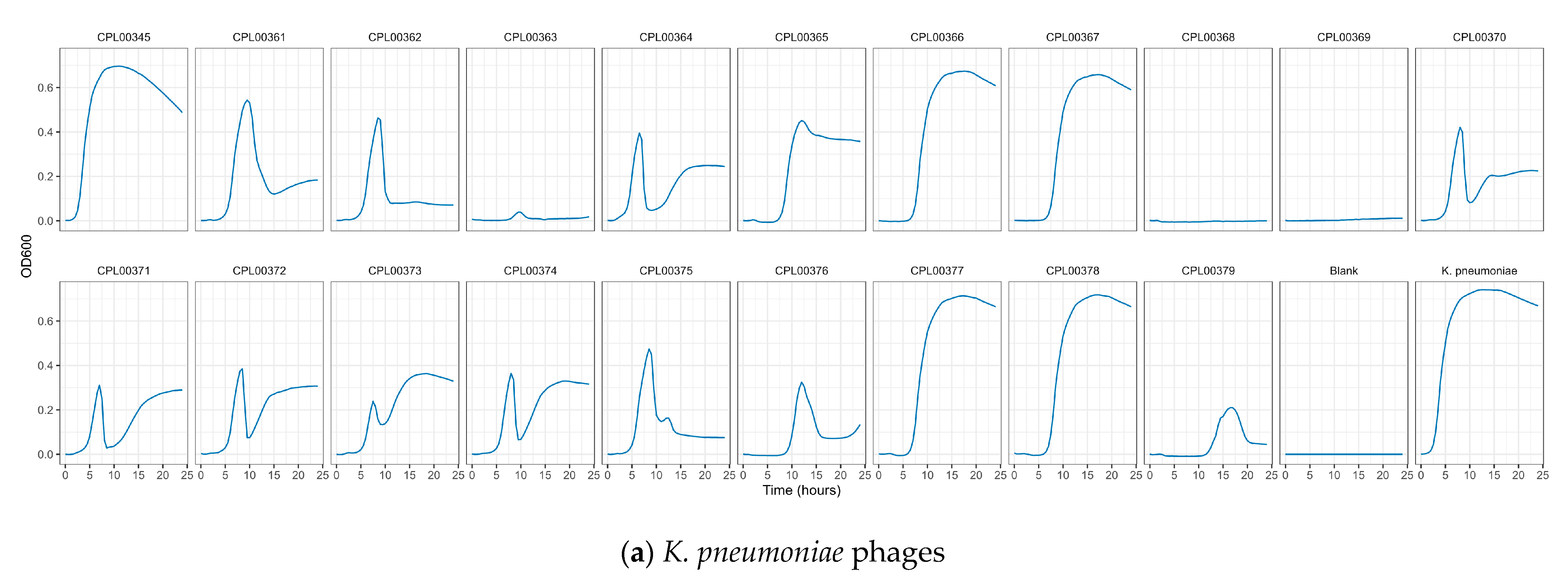
3.5. Phages Were Able to Suppress Bacterial Growth in Liquid Culture
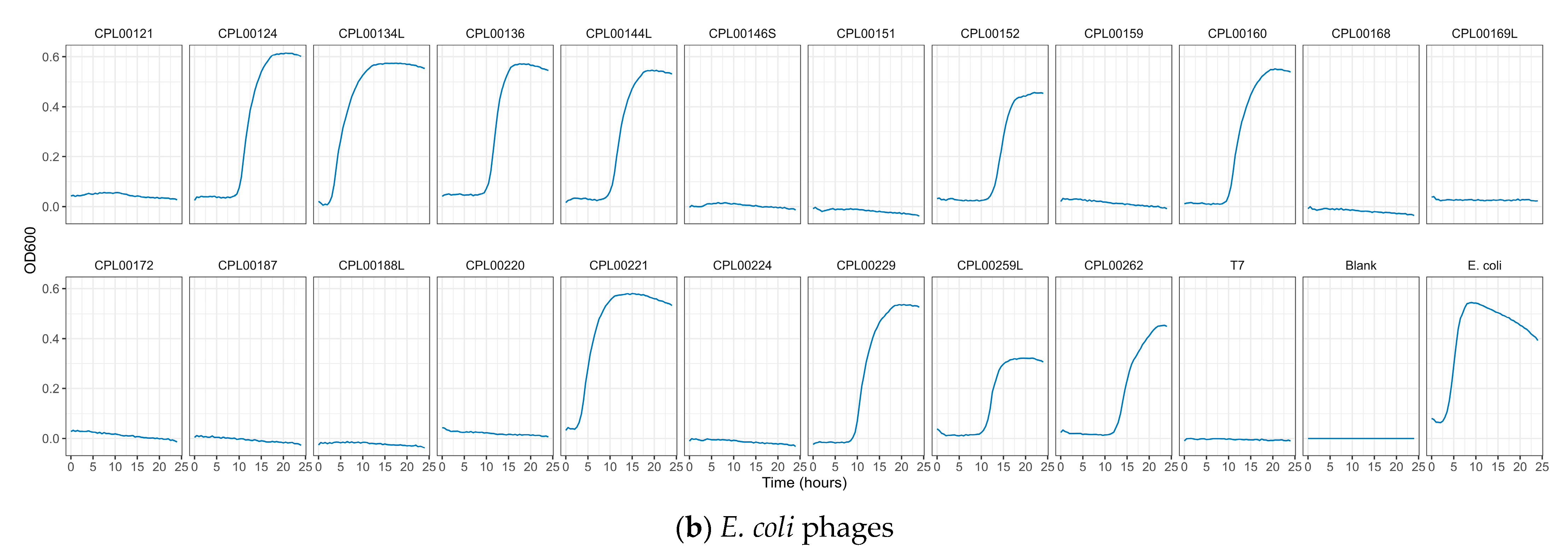
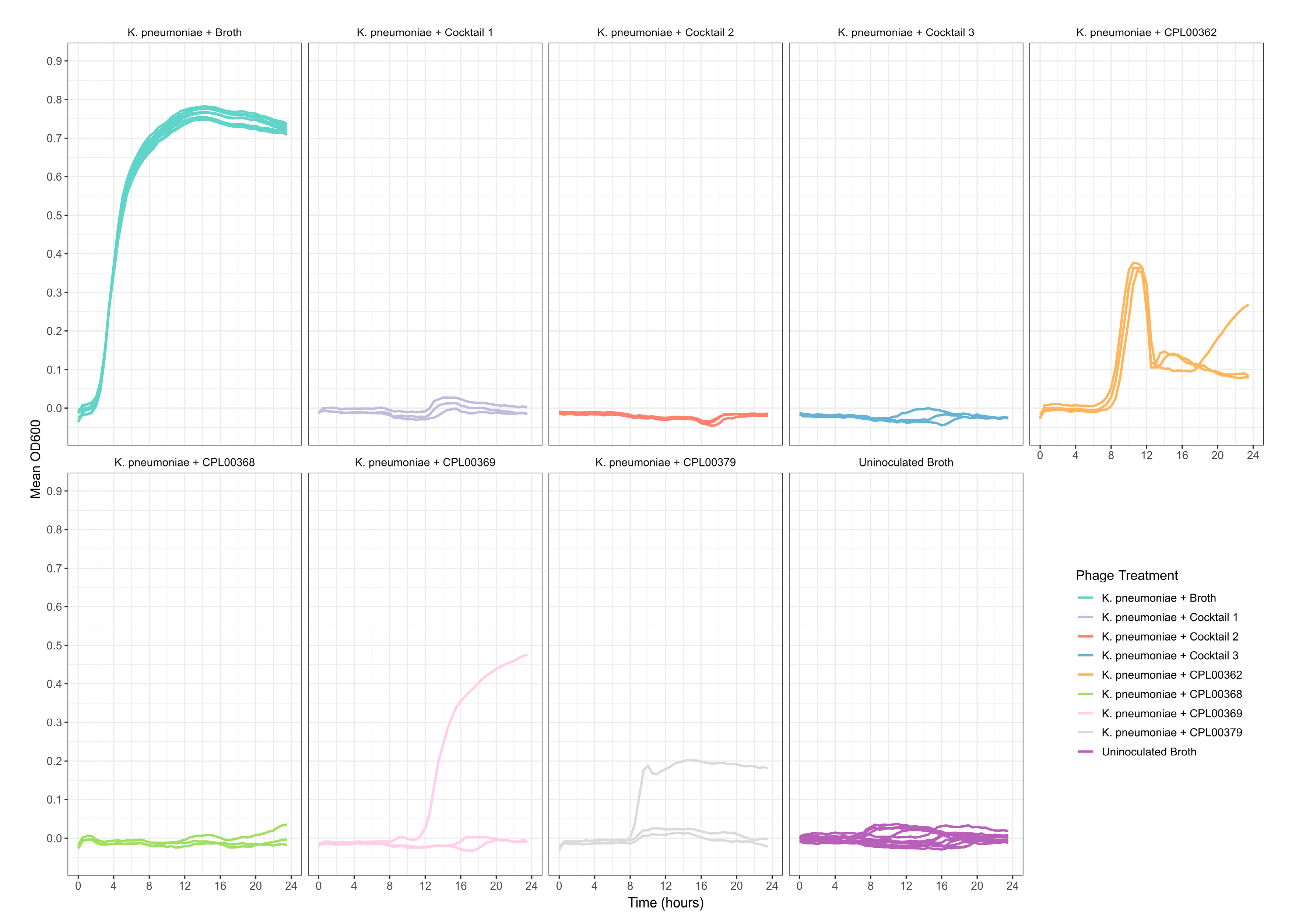
K. pneumoniae and E. coli phages were assessed for their ability to suppress the growth of the target strains in liquid culture. Growth was monitored by measuring OD600 at 30 min time intervals for 24 h (Figure 5a,b) and the Virulence Index (Vp) for each phage was calculated (Table S5) [48]. In the case of K. pneumoniae, two phages were able to completely inhibit all growth over the 24 h growth period, whilst for E. coli, 12 phages (including T7) caused complete inhibition of all growth (Figure 5a,b). In some cases, a temporary increase in OD600 was observed followed by a drop and flattening of the growth curve. This is likely due to bacterial resistance to the phage followed by phage adaptation and the restoration of killing activity. For the E. coli phages CPL00134L, CPL00221 and CPL00229, very little or no inhibition of growth was observed (Figure 5b), and this correlated with their lack of ability to propagate in the E. coli BPRG 1486 strain (Table 2). Ability to suppress growth over a 24 h period was considered a strong indicator for suitability for seed lot preparation. If a phage was able to suppress bacterial growth over a 24 h period and yet had a lower titre than other phages in the selection panel, then this phage was still considered of interest; for example, the K. pneumoniae phage CPL00368 had a relatively low titre compared to other phages in the selection panel but performed well in liquid culture over 24 h (Figure 5a). Three K. pneumoniae phage lysate cocktails were tested for their ability to inhibit growth in liquid culture: cocktail 1—CPL00362 and CPL00368; cocktail 2—CPL00369 and CPL00379 and cocktail 3—CPL00362, CPL00368, CPL00369 and CPL00379. All three cocktails were able to inhibit the growth of K. pneumoniae over the 24 h period (Figure 6).
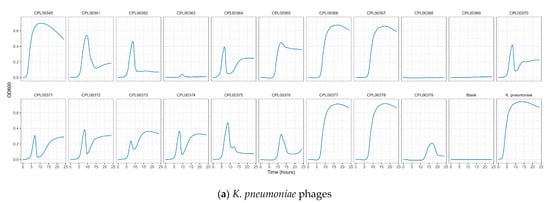
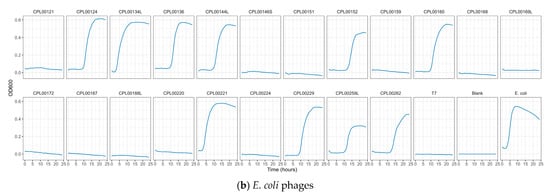
Figure 5.
Ability of phages to suppress growth of (a) K. pneumoniae BPRG 1484 and (b) E. coli BPRG 1486 as determined by measuring OD600 at 30 min intervals over 24 h.
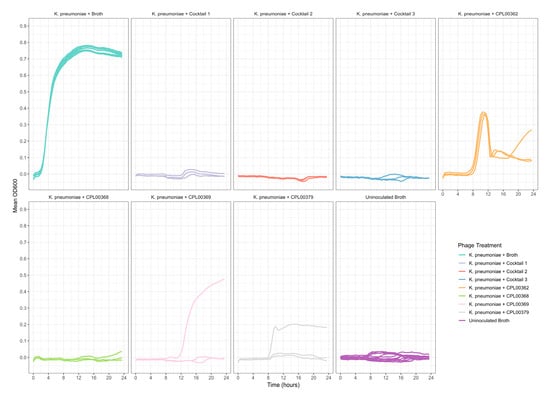
Figure 6.
Ability of phage lysate cocktails to suppress the growth of K. pneumoniae BPRG 1484 as determined by measuring OD600 at 30 min intervals over 24 h. Cocktail 1—CPL00362 and CPL00368; cocktail 2—CPL00369 and CPL00379; cocktail 3—CPL00362, CPL00368, CPL00369 and CPL00379. Control wells received 5 µL of sLB in place of phage lysates.
3.6. Phages against the Target Strains Exhibited Genomic Diversity and Clustering of Genomes into Distinct Groups
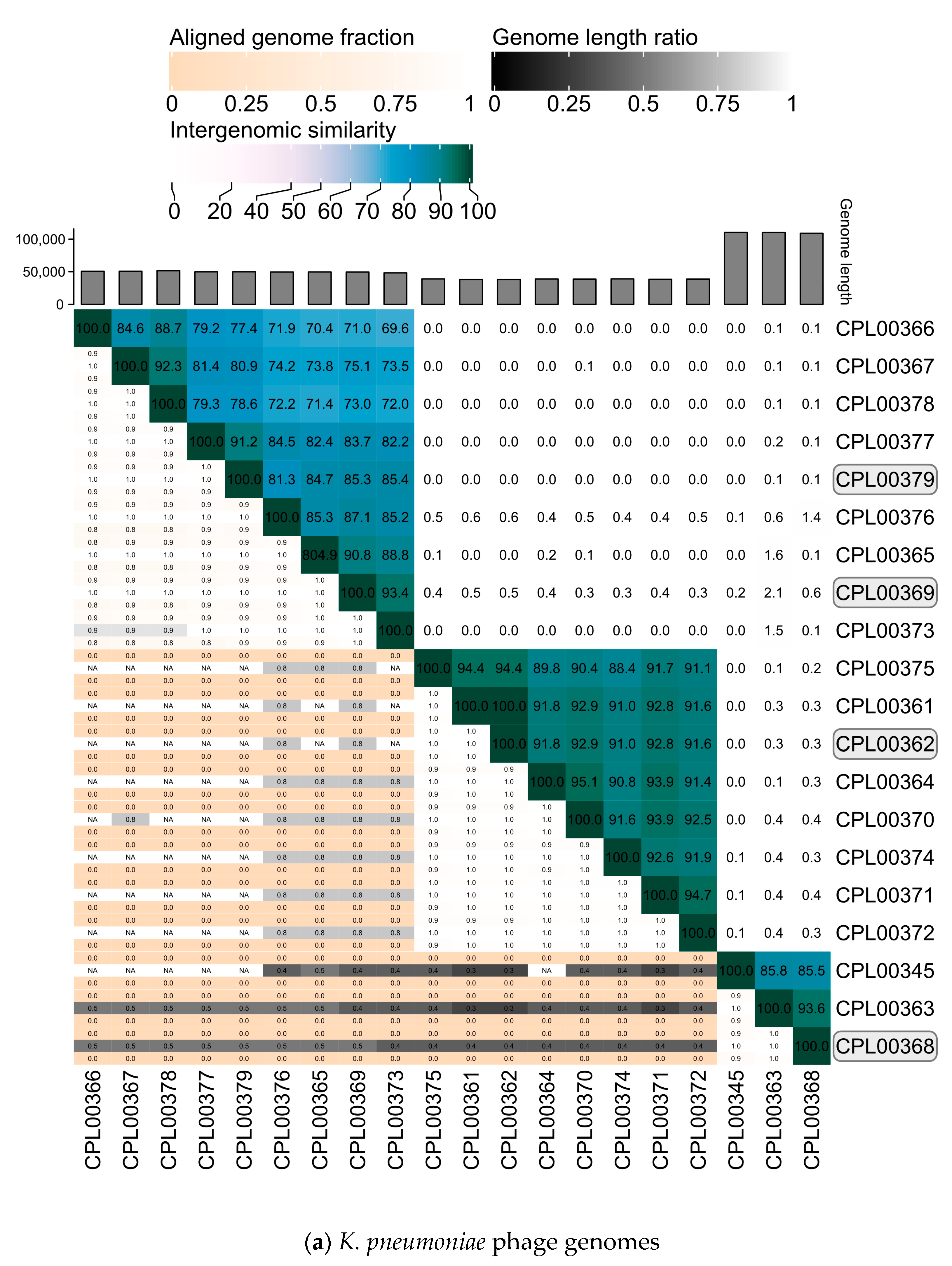
The K. pneumoniae phage genomes fell in to three distinct clusters whilst the E. coli phage were grouped into two large clusters, two small clusters of two phages and three phages which did not cluster with any other phages in the panel (Figure 7a,b).
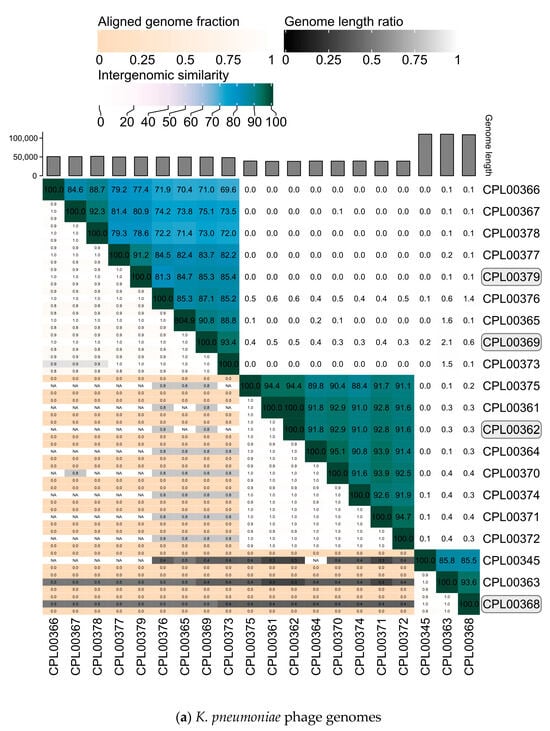
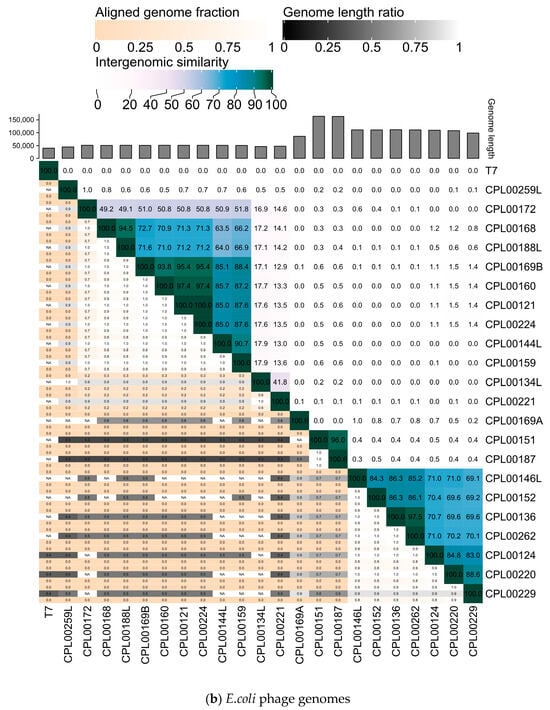
Figure 7.
Heatmaps for (a) K. pneumoniae and (b) E. coli phage genomes plotted using VIRIDIC showing intergenomic similarities amongst the viral genomes. Highlighted phages indicate phages that were taken forward for the preparation of seed lots.
3.7. K. pneumoniae Phages Were Selected for the Preparation of Seed Lots Based on Four Criteria
Phages were selected based on the absence of genes encoding unwanted characteristics, titre achieved on propagation (Table 2, ability to suppress growth in liquid culture over a 24 h growth period (Figure 5a,b) and genetic dissimilarity with other phages in the selection panel (Figure 7a,b). Four K. pneumoniae phages, namely CPL00362, CPL00368, CPL00369 and CPL00379, were chosen for the preparation of seed lots. The phage titre was determined at each step of the phage seed lot preparation (Table S6). In-house quality control was carried out and the results are presented in Table 3 (and Table S7). A phage dose of 109 PFU is often cited as a standard dose [14] and hence this has been used here to calculate a theoretical number of doses based on the titre and volume (1.4 mL) of the seed lot. The number of theoretical doses can be doubled (given in brackets) as only half of the PEG8000/NaCl precipitated phage suspension was applied to the CsCl gradient. It should be noted that lower phage doses are also often described [49]. The annotated genomes of the four phages can be viewed in Figure S5a–d.

Table 3.
In-house quality control of the K. pneumoniae seed lots.
3.8. Phage Cocktails Reduced the Melanisation of K. pneumoniae Infected G. mellonella Larvae
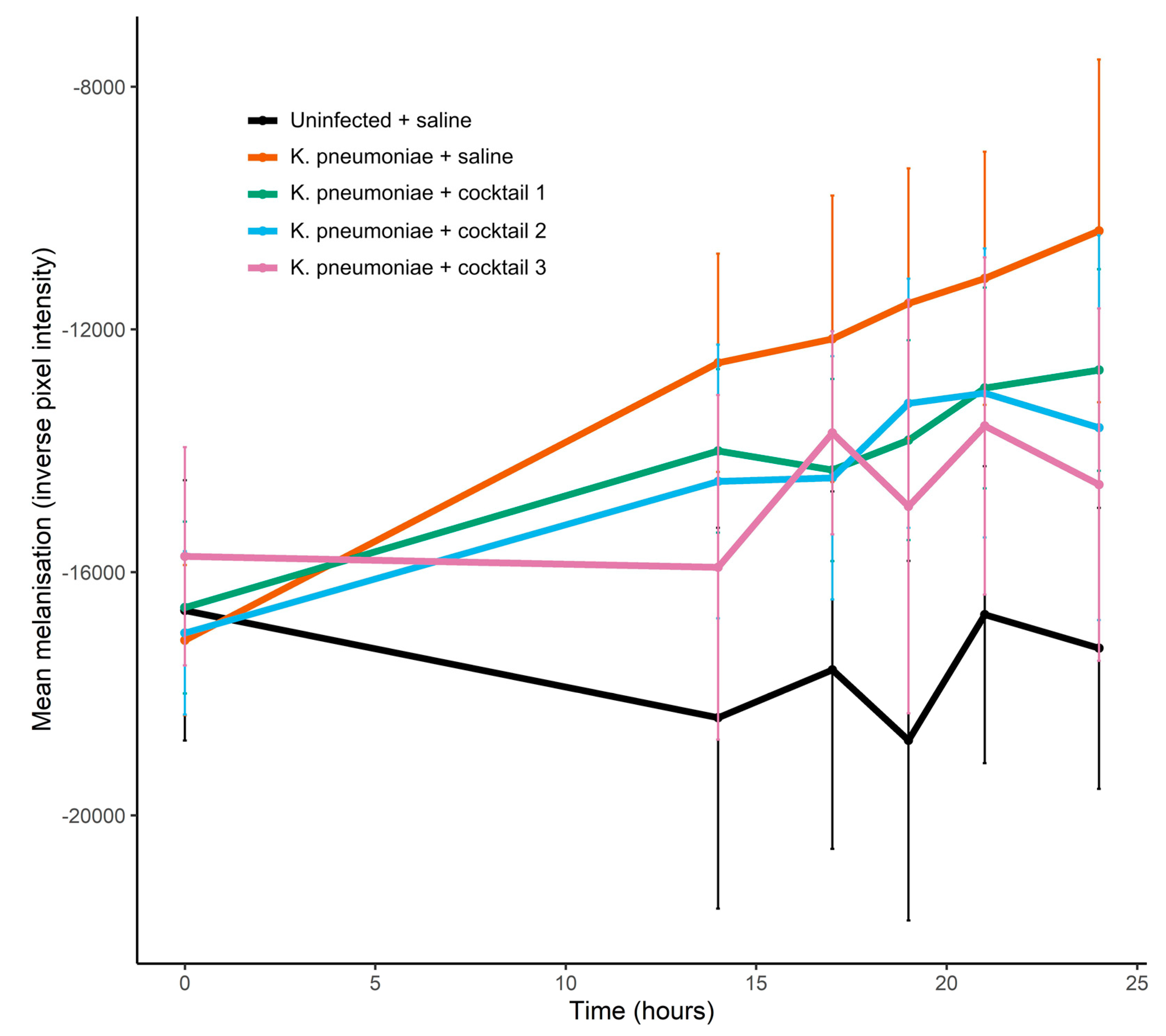
The first stage in the G. mellonella infection model experiments was to establish the dose of K. pneumoniae required to induce measurable signs of infection so that any rescue effect of the seed lot phage cocktails could be observed. A dose of 4.6 × 106 CFU induced the melanisation of the larvae and caused larval death (Figures S2 and S3). In the phage cocktail experiment, larvae receiving a dose of 4.0 × 106 CFU demonstrated significant melanisation over 24 h (Figure 8). Cocktail 3, which combined all four phages, was able to significantly reduce the melanisation of infected larvae and at 24 h, melanisation was significantly lower in this treatment group compared to infected larvae that had received saline in place of phage (linear model estimate ± s.e. = −5563.6 ± 2026.7, p = 0.006, Figure 8, Table S8). When treated with two-phage cocktails, melanisation was reduced by 20–40% in the K. pneumoniae infected larvae compared to infected larvae that had received no phage (Figure 8), though the differences were not significant (Table S8), with large standard error in melanisation observed even with 10 replicates per treatment. At 24 h, there was 100% survival in all treatment groups (Figure S4). After prolonged incubation (39 h), two deaths occurred in the K. pneumoniae infected larvae receiving saline in place of phage, but no deaths were observed at this point for infected larvae that had received phage cocktails (cocktails 1, 2 and 3). Uninfected larvae, that had received two saline injections or saline plus phage cocktail, demonstrated no melanisation over the course of the experiment.
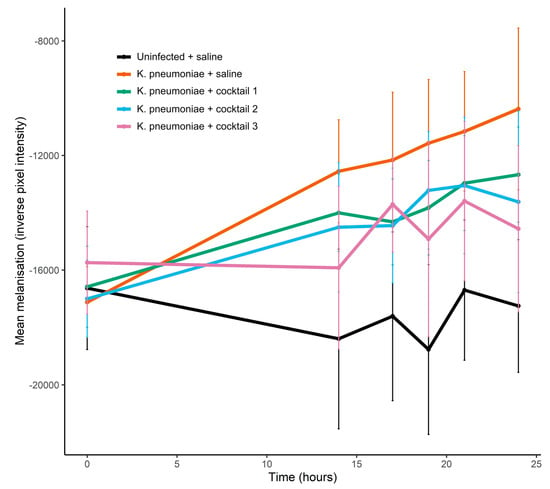
Figure 8.
Effect of seed lot phage cocktails on melanisation of K. pneumoniae infected G. mellonella larvae. Each larvae received 2 injections: 10 µL of K. pneumoniae bacterial cells or 10 µL of saline, and 10 µL of phage cocktail or 10 µL of saline. K. pneumoniae dose = 4.0 × 106 CFU delivered in a 10 µL volume. Phage cocktail dose = 2 × 107 PFU in saline delivered in a 10 µL volume (with 1 × 107 PFU for each phage in the two-phage cocktails and 5 × 106 PFU for each phage in the four-phage cocktail). Cocktail 1—CPL00362 and CPL00368; cocktail 2—CPL00369 and CPL00379; cocktail 3—CPL00362, CPL00368, CPL00369 and CPL00379. Data are expressed as the mean ± s.e., n = 10.
4. Discussion
The O’Neill report commissioned by the UK Government in 2014 predicted that without urgent action, there would be an estimated 10 million deaths per year globally due to AMR [50]. Despite recent funding, policy and legislative initiatives, the pipeline for new antibacterials remains sparse [6]. Phage therapy, with a proven track record of safety, could make a vital contribution to combatting MDR infections [51]. Jones et al. [52] present a comprehensive vision for phage therapy in the UK where off-the-shelf, licensed, GMP-manufactured cocktails are available for use throughout the NHS. In cases where a cocktail is ineffective against a patient’s strain, a personalised non-licensed phage formulation could be prepared from a bank of ready prepared GMP-manufactured phages. To achieve this goal will take many years and considerable investment. Named-patient use of non-GMP manufactured phage preparations is permitted in several countries [14,22,26,27] and such a pathway could be lifesaving for patients in the UK who might benefit from phage therapy in the immediate future. Currently in the UK, unlicensed non-GMP phage can, at least in principle, be used on a named-patient basis where a clinician deems that all licensed alternatives have been exhausted, though navigation to successful use remains challenging. Thus, whilst in theory there are no regulatory barriers to the appropriate use of phages in the UK, a well-defined pathway and the resources to facilitate their use is needed. In this study a workflow is described for the preparation of bespoke phage seed lots against MDR pathogens received from a UK hospital.
Named-patient phage therapy needs to be swiftly available, adaptive, effective and safe. Four phage seed lots active against K. pneumoniae were ready for quality control assessment within 40 days with the clock running continuously. In the protocol described by Luong et al. [34], the time taken to complete all of the elements of the workflow added up to 20.5 days. As the CPL continues to expand its phage collection it will be possible to reduce the timescales further as the need for environmental screening for new phages against a patient strain diminishes. Timescales could be further reduced if the most useful phages were available as high-quality seed lots [26] and by increasing the phage–bacteria characterisation level whereby their host range, putative receptors, and activity under different environmental conditions is known [53]. The provision of phage(s) also needs to be flexible to accommodate the occurrence of bacterial resistance to the phage(s), an adaptive immune response in the patient or co-infection with other species.
In order to maximise the potential effectiveness of phage cocktails, phages were selected for cocktail formulation based on a number of criteria: phage titre on propagation, ability to suppress growth in liquid culture and genetic dissimilarity with other phages in the selection panel. Bacterial growth was monitored over a 24 h period to evaluate the efficacy of each phage against the target strain and to assess the development of bacterial resistance over time. Using this method, phages were selected that were able to maintain their activity over an extended period and phages where the rapid emergence of resistance was observed could be excluded. Furthermore, phages that produce a ZOL in a spot assay but do not perform well in liquid culture over 24 h, could be eliminated as in the case of the E. coli phages CPL00134L, CPL00221 and CPL00229. Haines et al. [54] found that, of the four methods tested, planktonic killing assays, similar to that described here, were the best method for determining phage virulence and for selecting phages for inclusion in cocktails. In the current study, phage lysates held in the CPL collection, were screened for their ability to suppress growth in liquid culture without a prior standardisation of their titre. This method provided a rapid screening tool to identify phages within the library with sustained activity against the target strain. Further analysis of the growth kinetics in liquid culture would be desirable, including one-step growth curves to determine phage adsorption rates and the optimal phage–bacteria ratio (multiplicity of infection, MOI) [32,53,55,56]. The phages in this study were isolated and their in vitro activity assessed, under physiological temperature and pH. When utilising phages for phage therapy, there will be multiple environmental factors operating in vivo which may affect the activity of a phage against its target host. These include temperature and pH [57], the presence of biological materials such as mucus [58,59], host immune responses and the interaction with the host microbiome [57,60]. Additionally, bacteria within an infection may be present as biofilms or may exhibit altered phenotypes which may hinder the adsorption of the phage to the bacterial surface [61]. The incorporation of the G. mellonella model into the workflow enabled confirmation of the in vivo efficacy of the four-phage K. pneumoniae cocktail in a simple insect model with an innate immune system. When assessing phages for their suitability for use in phage therapy, other routine screening assays such as biofilm assays and serum neutralisation assays could be included. One particularly important factor that may affect the activity of phages and phage cocktails in vivo is the presence of antibiotics that are to be administered alongside phage therapy. Antibiotics are often shown to have a synergistic effect when combined with phages though this is not always the case and antagonistic effects have been observed [62,63]. In future, synograms [64] could be included as part of the routine CPL workflow to assess in vitro phage activity in combination with the relevant antibiotic(s). Phage cocktails that maintain their efficacy in vivo would ideally be composed of phages with different target receptors on the bacterial host surface and differing susceptibilities to host bacterial defence mechanisms. By plotting VIRIDIC heatmaps of the phage genomes, phages were selected from different genome clusters to ensure maximum genetic diversity in the phage cocktail. In future it may be possible to use other approaches, such as data mining of host-range datasets, to improve cocktail design [65]. An important aspect of the efficacy of a medicine is evidence of its stability and resulting shelf-life. Phage activity may be affected by the storage buffer, its pH and the storage temperature [66]. Work is ongoing within the CPL laboratory to assess these criteria.
In addition to optimising phage cocktails for maximum potential efficacy, named-patient phage therapy needs to be safe. All phages were sequenced early on in the workflow and phages containing lysogenic genes and genes encoding virulence factors or antibiotic resistance were eliminated. Quality control criteria for phage seed lots include adequate titre, safe endotoxin levels, phage purity and sterility [22,50]. An endotoxin limit of 5 EU per kg of body weight per hour (or 350 EU for a 70 kg adult) for any parenteral route of administration other than intrathecal is specified by the U.S. Pharmacopeia [67]. In the current study, phages were purified by PEG8000/NaCl precipitation followed by ultracentrifugation and extensive dialysis against sterile saline. The endotoxin levels of the resulting seed stocks exceeded the required limit, and the method was time consuming and technically demanding. Further work is required to determine the best method for obtaining high titre phage preparations with the lowest achievable endotoxin levels. Hietala et al. [68] found that the most effective method of endotoxin removal was ultrafiltration (which concentrated phage and allowed buffer exchange, but did not reduce endotoxin-phage-ratio) followed by passing the preparation through an EndoTrap HD column. This method resulted in an endotoxin concentration of 0.085 EU/109 PFU. Using cross-flow filtration, followed by LPS-affinity chromatography, Luong et al. [34] obtained an estimated 58–64,000 doses at 109 PFU from 6 L batches with endotoxin levels ranging from 0.00025 to 0.07 EU per 109 PFU dose. Purification steps such as endotoxin removal (Stage 4, Figure 1) would ideally be carried out in an environment with specified air quality and cleanliness. Whilst defined limits are specified for endotoxin levels, consideration should also be given to other toxins that may be produced by the propagation strain, such as the heat stable enterotoxins [69] and colibactin [70] produced by K. pneumoniae and E. coli. Purification protocols should in theory remove these alongside endotoxin, however evidence of the safety of a phage preparation could be confirmed with the use of cell viability assays [34]. The use of avirulent, low endotoxin propagation strains devoid of prophages where possible, would provide a more convenient route to achieving safe, non-toxic preparations [71]. Sequencing of the four K. pneumoniae seed lots was carried out and confirmed that the preparations were pure axenic phage with no evidence of cross contamination with other phages including prophages. Two of the seed lots contained significant amounts of host DNA, which could be reduced by introducing a second DNase/RNase treatment step after phage precipitation and prior to ultracentrifugation. Sterility was assessed by spreading an aliquot of each seed lot onto LB agar. This is perhaps adequate at this stage, as full sterility testing would be carried out by accredited laboratories if the seed lots were to progress to the preparation of phage active pharmaceutical ingredients (phage APIs) [26]. However, in future, in-house sterility testing will be assessed using bioburden testing on trypticase soya agar (Total Aerobic Microbial Count) and Sabouraud dextrose agar (Total Yeast and Mould Count) or by direct inoculation into tryptic soy broth and thioglycolate medium to ensure screening for the widest range of microorganisms. The workflow presented here ends with the in-house quality control of the purified seed lots. The continuation of this workflow to achieve high quality, non-GMP magistral standard APIs has been proposed in the final box in Figure 1, based on regimes utilised by Belgium and Australia [49,72].
This study has demonstrated that it was possible to identify phages targeted against multiple strains in a short time frame at a relatively low cost that are effective in vitro and in an in vivo G. mellonella infection model. Access to an extensive collection of water samples and biobanked phages expedited the screening process. In addition to the educational benefits, citizen science has proven highly productive in generating a diverse range of clinically suitable phages. An outline of how high quality non-GMP phages could be produced in the UK has been proposed and this, together with a clearly defined regulatory framework and adequate resources, could be used to provide phages for named-patient use in situations where all other therapeutic options have been exhausted. Further work is required to reduce the endotoxin levels of phage seed lots and to determine their stability over time. More detailed kinetic assays and phage–antibiotic synograms could be added to the workflow to further characterise the phage and enhance the selection process. The Citizen Phage Library will continue to engage with communities and expand its library of clinically suitable, fully characterised phages which can be provided for phage therapy globally. Recently, 175 fully characterised phages from the CPL were shared with PhageAustralia and a further 60 were shared with Canada to assist in their efforts, with additional shipments planned to support efforts in the US. The Citizen Phage Library will continue to work with the MHRA, clinicians and hospital pharmacies to establish a clear route to enable timely access to named-patient phage therapy in the UK.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12020253/s1, Table S1: Antibiotic susceptibility profiles for K. pneumoniae, E. cloacae and E. coli strains; Table S2: Treatment groups for the experiment to determine effect of phage cocktails on the melanisation of K. pneumoniae infected G. mellonella larvae; Table S3: Plate layouts for the 8 treatment groups for the experiment to determine effect of phage cocktails on the survival and melanisation of K. pneumoniae BPRG 1484 infected G. mellonella larvae; Figure S1: 3D printed plastic trays with wells to separate and contain individual G. mellonella larvae; Table S4a: Characteristics of the 20 phages active against the K. pneumoniae BPRG 1484 strain and their citizen scientist given names; Table S4b: Characteristics of the 21 phages active against the E. coli BPRG 1486 strain and their citizen scientist given names; Table S5: Virulence Index (Vp) for each of the phages against the target strains K. pneumoniae BPRG 1484 and E. coli BPRG 1486; Table S6: Titre (PFU/mL) of phage lysates at each stage of the preparation of the four K. pneumoniae seed lots; Table S7: Purity of the four K. pneumoniae phage seed lots as determined by short-read DNA sequencing; Figure S2: Effect of K. pneumoniae dose (CFU/mL) on melanisation of G. mellonella larvae; Figure S3: Effect of K. pneumoniae dose (CFU/mL) on survival of G. mellonella larvae; Figure S4: Effect of phage cocktails on survival of K. pneumoniae infected G. mellonella larvae; Table S8: Full linear model showing the effect of treatment (saline, phage cocktail 1, phage cocktail 2, phage cocktail 3) and time on the melanisation of K. pneumoniae infected larvae; Figure S5: K. pneumoniae phage genomes for (a) CPL00362 (b) CPL00368 (c) CPL00369 and (d) CPL00379 annotated using pharokka; Figure S6. Dendrogram of phage genomes isolated in this study, with 3952 closely related phage genomes from Virus-Host DB: RefSeq release 220.
Author Contributions
Conceptualization, B.T., J.F., J.S., A.M. and N.W.; Methodology, B.T., J.F., R.M., C.F., C.B., M.M., K.M., A.F. and S.A.; Validation, B.T., J.F., R.M. and C.F.; Formal Analysis, B.T., R.M. and C.F.; Investigation, B.T., J.F., R.M., C.F., C.B., M.M., K.M., A.F. and S.A.; Resources, B.T., S.M., J.F., C.B. and N.S.; Data Curation, B.T., J.F., R.M. and C.F.; Writing—Original Draft Preparation, J.F.; Writing—Review & Editing, J.F., B.T., S.A., C.B., A.F., C.F., R.M., M.M., A.M., K.M., N.S., J.S., N.W. and S.M.; Visualization, B.T., J.F., R.M. and C.F.; Supervision, B.T.; J.F. and R.M.; Project Administration, B.T., J.F. and C.B.; Funding Acquisition, B.T. and S.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a Confidence in Concept (MRC) grant awarded to B.T. and in part by grant MR/N0137941/1 for the GW4 BIOMED MRC DTP, awarded to the Universities of Bath, Bristol, Cardiff and Exeter from the MRC/UKRI. The project utilised equipment funded by the Wellcome Trust (Multi-User Equipment Grant award number 218247/Z/19/Z). For the purpose of open access, the author has applied a Creative Commons Attribution (CC BY) licence to any Author Accepted Manuscript version arising.
Data Availability Statement
Bacterial and phage genome data, and associated reads, are available on NCBI under BioProject PRJNA993854. Data for the growth curve experiments, endotoxin assays and G. mellonella infection model assays are available on request.
Acknowledgments
We are grateful to all of the citizen scientists who took part in outreach events and collected water samples on behalf of the project. We also thank Nick Evens and the team at the Environment Agency for providing wastewater samples. We are grateful to Jennie Campbell and James Pearce from the Galleria mellonella Research Centre at the University of Exeter for the wax moth larvae. We thank Vicki Gold and Kelly Sanders for advice on the use of the ultracentrifuge and Robin Wright for help with the dialysis of the phage seed lots.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Antimicrobial Resistance: A Top Ten Global Public Health Threat. EClinicalMedicine 2021, 41, 101221. [CrossRef] [PubMed]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Catacutan, D.B.; Rathod, K.; Swanson, K.; Jin, W.; Mohammed, J.C.; Chiappino-Pepe, A.; Syed, S.A.; Fragis, M.; Rachwalski, K.; et al. Deep Learning-Guided Discovery of an Antibiotic Targeting Acinetobacter baumannii. Nat. Chem. Biol. 2023, 19, 1342–1350. [Google Scholar] [CrossRef]
- Roberts, A.P. Swab and Send: A Citizen Science, Antibiotic Discovery Project. Future Sci. OA 2020, 6, FSO477. [Google Scholar] [CrossRef] [PubMed]
- Barlow, E.; Morton, A.; Megiddo, I.; Colson, A. Optimal Subscription Models to Pay for Antibiotics. Soc. Sci. Med. 2022, 298, 114818. [Google Scholar] [CrossRef]
- Butler, M.S.; Henderson, I.R.; Capon, R.J.; Blaskovich, M.A.T. Antibiotics in the Clinical Pipeline as of December 2022. J. Antibiot. 2023, 76, 431–473. [Google Scholar] [CrossRef]
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef]
- Mu, A.; McDonald, D.; Jarmusch, A.K.; Martino, C.; Brennan, C.; Bryant, M.; Humphrey, G.C.; Toronczak, J.; Schwartz, T.; Nguyen, D.; et al. Assessment of the Microbiome during Bacteriophage Therapy in Combination with Systemic Antibiotics to Treat a Case of Staphylococcal Device Infection. Microbiome 2021, 9, 92. [Google Scholar] [CrossRef]
- Międzybrodzki, R.; Hoyle, N.; Zhvaniya, F.; Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Łobocka, M.; Borysowski, J.; Alavidze, Z.; Kutter, E.; Górski, A.; et al. Current Updates from the Long-Standing Phage Research Centers in Georgia, Poland, and Russia. In Bacteriophages; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M.L., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 1–31. [Google Scholar] [CrossRef]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage Treatment of Human Infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Van Belleghem, J.D.; de Vries, C.R.; Burgener, E.; Chen, Q.; Manasherob, R.; Aronson, J.R.; Amanatullah, D.F.; Tamma, P.D.; Suh, G.A. The Safety and Toxicity of Phage Therapy: A Review of Animal and Clinical Studies. Viruses 2021, 13, 1268. [Google Scholar] [CrossRef] [PubMed]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage Therapy: From Biological Mechanisms to Future Directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Eskenazi, A.; Lood, C.; Wubbolts, J.; Hites, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Kvachadze, L.; van Noort, V.; Wagemans, J.; et al. Combination of Pre-Adapted Bacteriophage Therapy and Antibiotics for Treatment of Fracture-Related Infection Due to Pandrug-Resistant Klebsiella pneumoniae. Nat. Commun. 2022, 13, 302. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Smith, B.E.; Cristinziano, M.; Freeman, K.G.; Jacobs-Sera, D.; Belessis, Y.; Whitney Brown, A.; Cohen, K.A.; Davidson, R.M.; van Duin, D.; et al. Phage Therapy of Mycobacterium Infections: Compassionate Use of Phages in 20 Patients With Drug-resistant Mycobacterial Disease. Clin. Infect. Dis. 2023, 76, 103–112. [Google Scholar] [CrossRef] [PubMed]
- McCallin, S.; Sacher, J.C.; Zheng, J.; Chan, B.K. Current State of Compassionate Phage Therapy. Viruses 2019, 11, 343. [Google Scholar] [CrossRef] [PubMed]
- Ferry, T.; Kolenda, C.; Laurent, F.; Leboucher, G.; Merabischvilli, M.; Djebara, S.; Gustave, C.A.; Perpoint, T.; Barrey, C.; Pirnay, J.P.; et al. Personalized Bacteriophage Therapy to Treat Pandrug-Resistant Spinal Pseudomonas aeruginosa Infection. Nat. Commun. 2022, 13, 4239. [Google Scholar] [CrossRef]
- Aslam, S.; Lampley, E.; Wooten, D.; Karris, M.; Benson, C.; Strathdee, S.; Schooley, R.T. Lessons Learned from the First 10 Consecutive Cases of Intravenous Bacteriophage Therapy to Treat Multidrug-Resistant Bacterial Infections at a Single Center in the United States. Open Forum. Infect. Dis. 2020, 7, 389. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.Gov. Available online: https://clinicaltrials.gov (accessed on 1 August 2023).
- Oromí-Bosch, A.; Antani, J.D.; Turner, P.E. Developing Phage Therapy That Overcomes the Evolution of Bacterial Resistance. Annu. Rev. Virol. 2023, 10, 503–524. [Google Scholar] [CrossRef]
- Georjon, H.; Bernheim, A. The Highly Diverse Antiphage Defence Systems of Bacteria. Nat. Rev. Microbiol. 2023, 21, 686–700. [Google Scholar] [CrossRef]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Wozniak, D.J. Phage Cocktail Development for Bacteriophage Therapy: Toward Improving Spectrum of Activity Breadth and Depth. Pharmaceuticals 2021, 14, 1019. [Google Scholar] [CrossRef]
- Verbeken, G.; Pirnay, J.P. European Regulatory Aspects of Phage Therapy: Magistral Phage Preparations. Curr. Opin. Virol. 2022, 52, 24–29. [Google Scholar] [CrossRef]
- Green, S.I.; Clark, J.R.; Santos, H.H.; Weesner, K.E.; Salazar, K.C.; Aslam, S.; Campbell, J.W.; Doernberg, S.B.; Blodget, E.; Morris, M.I.; et al. A Retrospective, Observational Study of 12 Cases of Expanded-Access Customized Phage Therapy: Production, Characteristics, and Clinical Outcomes. Clin. Infect. Dis. 2023, 77, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Ferry, T.; Kolenda, C.; Briot, T.; Lustig, S.; Leboucher, G.; Laurent, F.; Valour, F.; Perpoint, T.; Ader, F.; Roux, S.; et al. Implementation of a Complex Bone and Joint Infection Phage Therapy Centre in France: Lessons to Be Learned after 4 Years’ Experience. Clin. Microbiol. Infect. 2022, 28, 145–146. [Google Scholar] [CrossRef] [PubMed]
- Suh, G.A.; Lodise, T.P.; Tamma, P.D.; Knisely, J.M.; Alexander, J.; Aslam, S.; Barton, K.D.; Bizzell, E.; Totten, K.M.C.; Campbell, J.L.; et al. Considerations for the Use of Phage Therapy in Clinical Practice. Antimicrob. Agents Chemother. 2022, 66, e0207121. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.P.; Verbeken, G.; Ceyssens, P.J.; Huys, I.; de Vos, D.; Ameloot, C.; Fauconnier, A. The Magistral Phage. Viruses 2018, 10, 64. [Google Scholar] [CrossRef]
- Khatami, A.; Foley, D.A.; Warner, M.S.; Barnes, E.H.; Peleg, A.Y.; Li, J.; Stick, S.; Burke, N.; Lin, R.C.Y.; Warning, J.; et al. Standardised Treatment and Monitoring Protocol to Assess Safety and Tolerability of Bacteriophage Therapy for Adult and Paediatric Patients (STAMP Study): Protocol for an Open-Label, Single-Arm Trial. BMJ Open 2022, 12, e065401. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered Bacteriophages for Treatment of a Patient with a Disseminated Drug-Resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Young, M.J.; Hall, L.M.L.; Merabishvilli, M.; Pirnay, J.-P.; Clark, J.R.; Jones, J.D. Phage Therapy for Diabetic Foot Infection: A Case Series. Clin. Ther. 2023, 45, 797–801. [Google Scholar] [CrossRef]
- Nagel, T.; Musila, L.; Muthoni, M.; Nikolich, M.; Nakavuma, J.L.; Clokie, M.R. Phage Banks as Potential Tools to Rapidly and Cost-Effectively Manage Antimicrobial Resistance in the Developing World. Curr. Opin. Virol. 2022, 53, 101208. [Google Scholar] [CrossRef]
- Hanauer, D.I.; Graham, M.J.; Betancur, L.; Bobrownicki, A.; Cresawn, S.G.; Garlena, R.A.; Jacobs-Sera, D.; Kaufmann, N.; Pope, W.H.; Russell, D.A.; et al. An Inclusive Research Education Community (IREC): Impact of the SEA-PHAGES Program on Research Outcomes and Student Learning. Proc. Natl. Acad. Sci. USA 2017, 114, 13531–13536. [Google Scholar] [CrossRef] [PubMed]
- Glonti, T.; Pirnay, J.P. In Vitro Techniques and Measurements of Phage Characteristics That Are Important for Phage Therapy Success. Viruses 2022, 14, 1490. [Google Scholar] [CrossRef] [PubMed]
- Olsen, N.S.; Hendriksen, N.B.; Hansen, L.H.; Kot, W. A New High-Throughput Screening Method for Phages: Enabling Crude Isolation and Fast Identification of Diverse Phages with Therapeutic Potential. Phage 2020, 1, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.; Salabarria, A.C.; Edwards, R.A.; Roach, D.R. Standardized Bacteriophage Purification for Personalized Phage Therapy. Nat. Protoc. 2020, 15, 2867–2890. [Google Scholar] [CrossRef] [PubMed]
- Cook, R.; Brown, N.; Redgwell, T.; Rihtman, B.; Barnes, M.; Clokie, M.; Stekel, D.J.; Hobman, J.; Jones, M.A.; Millard, A. INfrastructure for a PHAge REference Database: Identification of Large-Scale Biases in the Current Collection of Cultured Phage Genomes. Phage 2021, 2, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The Viral Proteomic Tree Server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.J.; Vreugde, S. Pharokka: A Fast Scalable Bacteriophage Annotation Tool. Bioinformatics 2023, 39, btac776. [Google Scholar] [CrossRef] [PubMed]
- Tynecki, P.; Guziński, A.; Kazimierczak, J.; Jadczuk, M.; Dastych, J.; Onisko, A. PhageAI-Bacteriophage Life Cycle Recognition with Machine Learning and Natural Language Processing. BioRxiv 2020, 11, 1198606. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, 517–525. [Google Scholar] [CrossRef]
- Yukgehnaish, K.; Rajandas, H.; Parimannan, S.; Manickam, R.; Marimuthu, K.; Petersen, B.; Clokie, M.R.J.; Millard, A.; Sicheritz-Pontén, T. PhageLeads: Rapid Assessment of Phage Therapeutic Suitability Using an Ensemble Machine Learning Approach. Viruses 2022, 14, 342. [Google Scholar] [CrossRef]
- Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages Using the Small Drop Plaque Assay System. In Bacteriphages Methods and Protocols; Clokie, R.J., Kropinski, A.M., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 501, pp. 81–85. [Google Scholar]
- Green, S. How To Purify a Phage. Available online: https://www.youtube.com/watch?v=-t85C04Ueio (accessed on 21 October 2022).
- Champion, O.L.; Cooper, I.A.M.; James, S.L.; Ford, D.; Karlyshev, A.; Wren, B.W.; Duffield, M.; Oyston, P.C.F.; Titball, R.W. Galleria mellonella as an Alternative Infection Model for Yersinia pseudotuberculosis. Microbiology 2009, 155, 1516–1522. [Google Scholar] [CrossRef]
- Therneau, T. A Package for Survival Analysis in R. R Package Version 3.5-5. Available online: https://CRAN.R-project.org/package=survival (accessed on 17 July 2023).
- Bates, D.; Mächler, M.; Bolker, B.M.; Walker, S.C. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Storms, Z.J.; Teel, M.R.; Mercurio, K.; Sauvageau, D. The Virulence Index: A Metric for Quantitative Analysis of Phage Virulence. Phage 2020, 1, 27–36. [Google Scholar] [CrossRef]
- Pirnay, J.P.; Blasdel, B.G.; Bretaudeau, L.; Buckling, A.; Chanishvili, N.; Clark, J.R.; Corte-Real, S.; Debarbieux, L.; Dublanchet, A.; De Vos, D.; et al. Quality and Safety Requirements for Sustainable Phage Therapy Products. Pharm. Res. 2015, 32, 2173–2179. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. The Review on Antimicrobial Resistance. 2014. Available online: https://wellcomecollection.org/works/rdpck35v (accessed on 1 August 2023).
- Uyttebroek, S.; Chen, B.; Onsea, J.; Ruythooren, F.; Debaveye, Y.; Devolder, D.; Spriet, I.; Depypere, M.; Wagemans, J.; Lavigne, R.; et al. Safety and Efficacy of Phage Therapy in Difficult-to-Treat Infections: A Systematic Review. Lancet Infect. Dis. 2022, 22, e208–e220. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Trippett, C.; Suleman, M.; Clokie, M.R.J.; Clark, J.R. The Future of Clinical Phage Therapy in the United Kingdom. Viruses 2023, 15, 721. [Google Scholar] [CrossRef] [PubMed]
- Mutalik, V.K.; Arkin, A.P. A Phage Foundry Framework to Systematically Develop Viral Countermeasures to Combat Antibiotic-Resistant Bacterial Pathogens. iScience 2022, 25, 104121. [Google Scholar] [CrossRef] [PubMed]
- Haines, M.E.K.; Hodges, F.E.; Nale, J.Y.; Mahony, J.; van Sinderen, D.; Kaczorowska, J.; Alrashid, B.; Akter, M.; Brown, N.; Sauvageau, D.; et al. Analysis of Selection Methods to Develop Novel Phage Therapy Cocktails Against Antimicrobial Resistant Clinical Isolates of Bacteria. Front. Microbiol. 2021, 12, 613529. [Google Scholar] [CrossRef] [PubMed]
- Gelman, D.; Yerushalmy, O.; Alkalay-Oren, S.; Rakov, C.; Ben-Porat, S.; Khalifa, L.; Adler, K.; Abdalrhman, M.; Coppenhagen-Glazer, S.; Aslam, S.; et al. Clinical Phage Microbiology: A Suggested Framework and Recommendations for the in-Vitro Matching Steps of Phage Therapy. Lancet Microbe 2021, 2, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, A.; Clark, J.; Karris, M.; Hernandez-Santos, H.; Green, S.; Aslam, S.; Maresso, A. Phage Therapy Related Microbial Succession Associated with Successful Clinical Outcome for a Recurrent Urinary Tract Infection. Viruses 2021, 13, 2049. [Google Scholar] [CrossRef]
- Dąbrowska, K. Phage Therapy: What Factors Shape Phage Pharmacokinetics and Bioavailability? Systematic and Critical Review. Med. Res. Rev. 2019, 39, 2000–2025. [Google Scholar] [CrossRef]
- Ling, K.M.; Stick, S.M.; Kicic, A. Pulmonary Bacteriophage and Cystic Fibrosis Airway Mucus: Friends or Foes? Front. Med. 2023, 10, 1088494. [Google Scholar] [CrossRef] [PubMed]
- Mutti, M.; Moreno, D.S.; Restrepo-Córdoba, M.; Visram, Z.; Resch, G.; Corsini, L. Phage Activity against Staphylococcus aureus is Impaired in Plasma and Synovial Fluid. Sci. Rep. 2023, 13, 18204. [Google Scholar] [CrossRef] [PubMed]
- Nang, S.C.; Lin, Y.W.; Petrovic Fabijan, A.; Chang, R.Y.K.; Rao, G.G.; Iredell, J.; Chan, H.K.; Li, J. Pharmacokinetics/Pharmacodynamics of Phage Therapy: A Major Hurdle to Clinical Translation. Clin. Microbiol. Infect. 2023, 29, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Meneses, L.; Brandão, A.C.; Azeredo, J. An Overview of the Current State of Phage Therapy for the Treatment of Biofilm-Related Infections. Curr. Opin. Virol. 2022, 53, 101209. [Google Scholar] [CrossRef] [PubMed]
- Diallo, K.; Dublanchet, A. Benefits of Combined Phage–Antibiotic Therapy for the Control of Antibiotic-Resistant Bacteria: A Literature Review. Antibiotics 2022, 11, 839. [Google Scholar] [CrossRef]
- Łusiak-Szelachowska, M.; Międzybrodzki, R.; Drulis-Kawa, Z.; Cater, K.; Knežević, P.; Winogradow, C.; Amaro, K.; Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Rękas, J.; et al. Bacteriophages and Antibiotic Interactions in Clinical Practice: What We Have Learned so Far. J. Biomed. Sci. 2022, 29, 23. [Google Scholar] [CrossRef]
- Liu, C.G.; Green, S.I.; Min, L.; Clark, J.R.; Salazar, K.C.; Terwilliger, A.L.; Kaplan, H.B.; Trautner, B.W.; Ramig, R.F.; Maresso, A.W. Phage-Antibiotic Synergy Is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mBio 2020, 11, 1–19. [Google Scholar] [CrossRef]
- Lood, C.; Haas, P.J.; van Noort, V.; Lavigne, R. Shopping for Phages? Unpacking Design Rules for Therapeutic Phage Cocktails. Curr. Opin. Virol. 2022, 52, 236–243. [Google Scholar] [CrossRef]
- Duyvejonck, H.; Merabishvili, M.; Vaneechoutte, M.; de Soir, S.; Wright, R.; Friman, V.P.; Verbeken, G.; De Vos, D.; Pirnay, J.P.; Van Mechelen, E.; et al. Evaluation of the Stability of Bacteriophages in Different Solutions Suitable for the Production of Magistral Preparations in Belgium. Viruses 2021, 13, 865. [Google Scholar] [CrossRef]
- U.S. Pharmocopeia (USP) <85> Bacterial Endotoxins, Bacterial Endotoxins Monograph. Available online: https://www.usp.org/harmonization-standards/pdg/general-methods/bacterial-endotoxins (accessed on 9 August 2023).
- Hietala, V.; Horsma-Heikkinen, J.; Carron, A.; Skurnik, M.; Kiljunen, S. The Removal of Endo- and Enterotoxins from Bacteriophage Preparations. Front. Microbiol. 2019, 10, 1674. [Google Scholar] [CrossRef] [PubMed]
- Albano, F.; Thompson, M.R.; Orrú, S.; Orrú, O.; Scaloni, A.; Musetta, A.; Pucci, P.; Guarino, A. Structural and Functional Features of Modified Heat-Stable Toxins Produced by Enteropathogenic Klebsiella Cells. Pediatr. Res. 2000, 48, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Strakova, N.; Korena, K.; Karpiskova, R. Klebsiella pneumoniae Producing Bacterial Toxin Colibactin as a Risk of Colorectal Cancer Development—A Systematic Review. Toxicon 2021, 197, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Mamat, U.; Wilke, K.; Bramhill, D.; Schromm, A.B.; Lindner, B.; Kohl, T.A.; Corchero, J.L.; Villaverde, A.; Schaffer, L.; Head, S.R.; et al. Detoxifying Escherichia coli for Endotoxin-Free Production of Recombinant Proteins. Microb. Cell Fact. 2015, 14, 57. [Google Scholar] [CrossRef]
- Sacher, J. How Our Phage Therapy System Works at Phage Australia. Available online: https://phage.directory/capsid/phage-aus-process-overview (accessed on 7 July 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).