Analysis of the Respiratory Activity in the Antarctic Yeast Rhodotorula mucilaginosa M94C9 Reveals the Presence of Respiratory Supercomplexes and Alternative Elements
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Dry Weight Determination
2.3. Glucose Determination
2.4. Oxygen Consumption
2.5. Cell Permeabilization
2.6. Mitochondria Isolation
2.7. Competition Plot
2.8. Bioinformatic Analysis
2.9. Blue Native-PAGE and in-Gel Catalytic Activity Assays
2.10. 2D SDS-PAGE
2.11. Statistical Analysis
3. Results
3.1. Bioinformatic Results
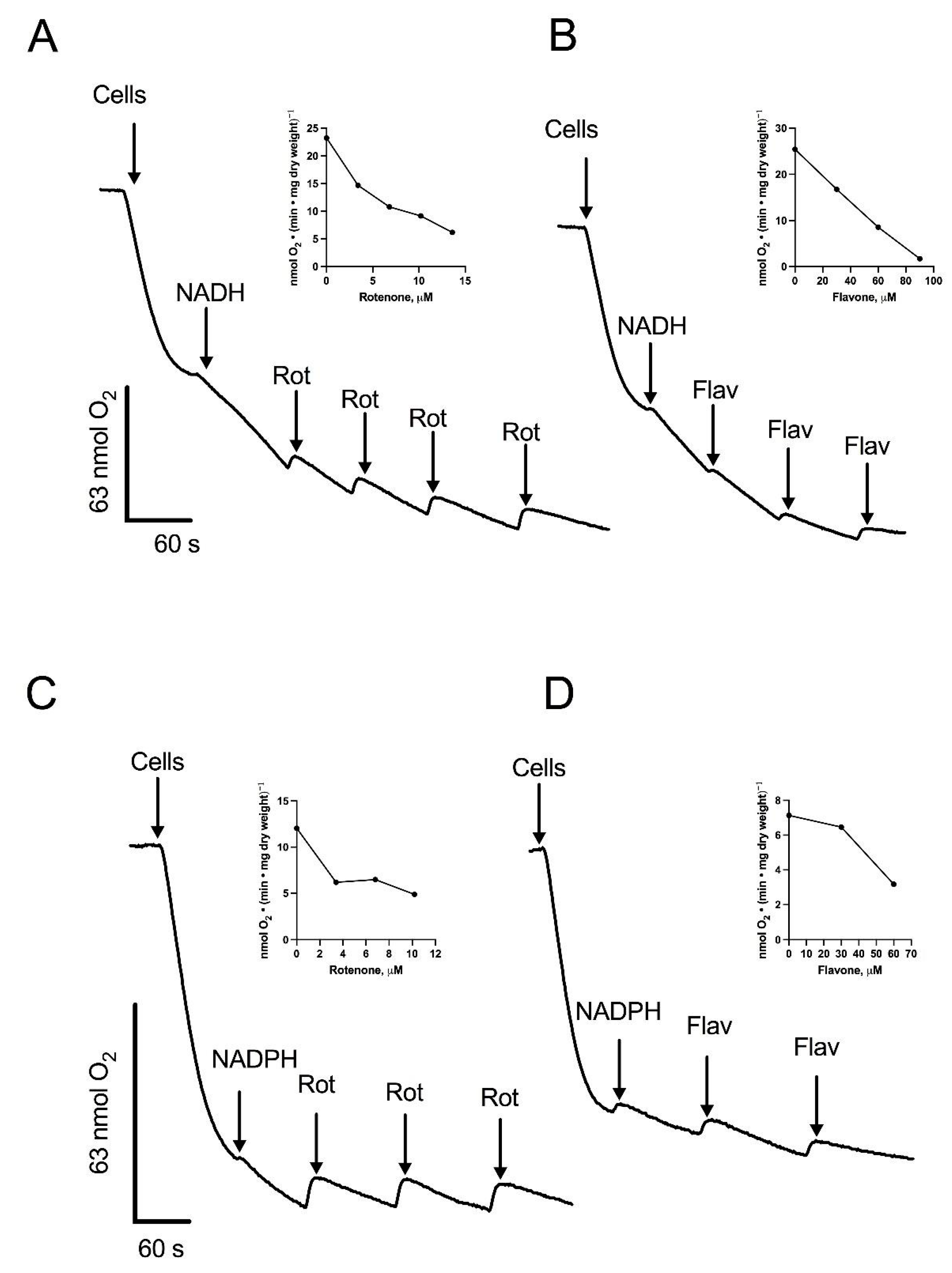
3.2. Oxygen Consumption by Intact Cells: The Cytochrome c Oxidase and the AOX Are the Terminal Oxidases
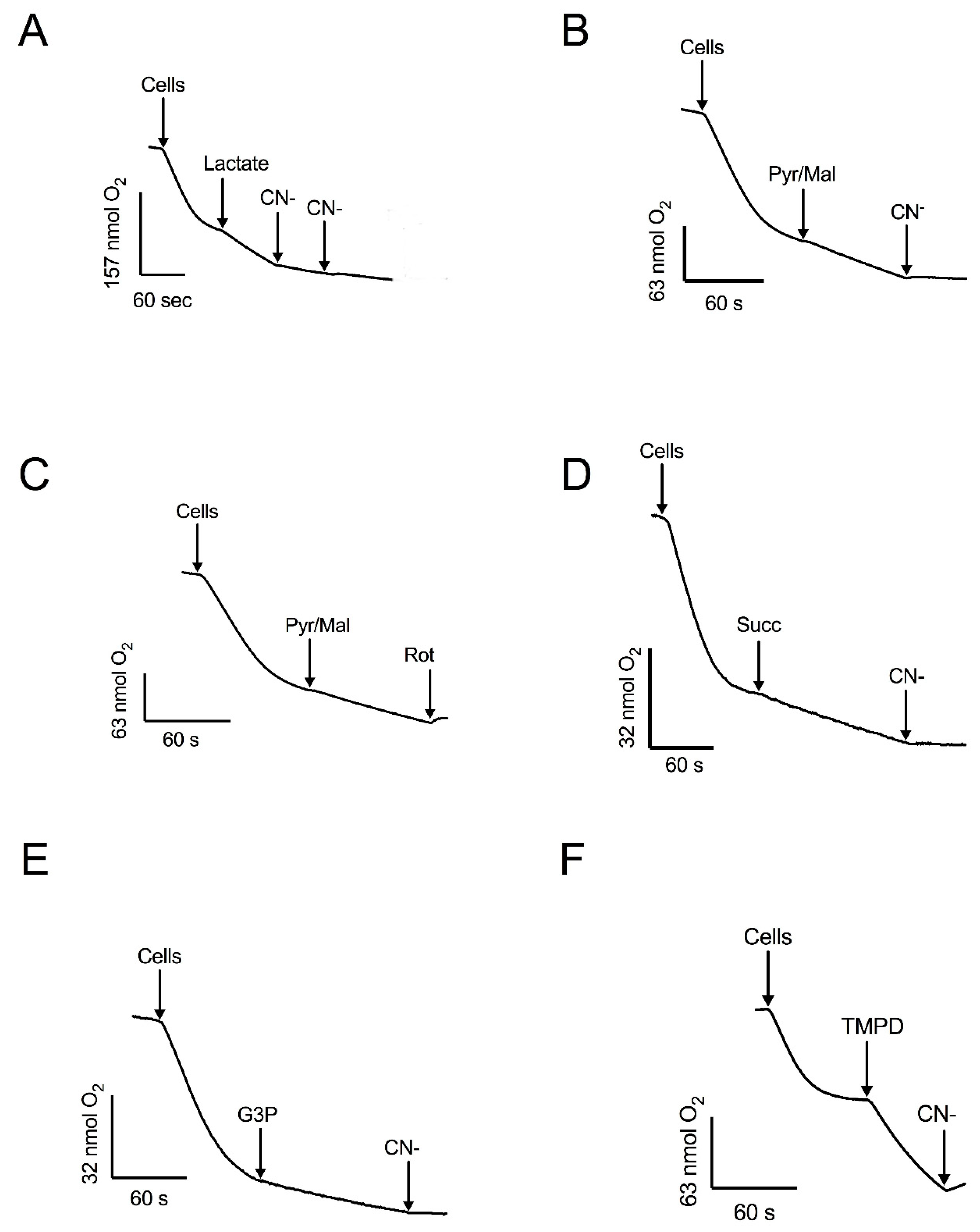
3.3. Respiratory Activities in Permeabilized Cells: Mitochondria Contain an Alternative NADPH Dehydrogenase
3.4. Respiratory Activities in Isolated Mitochondria
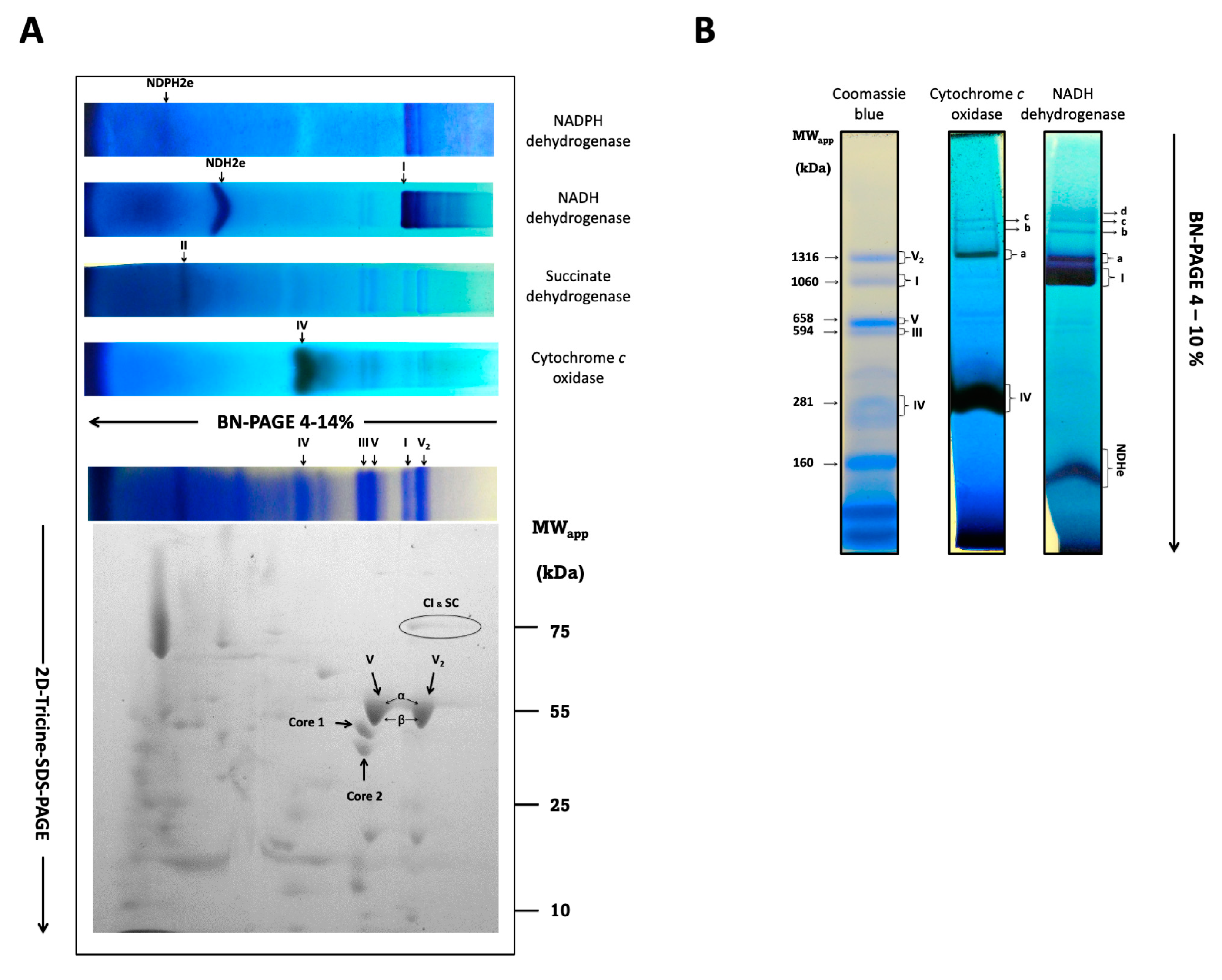
3.5. Respiratory Components of R. mucilaginosa Are Organized in Complexes and Supercomplexes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garcia-Cortes, A.; Garcia-Vásquez, J.A.; Aranguren, Y.; Ramirez-Castrillon, M. Pigment Production Improvement in Rhodotorula Mucilaginosa AJB01 Using Design of Experiments. Microorganisms 2021, 9, 387. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, C.; Liu, D.; He, N.; Deng, X. Hg Tolerance and Biouptake of an Isolated Pigmentation Yeast Rhodotorula Mucilaginosa. PLoS ONE 2017, 12, e0172984. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Shi, Y.; Peng, C.; Tang, L.; Chen, Y.; Wang, T.; Wang, Z.; Wang, S.; Li, Z. Transcriptome Analysis on Key Metabolic Pathways in Rhodotorula Mucilaginosa Under Pb(II) Stress. Appl. Environ. Microbiol. 2022, 88, e02215-21. [Google Scholar] [CrossRef]
- Hirano, R.; Mitsuhashi, T.; Osanai, K. Rhodotorula Mucilaginosa Fungemia, a Rare Opportunistic Infection without Central Venous Catheter Implantation, Successfully Treated by Liposomal Amphotericin B. Case Rep. Infect. Dis. 2022, 2022, 7830126. [Google Scholar] [CrossRef] [PubMed]
- Castañeda-Tamez, P.; Chiquete-Félix, N.; Uribe-Carvajal, S.; Cabrera-Orefice, A. The Mitochondrial Respiratory Chain from Rhodotorula Mucilaginosa, an Extremophile Yeast. Biochim. Biophys. Acta Bioenerg. 2024, 1865, 149035. [Google Scholar] [CrossRef]
- Troncoso, E.; Barahona, S.; Carrasco, M.; Villarreal, P.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Identification and Characterization of Yeasts Isolated from the South Shetland Islands and the Antarctic Peninsula. Polar Biol. 2017, 40, 649–658. [Google Scholar] [CrossRef]
- Romero-Aguilar, L.; Hernández-Morfín, K.D.; Guerra-Sánchez, G.; Pardo, J.P. Metabolic Changes and Antioxidant Response in Ustilago Maydis Grown in Acetate. J. Fungi 2023, 9, 749. [Google Scholar] [CrossRef]
- Juárez, O.; Guerra, G.; Martínez, F.; Pardo, J.P. The Mitochondrial Respiratory Chain of Ustilago Maydis. Biochim. Biophys. Acta 2004, 1658, 244–251. [Google Scholar] [CrossRef][Green Version]
- Robles-Martínez, L.; Guerra-Sánchez, M.G.; Flores-Herrera, O.; Hernández-Lauzardo, A.N.; Velázquez-Del Valle, M.G.; Pardo, J.P. The Mitochondrial Respiratory Chain of Rhizopus Stolonifer (Ehrenb.:Fr.) Vuill. Arch. Microbiol. 2013, 195, 51–61. [Google Scholar] [CrossRef]
- Matuz-Mares, D.; Flores-Herrera, O.; Guerra-Sánchez, G.; Romero-Aguilar, L.; Vázquez-Meza, H.; Matus-Ortega, G.; Martínez, F.; Pardo, J.P. Carbon and Nitrogen Sources Have No Impact on the Organization and Composition of Ustilago Maydis Respiratory Supercomplexes. J. Fungi 2021, 7, 42. [Google Scholar] [CrossRef]
- Pardo, J.P.; Guerra-Sánchez, G.; Flores-Herrera, O.; Romero-Aguilar, L. Isolation of Mitochondria from Ustilago Maydis Protoplasts. Bio Protoc. 2022, 12, e4277. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with The Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Chevillard, C.; Cárdenas, M.L.; Cornish-Bowden, A. The Competition Plot: A Simple Test of Whether Two Reactions Occur at the Same Active Site. Biochem. J. 1993, 289, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Thumuluri, V.; Almagro Armenteros, J.J.; Johansen, A.R.; Nielsen, H.; Winther, O. DeepLoc 2.0: Multi-Label Subcellular Localization Prediction Using Protein Language Models. Nucleic Acids Res. 2022, 50, W228–W234. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting Subcellular Localization of Proteins Based on Their N-Terminal Amino Acid Sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Nübel, E.; Wittig, I.; Kerscher, S.; Brandt, U.; Schägger, H. Two-Dimensional Native Electrophoretic Analysis of Respiratory Supercomplexes from Yarrowia Lipolytica. Proteomics 2009, 9, 2408–2418. [Google Scholar] [CrossRef]
- Carroll, J.; Fearnley, I.M.; Skehel, J.M.; Shannon, R.J.; Hirst, J.; Walker, J.E. Bovine Complex I Is a Complex of 45 Different Subunits. J. Biol. Chem. 2006, 281, 32724–32727. [Google Scholar] [CrossRef]
- Bridges, H.R.; Fearnley, I.M.; Hirst, J. The Subunit Composition of Mitochondrial NADH:Ubiquinone Oxidoreductase (Complex I) from Pichia Pastoris. Mol. Cell Proteom. 2010, 9, 2318–2326. [Google Scholar] [CrossRef]
- Morgner, N.; Zickermann, V.; Kerscher, S.; Wittig, I.; Abdrakhmanova, A.; Barth, H.-D.; Brutschy, B.; Brandt, U. Subunit Mass Fingerprinting of Mitochondrial Complex I. Biochim. Biophys. Acta 2008, 1777, 1384–1391. [Google Scholar] [CrossRef][Green Version]
- Marques, I.; Duarte, M.; Assunção, J.; Ushakova, A.V.; Videira, A. Composition of Complex I from Neurospora Crassa and Disruption of Two “Accessory” Subunits. Biochim. Biophys. Acta 2005, 1707, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Dejean, L.; Beauvoit, B.; Guérin, B.; Rigoulet, M. Growth of the Yeast Saccharomyces Cerevisiae on a Non-Fermentable Substrate: Control of Energetic Yield by the Amount of Mitochondria. Biochim. Biophys. Acta 2000, 1457, 45–56. [Google Scholar] [CrossRef]
- Kamzolova, S.V.; Shishkanova, N.V.; Morgunov, I.G.; Finogenova, T.V. Oxygen Requirements for Growth and Citric Acid Production of Yarrowia Lipolytica. FEMS Yeast Res. 2003, 3, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Copsey, A.C.; Young, L.; Barsottini, M.R.O.; Albury, M.S.; Moore, A.L. Comparison of the Kinetic Parameters of Alternative Oxidases From Trypanosoma Brucei and Arabidopsis Thaliana—A Tale of Two Cavities. Front. Plant Sci. 2021, 12, 744218. [Google Scholar] [CrossRef] [PubMed]
- Hoefnagel, M.H.N.; Rich, P.R.; Zhang, Q.; Wiskich, J.T. Substrate Kinetics of the Plant Mitochondrial Alternative Oxidase and the Effects of Pyruvate. Plant Physiol. 1997, 115, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Barsottini, M.R.O.; Copsey, A.; Young, L.; Baroni, R.M.; Cordeiro, A.T.; Pereira, G.A.G.; Moore, A.L. Biochemical Characterization and Inhibition of the Alternative Oxidase Enzyme from the Fungal Phytopathogen Moniliophthora Perniciosa. Commun. Biol. 2020, 3, 263. [Google Scholar] [CrossRef]
- Medentsev, A.G.; Arinbasarova, A.Y.; Golovchenko, N.P.; Akimenko, V.K. Involvement of the Alternative Oxidase in Respiration of Yarrowia Lipolytica Mitochondria Is Controlled by the Activity of the Cytochrome Pathway. FEMS Yeast Res. 2002, 2, 519–524. [Google Scholar] [CrossRef][Green Version]
- Cárdenas-Monroy, C.A.; Pohlmann, T.; Piñón-Zárate, G.; Matus-Ortega, G.; Guerra, G.; Feldbrügge, M.; Pardo, J.P. The Mitochondrial Alternative Oxidase Aox1 Is Needed to Cope with Respiratory Stress but Dispensable for Pathogenic Development in Ustilago Maydis. PLoS ONE 2017, 12, e0173389. [Google Scholar] [CrossRef]
- Brand, M.D.; Nicholls, D.G. Assessing Mitochondrial Dysfunction in Cells. Biochem. J. 2011, 435, 297–312. [Google Scholar] [CrossRef]
- Orij, R.; Postmus, J.; Ter Beek, A.; Brul, S.; Smits, G.J. In Vivo Measurement of Cytosolic and Mitochondrial pH Using a pH-Sensitive GFP Derivative in Saccharomyces Cerevisiae Reveals a Relation between Intracellular pH and Growth. Microbiology 2009, 155, 268–278. [Google Scholar] [CrossRef]
- Sanders, D.; Slayman, C.L. Control of Intracellular pH. Predominant Role of Oxidative Metabolism, Not Proton Transport, in the Eukaryotic Microorganism Neurospora. J. Gen. Physiol. 1982, 80, 377–402. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Carrada, M.; Feldbrügge, M.; Olicón-Hernández, D.R.; Guerra-Sánchez, G.; Pardo, J.P. Functional Analysis of the Plasma Membrane H+-ATPases of Ustilago Maydis. J. Fungi 2022, 8, 550. [Google Scholar] [CrossRef] [PubMed]
- de VRIES, S.; Grivell, L.A. Purification and Characterization of a Rotenone-Insensitive NADH: Q6 Oxidoreductase from Mitochondria of Saccharomyces Cerevisiae. Eur. J. Biochem. 1988, 176, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, I.; Pardo, J.P. Kinetic Characterization of the Rotenone-Insensitive Internal NADH: Ubiquinone Oxidoreductase of Mitochondria from Saccharomyces Cerevisiae. Arch. Biochem. Biophys. 2001, 389, 7–14. [Google Scholar] [CrossRef]
- Saks, V.A.; Kuznetsov, A.V.; Khuchua, Z.A.; Vasilyeva, E.V.; Belikova, J.O.; Kesvatera, T.; Tiivel, T. Control of Cellular Respiration in Vivo by Mitochondrial Outer Membrane and by Creatine Kinase. A New Speculative Hypothesis: Possible Involvement of Mitochondrial-Cytoskeleton Interactions. J. Mol. Cell Cardiol. 1995, 27, 625–645. [Google Scholar] [CrossRef]
- Avéret, N.; Fitton, V.; Bunoust, O.; Rigoulet, M.; Guérin, B. Yeast Mitochondrial Metabolism: From in Vitro to in Situ Quantitative Study. Mol. Cell Biochem. 1998, 184, 67–79. [Google Scholar] [CrossRef]
- Matsunaka, S.; Morita, S.; Conti, S.F. Respiratory System of Rhodotorula Glutinis. I. Inhibitor Tolerance and Cytochrome Components. Plant Physiol. 1966, 41, 1364–1369. [Google Scholar] [CrossRef]
- Matsunaka, S.; Conti, S.F. The Respiratory System of Rhodotorula Glutinis. II. Mechanism of Inhibitor Tolerant Respiration. Plant Physiol. 1966, 41, 1370–1375. [Google Scholar] [CrossRef]
- Henry, M.F.; Hamaide-Deplus, M.C.; Nyns, E.J. Cyanide-Insensitive Respiration of Candida Lipolytica. Antonie Van Leeuwenhoek 1974, 40, 79–91. [Google Scholar] [CrossRef]
- Johnson, K.L. Turning Up the Heat: The Alternative Oxidase Pathway Drives Thermogenesis in Cycad Cones. Plant Physiol. 2019, 180, 689–690. [Google Scholar] [CrossRef]
- Lin, Z.; Wu, J.; Jamieson, P.A.; Zhang, C. Alternative Oxidase Is Involved in the Pathogenicity, Development, and Oxygen Stress Response of Botrytis cinerea. Phytopathology® 2019, 109, 1679–1688. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Zhao, M.; Huang, C.; He, Q.; Zhang, L.; Zhang, J. Alternative Oxidase Gene Induced by Nitric Oxide Is Involved in the Regulation of ROS and Enhances the Resistance of Pleurotus Ostreatus to Heat Stress. Microb. Cell Fact. 2021, 20, 137. [Google Scholar] [CrossRef] [PubMed]
- Szibor, M.; Schenkl, C.; Barsottini, M.R.O.; Young, L.; Moore, A.L. Targeting the Alternative Oxidase (AOX) for Human Health and Food Security, a Pharmaceutical and Agrochemical Target or a Rescue Mechanism? Biochem. J. 2022, 479, 1337–1359. [Google Scholar] [CrossRef]
- Pérez-Sánchez, A.; Uribe-Carvajal, S.; Cabrera-Orefice, A.; Barrios-González, J. Key Role of Alternative Oxidase in Lovastatin Solid-State Fermentation. Appl. Microbiol. Biotechnol. 2017, 101, 7347–7356. [Google Scholar] [CrossRef] [PubMed]
- Kerscher, S.; Dröse, S.; Zwicker, K.; Zickermann, V.; Brandt, U. Yarrowia Lipolytica, a Yeast Genetic System to Study Mitochondrial Complex I. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2002, 1555, 83–91. [Google Scholar] [CrossRef]
- Matuz-Mares, D.; Matus-Ortega, G.; Cárdenas-Monroy, C.; Romero-Aguilar, L.; Villalobos-Rocha, J.C.; Vázquez-Meza, H.; Guerra-Sánchez, G.; Peña-Díaz, A.; Pardo, J.P. Expression of Alternative NADH Dehydrogenases (NDH-2) in the Phytopathogenic Fungus Ustilago Maydis. FEBS Open Bio 2018, 8, 1267–1279. [Google Scholar] [CrossRef]
- Guerrero-Castillo, S.; Vázquez-Acevedo, M.; González-Halphen, D.; Uribe-Carvajal, S. In Yarrowia Lipolytica Mitochondria, the Alternative NADH Dehydrogenase Interacts Specifically with the Cytochrome Complexes of the Classic Respiratory Pathway. Biochim. Biophys. Acta 2009, 1787, 75–85. [Google Scholar] [CrossRef]
- Melo, A.M.; Duarte, M.; Møller, I.M.; Prokisch, H.; Dolan, P.L.; Pinto, L.; Nelson, M.A.; Videira, A. The External Calcium-Dependent NADPH Dehydrogenase from Neurospora Crassa Mitochondria. J. Biol. Chem. 2001, 276, 3947–3951. [Google Scholar] [CrossRef]
- Rasmusson, A.G.; Soole, K.L.; Elthon, T.E. Alternative NAD(P)H Dehydrogenases of Plant Mitochondria. Annu. Rev. Plant Biol. 2004, 55, 23–39. [Google Scholar] [CrossRef]
- Boubacar, A.K.O.; Pethe, S.; Mahy, J.-P.; Lederer, F. Flavocytochrome B2: Reactivity of Its Flavin with Molecular Oxygen. Biochemistry 2007, 46, 13080–13088. [Google Scholar] [CrossRef]
- Bakker, B.M.; Overkamp, K.M.; van Maris, A.J.A.; Kötter, P.; Luttik, M.A.H.; van Dijken, J.P.; Pronk, J.T. Stoichiometry and Compartmentation of NADH Metabolism in Saccharomyces Cerevisiae. FEMS Microbiol. Rev. 2001, 25, 15–37. [Google Scholar] [CrossRef] [PubMed]
- Kerscher, S.J. Diversity and Origin of Alternative NADH:Ubiquinone Oxidoreductases. Biochim. Biophys. Acta 2000, 1459, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Orefice, A.; Chiquete-Félix, N.; Espinasa-Jaramillo, J.; Rosas-Lemus, M.; Guerrero-Castillo, S.; Peña, A.; Uribe-Carvajal, S. The Branched Mitochondrial Respiratory Chain from Debaryomyces Hansenii: Components and Supramolecular Organization. Biochim. Biophys. Acta 2014, 1837, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Marques, I.; Dencher, N.A.; Videira, A.; Krause, F. Supramolecular Organization of the Respiratory Chain in Neurospora Crassa Mitochondria. Eukaryot. Cell 2007, 6, 2391–2405. [Google Scholar] [CrossRef][Green Version]
- Berndtsson, J.; Kohler, A.; Rathore, S.; Marin-Buera, L.; Dawitz, H.; Diessl, J.; Kohler, V.; Barrientos, A.; Büttner, S.; Fontanesi, F.; et al. Respiratory Supercomplexes Enhance Electron Transport by Decreasing Cytochrome c Diffusion Distance. EMBO Rep. 2020, 21, e51015. [Google Scholar] [CrossRef]
- Reyes-Galindo, M.; Suarez, R.; Esparza-Perusquía, M.; de Lira-Sánchez, J.; Pardo, J.P.; Martínez, F.; Flores-Herrera, O. Mitochondrial Respirasome Works as a Single Unit and the Cross-Talk between Complexes I, III2 and IV Stimulates NADH Dehydrogenase Activity. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2019, 1860, 618–627. [Google Scholar] [CrossRef]
- Acín-Pérez, R.; Bayona-Bafaluy, M.P.; Fernández-Silva, P.; Moreno-Loshuertos, R.; Pérez-Martos, A.; Bruno, C.; Moraes, C.T.; Enríquez, J.A. Respiratory Complex III Is Required to Maintain Complex I in Mammalian Mitochondria. Mol. Cell 2004, 13, 805–815. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Norberg, F.E.B.; Szilágyi, A.; De Paepe, R.; Åkerlund, H.-E.; Rasmusson, A.G. The Mitochondrial External NADPH Dehydrogenase Modulates the Leaf NADPH/NADP+ Ratio in Transgenic Nicotiana Sylvestris. Plant Cell Physiol. 2008, 49, 251–263. [Google Scholar] [CrossRef]
- Hagman, A.; Säll, T.; Piškur, J. Analysis of the Yeast Short-Term Crabtree Effect and Its Origin. FEBS J. 2014, 281, 4805–4814. [Google Scholar] [CrossRef]
- Sakoda, Y.; Matsumoto, T.; Kudo, A.; Yoshida, K.; Ishibashi, K.; Saruwatari, A.; Ogata, T.; Honda, J. Asymptomatic Fungemia Due to Rhodotorula spp. Caused by a Subcutaneously Implanted Central Venous Port Catheter. Intern. Med. 2022, 61, 2677–2680. [Google Scholar] [CrossRef]
- Jarros, I.C.; Veiga, F.F.; Corrêa, J.L.; Barros, I.L.E.; Gadelha, M.C.; Voidaleski, M.F.; Pieralisi, N.; Pedroso, R.B.; Vicente, V.A.; Negri, M.; et al. Microbiological and Virulence Aspects of Rhodotorula Mucilaginosa. EXCLI J. 2020, 19, 687–704. [Google Scholar] [PubMed]



| CI | CII | CIII | CIV | CV | |
|---|---|---|---|---|---|
| Mitochondrial encoded | 7 | 0 | 1 | 3 | 3 |
| Nuclear encoded | 30 | 4 | 9 | 8 | 13 |
| Molecular mass (kDa) | 937.4 | 123.2 | 274 | 208.3 | 622.2 |
| Protein/accession number | Molecular mass (kDa) | ||||
| Alternative oxidase KAG0654415.1 | 43.2 | ||||
| Glycerol 3-P dehydrogenase KAG0655011.1 | 96.4 | ||||
| Lactate cyt b2 dehydrogenase KAG0667662.1 | 57.4 | ||||
| NAD(P)H dehydrogenases: | |||||
| KAG0665032.1 | 62.3 | ||||
| KAG0656515.1 | 49.8 | ||||
| KAG0656514.1 | 51.4 | ||||
| KAG0663112.1 | 74.1 | ||||
| KAG0663538.1 | 41.8 | ||||
| Respiratory Activity [nmol O2 min−1 mg Dry Weight −1] * | Substrate (mM) |
|---|---|
| 102 ± 15 | Initial |
| 117 ± 23 | KCN |
| 97 ± 18 | nOG |
| Permeabilized cells * | |
| 22.3 ± 5.9 | NADH |
| 14.4 ± 4.0 | NADPH |
| 11.6 ± 1.9 | Succinate |
| 13.9 ± 1.2 | Lactate |
| 6.9 ± 3.8 | Pyruvate-malate |
| 3.0 ± 1.2 | Glycerol-3-phosphate |
| 48.0 ± 23.9 | TMPD |
| Isolated Mitochondria | |
| [nmol O2 min−1 mg protein−1] | |
| 241.5 ± 29.7 | NADH |
| 192.9 ± 25.4 | NADPH |
| 53.6 ± 12. 2 | Pyruvate-Malate |
| 60.1 ± 10.9 | Succinate |
| 69.2 ± 1.1 | Lactate |
| 294.3 ± 81.1 | TMPD |
| Condition/Units | Substrate | |
|---|---|---|
| Permeabilized cells | ||
| K0.5 (μM) | Vmax (nmol·min−1·mg dry weight −1) | |
| 61 ± 42 | 28 ± 6 | NADH |
| 149 ± 37 * | 21 ± 4 | NADPH |
| 108 ± 26 | 10 ± 1 | Succinate |
| 41 ± 28 | 17 ± 1 | DL-Lactate |
| Isolated Mitochondria | ||
| K0.5 (µM) | Vmax (nmol·min−1·mg protein−1) | |
| 54 ± 31 | 299 ± 82 | NADH |
| 56 ± 16 | 206 ± 49 | NADPH |
| 64 ± 47 | 52 ± 6 | Succinate |
| 20 ± 13 | 73 ± 2 | DL-Lactate |
| Band | Apparent Mass (KDa) | Calculated Mass (KDa) | Tentative Stoichiometry |
|---|---|---|---|
| a | 1254 | 1145 | I1IV1 |
| b | 1691 | 1693 | I1III2IV1 |
| c | 1937 | 1901 | I1III2IV2 |
| d | 2046 | 2055 * | I1III2IV1X* |
| Complexes | R. mucilaginosa | R. mucilaginosa * | U. maydis ** | Y. lipolytica *** | |
|---|---|---|---|---|---|
| Experimental | Predicted | ||||
| I | 1061 | 937 | 1000 | 980 | 960 |
| II | 130 | 123 | 100 | 139 | ND |
| III | 594 | 548 | 580 | 510 | 458 |
| IV | 281 | 208 | 200 | 240 | 189 |
| V | 658 | 622 | 580 | 640 | 543 |
| V2 | 1316 | 1244 | 1130 | 1280 | 1231 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes-Rosario, D.; Pardo, J.P.; Guerra-Sánchez, G.; Vázquez-Meza, H.; López-Hernández, G.; Matus-Ortega, G.; González, J.; Baeza, M.; Romero-Aguilar, L. Analysis of the Respiratory Activity in the Antarctic Yeast Rhodotorula mucilaginosa M94C9 Reveals the Presence of Respiratory Supercomplexes and Alternative Elements. Microorganisms 2024, 12, 1931. https://doi.org/10.3390/microorganisms12101931
Reyes-Rosario D, Pardo JP, Guerra-Sánchez G, Vázquez-Meza H, López-Hernández G, Matus-Ortega G, González J, Baeza M, Romero-Aguilar L. Analysis of the Respiratory Activity in the Antarctic Yeast Rhodotorula mucilaginosa M94C9 Reveals the Presence of Respiratory Supercomplexes and Alternative Elements. Microorganisms. 2024; 12(10):1931. https://doi.org/10.3390/microorganisms12101931
Chicago/Turabian StyleReyes-Rosario, Daniel, Juan Pablo Pardo, Guadalupe Guerra-Sánchez, Héctor Vázquez-Meza, Georgina López-Hernández, Genaro Matus-Ortega, James González, Marcelo Baeza, and Lucero Romero-Aguilar. 2024. "Analysis of the Respiratory Activity in the Antarctic Yeast Rhodotorula mucilaginosa M94C9 Reveals the Presence of Respiratory Supercomplexes and Alternative Elements" Microorganisms 12, no. 10: 1931. https://doi.org/10.3390/microorganisms12101931
APA StyleReyes-Rosario, D., Pardo, J. P., Guerra-Sánchez, G., Vázquez-Meza, H., López-Hernández, G., Matus-Ortega, G., González, J., Baeza, M., & Romero-Aguilar, L. (2024). Analysis of the Respiratory Activity in the Antarctic Yeast Rhodotorula mucilaginosa M94C9 Reveals the Presence of Respiratory Supercomplexes and Alternative Elements. Microorganisms, 12(10), 1931. https://doi.org/10.3390/microorganisms12101931






