U2-Net and ResNet50-Based Automatic Pipeline for Bacterial Colony Counting
Abstract
1. Introduction
2. Materials and Methods
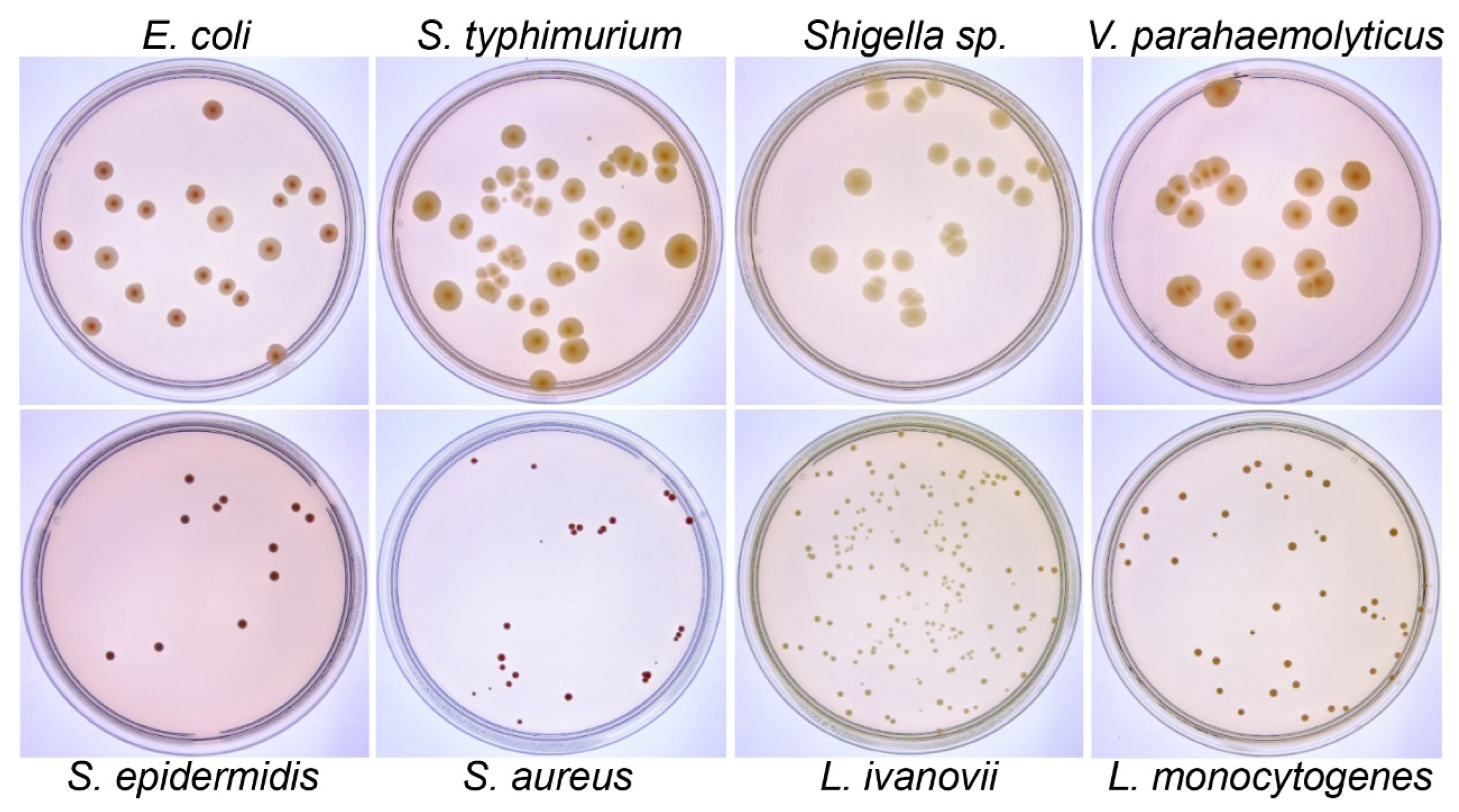
2.1. Bacterial Culture
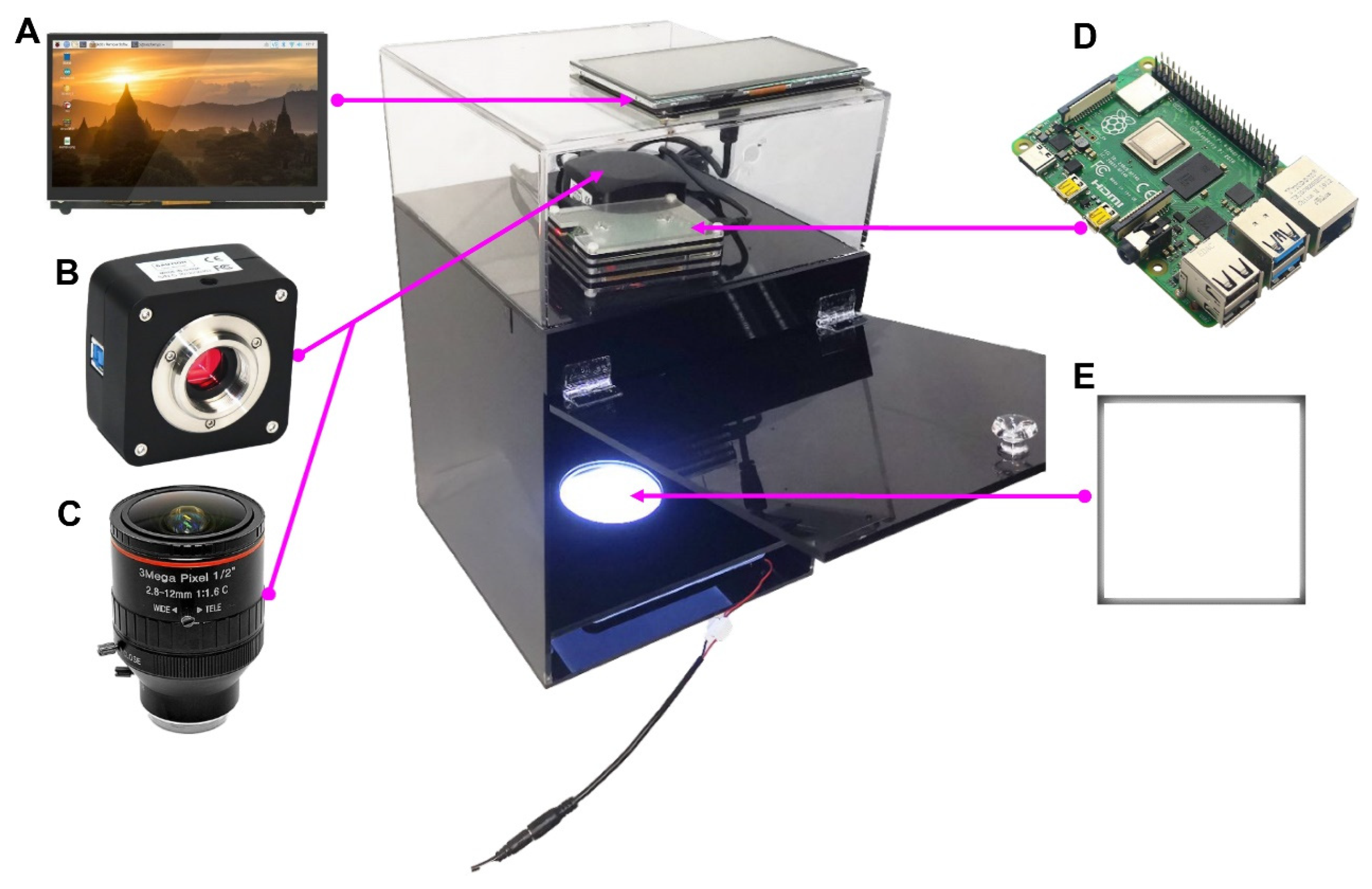
2.2. Assembly of the Imaging Device
2.3. Raw Image Acquisition
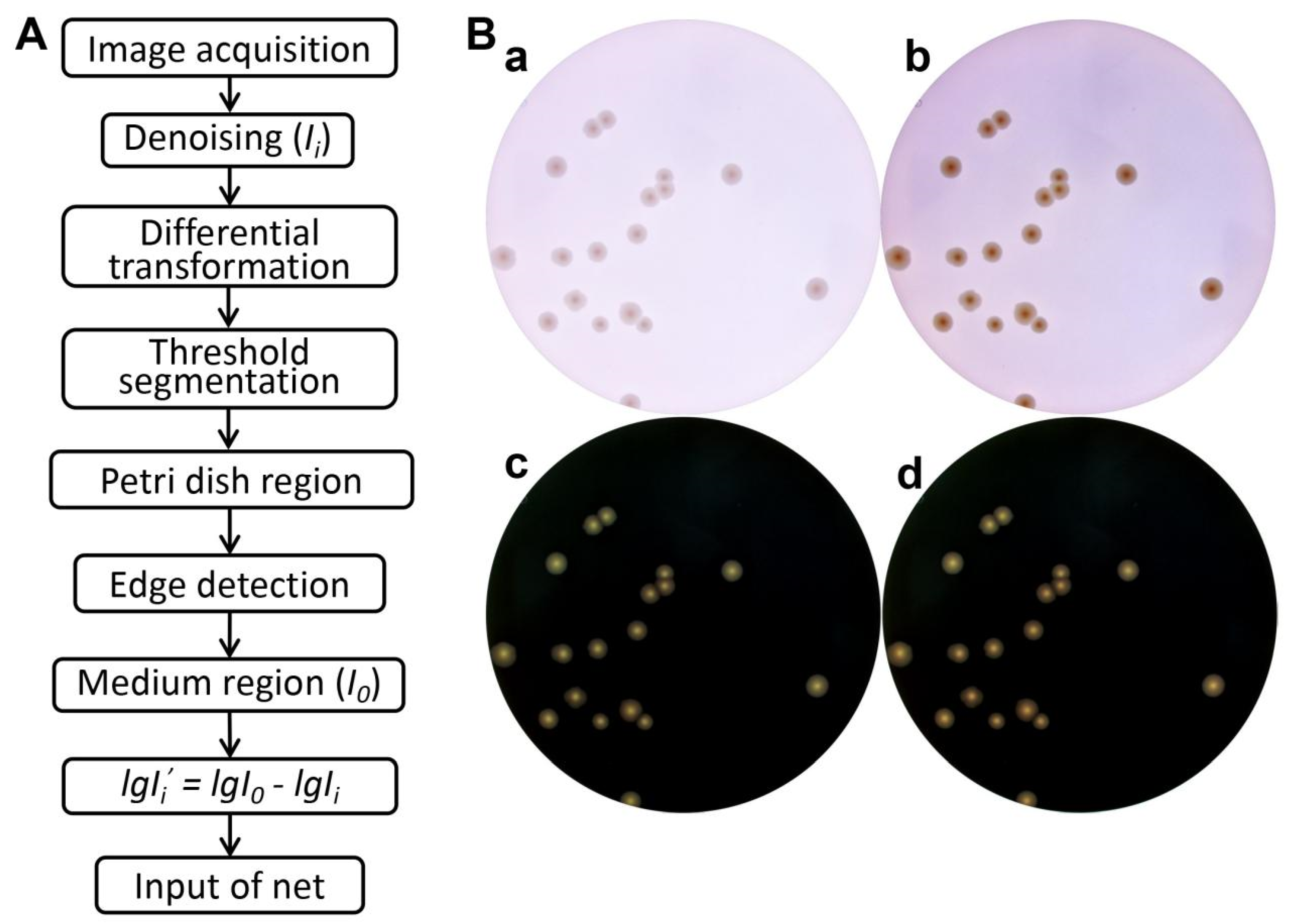
2.4. Image Preprocessing
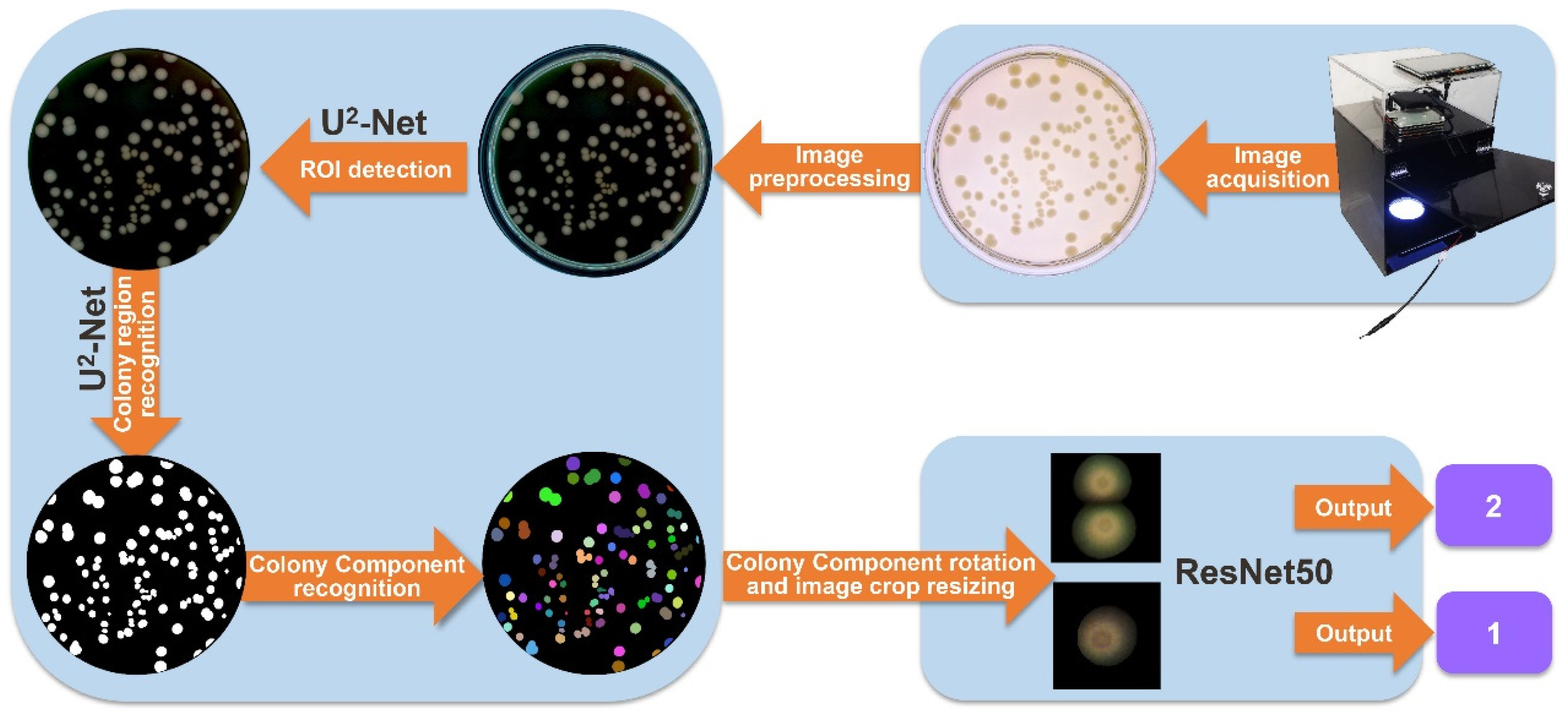
2.5. Colony Identification and Counting
2.5.1. Culture Dish Edge Segmentation
Dataset Preparation and Network Construction
Network Training
2.5.2. Colony Region Separation
Dataset Preparation
Network Training
2.5.3. Colony Counting
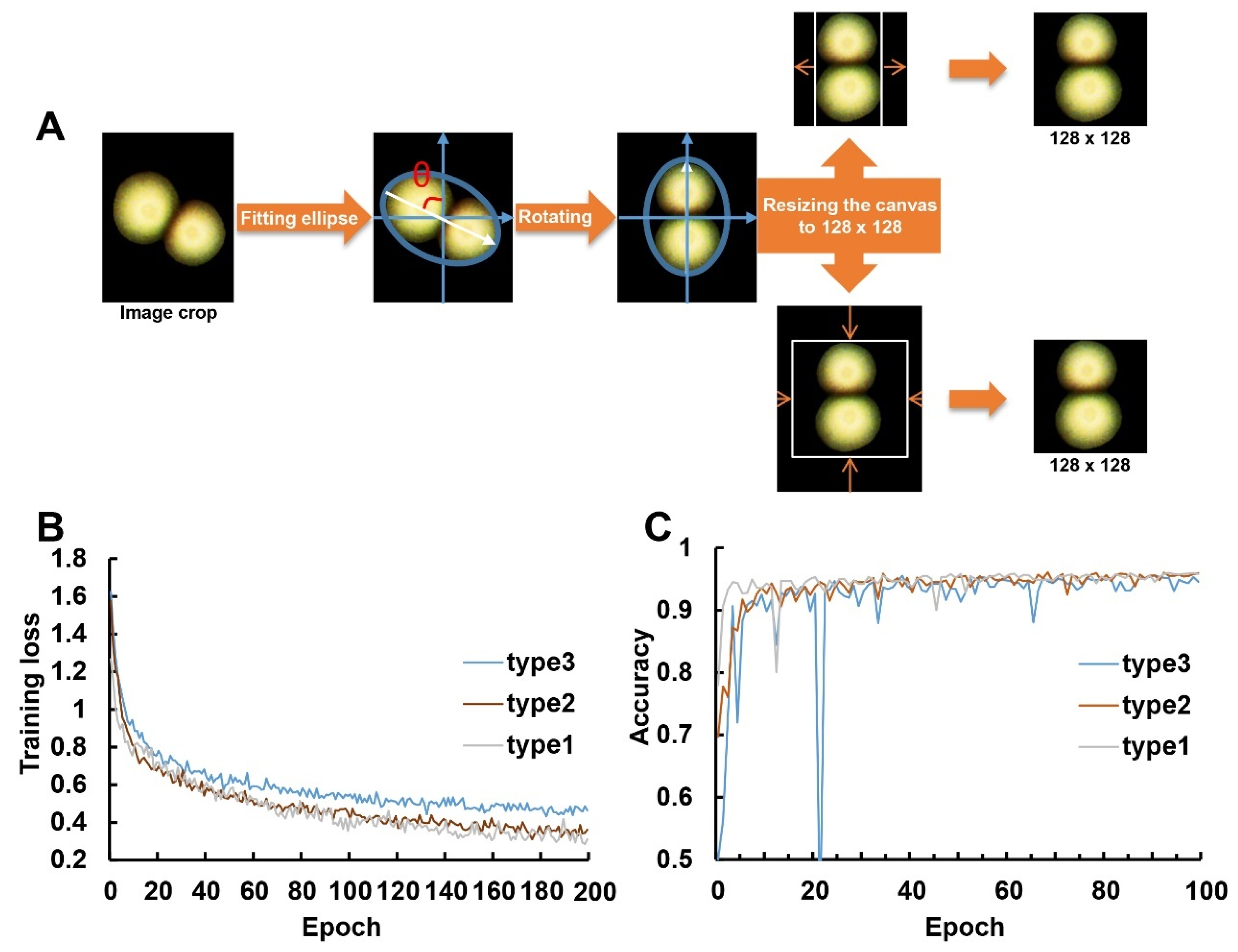
Dataset Preparation
Network Architecture
Network Training
2.6. Training Environment
3. Results
3.1. The Imaging System Captures High-Quality Colony Images
3.2. Image Preprocessing
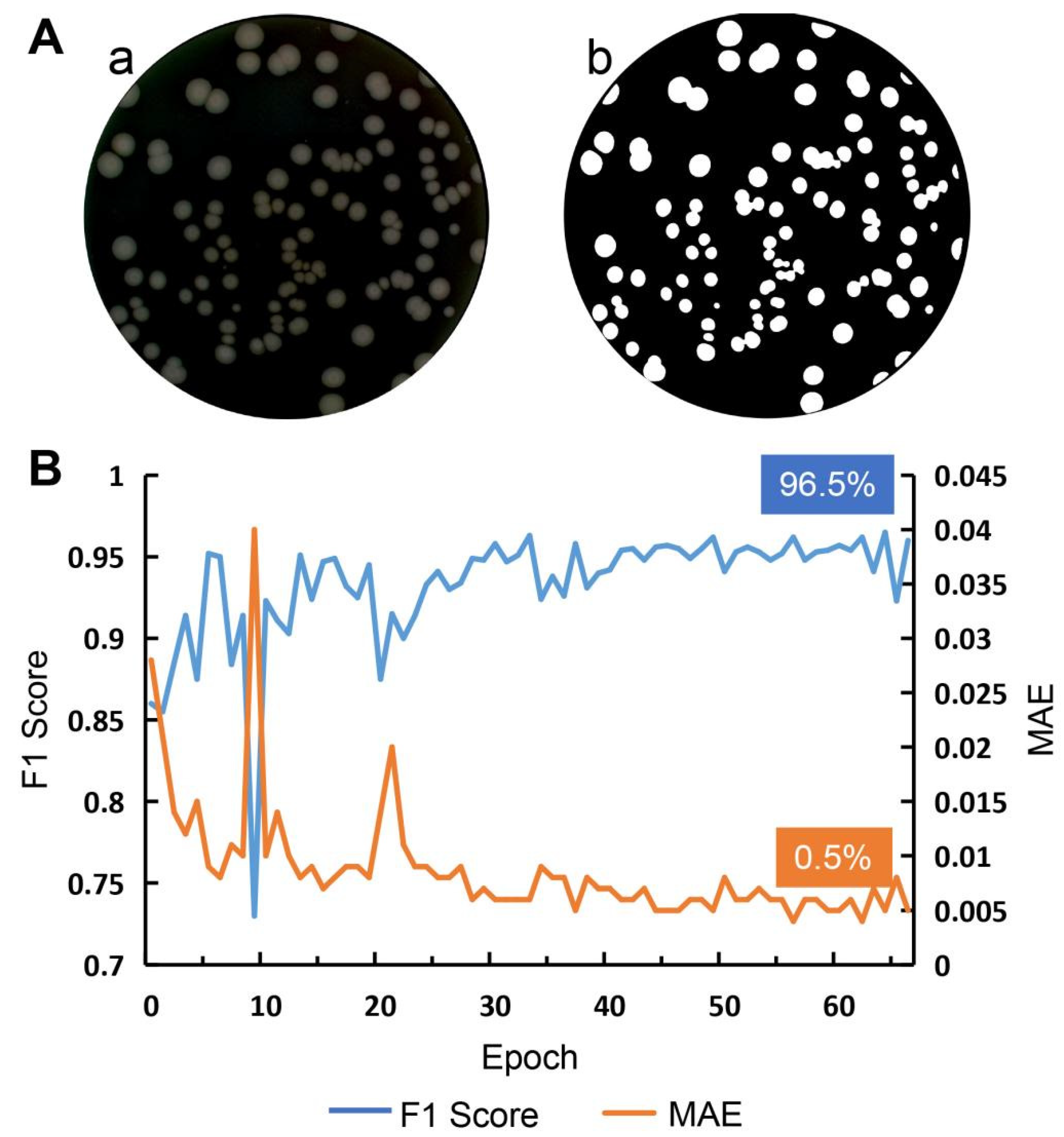
3.3. Petri Dish Edge Segmentation
3.4. Colony Region Separation
3.5. Training and Validation Performance of ResNet50 on Colony Counting
3.6. Test Performance of the Pipeline on Colony Counting of Entire Images
4. Discussion
4.1. U2-Net Is More Suitable for Locating the Edges of Petri Dishes and Extracting Bacterial Colony Areas Compared to Threshold Segmentation
4.2. The ResNet50 Model in Our Proposed Method Functions as an Interchangeable Module
4.3. Our Method Surpasses YOLO, the Segment Anything Model, and OpenCFU in Performance
4.4. The Impact of Bacterial Colony Quantity and Size on the Performance of Our Method
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gwimbi, P.; George, M.; Ramphalile, M. Bacterial contamination of drinking water sources in rural villages of Mohale Basin, Lesotho: Exposures through neighbourhood sanitation and hygiene practices. Environ. Health Prev. Med. 2019, 24, 33. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.L.; Burton, R.L.; Hill, A.N.; Litorja, M.; Nahm, M.H.; Hwang, J. Low-cost, high-throughput, automated counting of bacterial colonies. Cytom. Part A 2010, 77, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Liu, X.; Tian, Q.; Yue, W.; Zeng, J.; Chen, G.; Cai, X. Disposable bioluminescence-based biosensor for detection of bacterial count in food. Anal. Biochem. 2009, 394, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tillman, G.E.; Wasilenko, J.L.; Simmons, M.; Lauze, T.A.; Minicozzi, J.; Oakley, B.B.; Narang, N.; Fratamico, P.; Cray, A.C., Jr. Isolation of Shiga toxin-producing Escherichia coli serogroups O26, O45, O103, O111, O121, and O145 from ground beef using modified rainbow agar and post-immunomagnetic separation acid treatment. J. Food Prot. 2012, 75, 1548–1554. [Google Scholar] [CrossRef]
- Brugger, S.D.; Baumberger, C.; Jost, M.; Jenni, W.; Brugger, U.; Muhlemann, K. Automated counting of bacterial colony forming units on agar plates. PLoS ONE 2012, 7, e33695. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, W.B.; Liu, W.L.; Chen, C.B. An Automated Bacterial Colony Counting System. In Proceedings of the 2008 IEEE International Conference on Sensor Networks, Ubiquitous, and Trustworthy Computing (Sutc 2008), Taichung, Taiwan, 11–13 June 2008; pp. 233–240. [Google Scholar]
- Mukherjee, D.P.; Pal, A.; Sarma, S.E.; Majumder, D.D. Bacterial colony counting using distance transform. Int. J. Biomed. Comput. 1995, 38, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Coulthard, M.G. Defining urinary tract infection by bacterial colony counts: A case for 100,000 colonies/ml as the best threshold. Pediatr. Nephrol. 2019, 34, 1639–1649. [Google Scholar] [CrossRef]
- Ferrari, A.; Signoroni, A. Multistage classification for bacterial colonies recognition on solid agar images. In Proceedings of the 2014 IEEE International Conference on Imaging Systems and Techniques (IST) Proceedings, Santorini, Greece, 17 October 2014; pp. 101–106. [Google Scholar]
- Yoon, S.-C.; Lawrence, K.C.; Park, B. Automatic Counting and Classification of Bacterial Colonies Using Hyperspectral Imaging. Food Bioprocess Technol. 2015, 8, 2047–2065. [Google Scholar] [CrossRef]
- Kulwa, F.; Li, C.; Zhao, X.; Cai, B.; Xu, N.; Qi, S.; Chen, S.; Teng, Y. A State-of-the-Art Survey for Microorganism Image Segmentation Methods and Future Potential. IEEE Access 2019, 7, 100243–100269. [Google Scholar] [CrossRef]
- Goodswen, S.J.; Barratt, J.L.N.; Kennedy, P.J.; Kaufer, A.; Calarco, L.; Ellis, J.T. Machine learning and applications in microbiology. FEMS Microbiol. Rev. 2021, 45, fuab015. [Google Scholar] [CrossRef]
- Thakur, P.; Alaba, M.O.; Rauniyar, S.; Singh, R.N.; Saxena, P.; Bomgni, A.; Gnimpieba, E.Z.; Lushbough, C.; Goh, K.M.; Sani, R.K. Text-Mining to Identify Gene Sets Involved in Biocorrosion by Sulfate-Reducing Bacteria: A Semi-Automated Workflow. Microorganisms 2023, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, C.; Yin, Y.; Zhang, J.; Grzegorzek, M. Applications of artificial neural networks in microorganism image analysis: A comprehensive review from conventional multilayer perceptron to popular convolutional neural network and potential visual transformer. Artif. Intell. Rev. 2023, 56, 1013–1070. [Google Scholar] [CrossRef]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [PubMed]
- Deep bacteria: Robust deep learning data augmentation design for limited bacterial colony dataset. Int. J. Reason.-Based Intell. Syst. 2019, 11, 256–264. [CrossRef]
- Yu, W.; Xiang, Q.; Hu, Y.; Du, Y.; Kang, X.; Zheng, D.; Shi, H.; Xu, Q.; Li, Z.; Niu, Y.; et al. An improved automated diatom detection method based on YOLOv5 framework and its preliminary study for taxonomy recognition in the forensic diatom test. Front. Microbiol. 2022, 13, 963059. [Google Scholar] [CrossRef]
- Rani, P.; Kotwal, S.; Manhas, J.; Sharma, V.; Sharma, S. Machine Learning and Deep Learning Based Computational Approaches in Automatic Microorganisms Image Recognition: Methodologies, Challenges, and Developments. Arch. Comput. Methods Eng. 2022, 29, 1801–1837. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.; Lombardi, S.; Signoroni, A. Bacterial colony counting with Convolutional Neural Networks in Digital Microbiology Imaging. Pattern Recognit. 2017, 61, 629–640. [Google Scholar] [CrossRef]
- Carl, S.H.; Duempelmann, L.; Shimada, Y.; Buhler, M. A fully automated deep learning pipeline for high-throughput colony segmentation and classification. Biol. Open 2020, 9, bio052936. [Google Scholar] [CrossRef]
- Majchrowska, S.; Pawłowski, J.; Guła, G.; Bonus, T.; Hanas, A.; Loch, A.; Pawlak, A.; Roszkowiak, J.; Golan, T.; Drulis-Kawa, Z. AGAR a microbial colony dataset for deep learning detection. arXiv 2021, arXiv:2108.01234. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. In Proceedings of the 28th International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; Volume 1, pp. 91–99. [Google Scholar]
- Cai, Z.; Vasconcelos, N. Cascade R-CNN: Delving into High Quality Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 6154–6162. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar] [CrossRef]
- Zhang, B.; Zhou, Z.; Cao, W.; Qi, X.; Xu, C.; Wen, W. A New Few-Shot Learning Method of Bacterial Colony Counting Based on the Edge Computing Device. Biology 2022, 11, 156. [Google Scholar] [CrossRef]
- Qin, X.; Zhang, Z.; Huang, C.; Dehghan, M.; Zaiane, O.R.; Jagersand, M. U2-Net: Going deeper with nested U-structure for salient object detection. Pattern Recognit. 2020, 106, 107404. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Cham, Switzerland, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 936–944. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Nain, G.; Gupta, A. Automatic selection algorithm for region of interest of acne face image compression. Evol. Intell. 2023, 16, 711–717. [Google Scholar] [CrossRef]
- Singhal, P.; Verma, A.; Garg, A. A study in finding effectiveness of Gaussian blur filter over bilateral filter in natural scenes for graph based image segmentation. In Proceedings of the 2017 4th International Conference on Advanced Computing and Communication Systems (ICACCS), Coimbatore, India, 6–7 January 2017; pp. 1–6. [Google Scholar]
- Lewkowycz, A. How to decay your learning rate. arXiv 2021, arXiv:2103.12682. [Google Scholar] [CrossRef]
- Rezaei-Dastjerdehei, M.R.; Mijani, A.; Fatemizadeh, E. Addressing Imbalance in Multi-Label Classification Using Weighted Cross Entropy Loss Function. In Proceedings of the 2020 27th National and 5th International Iranian Conference on Biomedical Engineering (ICBME), Tehran, Iran, 26–27 November 2020; pp. 333–338. [Google Scholar]
- Huchtkoetter, J.; Reinhardt, A. On the Impact of Temporal Data Resolution on the Accuracy of Non-Intrusive Load Monitoring. In Proceedings of the 7th ACM International Conference on Systems for Energy-Efficient Buildings, Cities, and Transportation, Virtual Event, Japan, 18–20 November 2020; pp. 270–273. [Google Scholar]
- Lim, L.A.; Yalim Keles, H. Foreground segmentation using convolutional neural networks for multiscale feature encoding. Pattern Recognit. Lett. 2018, 112, 256–262. [Google Scholar] [CrossRef]
- Viola, P.; Jones, M. Rapid object detection using a boosted cascade of simple features. In Proceedings of the 2001 IEEE Computer Society Conference on Computer Vision and Pattern Recognition. CVPR 2001, Kauai, HI, USA, 8–14 December 2001; pp. I-511–I-518. [Google Scholar]
- Cheng, R.; Crouzier, M.; Hug, F.; Tucker, K.; Juneau, P.; McCreedy, E.; Gandler, W.; McAuliffe, M.J.; Sheehan, F.T. Automatic quadriceps and patellae segmentation of MRI with cascaded U2-Net and SASSNet deep learning model. Med. Phys. 2022, 49, 443–460. [Google Scholar] [CrossRef]
- Graczyk, K.M.; Pawłowski, J.; Majchrowska, S.; Golan, T. Self-normalized density map (SNDM) for counting microbiological objects. Sci. Rep. 2022, 12, 10583. [Google Scholar] [CrossRef]
- Nakao, H.; Magariyama, Y. Simple and rapid method for selective enumeration of lactic acid bacteria in commercially prepared yogurt by image analysis and K-means clustering. Anal. Sci. 2022, 38, 191–197. [Google Scholar] [CrossRef]
- Lin, Y.; Diao, Y.; Du, Y.; Zhang, J.; Li, L.; Liu, P. Automatic cell counting for phase-contrast microscopic images based on a combination of Otsu and watershed segmentation method. Microsc. Res. Tech. 2022, 85, 169–180. [Google Scholar] [CrossRef]
- Zagoruyko, S.; Komodakis, N. Wide Residual Networks. arXiv 2016, arXiv:1605.07146. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2019, arXiv:1905.11946. [Google Scholar] [CrossRef]
- Whipp, J.; Dong, A. YOLO-based Deep Learning to Automated Bacterial Colony Counting. In Proceedings of the 2022 IEEE Eighth International Conference on Multimedia Big Data (BigMM), Naples, Italy, 5–7 December 2022; pp. 120–124. [Google Scholar]
- Geissmann, Q. OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS ONE 2013, 8, e54072. [Google Scholar] [CrossRef] [PubMed]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.-Y.; et al. Segment Anything. arXiv 2023, arXiv:2304.02643. [Google Scholar] [CrossRef]
- Ma, J.; Wang, B. Segment Anything in Medical Images. arXiv 2023, arXiv:2304.12306. [Google Scholar] [CrossRef]



| Predict Class | Ground Truth | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| 0 | 485 | 36 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| 1 | 19 | 3002 | 10 | 3 | 1 | 0 | 1 | 0 | 0 | 0 |
| 2 | 0 | 37 | 458 | 15 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 1 | 4 | 97 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 58 | 6 | 0 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 1 | 3 | 56 | 5 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 71 | 2 | 1 | 0 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 103 | 1 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 61 | 3 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 75 |
| Recall | 96.23% | 97.59% | 97.03% | 83.62% | 89.23% | 90.32% | 87.65% | 98.10% | 96.83% | 96.15% |
| Precision | 92.56% | 98.88% | 89.80% | 95.10% | 90.63% | 86.15% | 95.95% | 95.37% | 95.31% | 99.99% |
| Recovery | 96.23% | 99.90% | 99.36% | 94.54% | 95.38% | 98.06% | 98.77% | 99.73% | 99.40% | 99.57% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, L.; Zeng, L.; Wang, Y.; Cao, J.; Han, Z.; Chen, Y.; Wang, Y.; Zhong, G.; Qiao, S. U2-Net and ResNet50-Based Automatic Pipeline for Bacterial Colony Counting. Microorganisms 2024, 12, 201. https://doi.org/10.3390/microorganisms12010201
Cao L, Zeng L, Wang Y, Cao J, Han Z, Chen Y, Wang Y, Zhong G, Qiao S. U2-Net and ResNet50-Based Automatic Pipeline for Bacterial Colony Counting. Microorganisms. 2024; 12(1):201. https://doi.org/10.3390/microorganisms12010201
Chicago/Turabian StyleCao, Libo, Liping Zeng, Yaoxuan Wang, Jiayi Cao, Ziyu Han, Yang Chen, Yuxi Wang, Guowei Zhong, and Shanlei Qiao. 2024. "U2-Net and ResNet50-Based Automatic Pipeline for Bacterial Colony Counting" Microorganisms 12, no. 1: 201. https://doi.org/10.3390/microorganisms12010201
APA StyleCao, L., Zeng, L., Wang, Y., Cao, J., Han, Z., Chen, Y., Wang, Y., Zhong, G., & Qiao, S. (2024). U2-Net and ResNet50-Based Automatic Pipeline for Bacterial Colony Counting. Microorganisms, 12(1), 201. https://doi.org/10.3390/microorganisms12010201








