Whole Genome Sequencing and Comparative Analysis of the First Ehrlichia canis Isolate in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of E. canis for Sequencing
2.2. Nucleic Acid Purification
2.3. Whole Genome Sequencing and Assembly
2.4. Ehrlichia canis YZ-1 Genome Analysis
2.4.1. Genomic Components Analysis
2.4.2. Genomic Function Annotation
2.5. Macroscopic Comparative Genomic and Phylogenetic Analysis
3. Results
3.1. E. canis YZ-1 Genome Assemblies and Characteristics
3.2. Genomic Functional Analysis
3.3. Similarity between E. canis YZ-1 and Other Ehrlichia Species/Strains
3.3.1. Synteny Comparative Genome Analysis
3.3.2. Insertion–Deletion Mutations Analysis
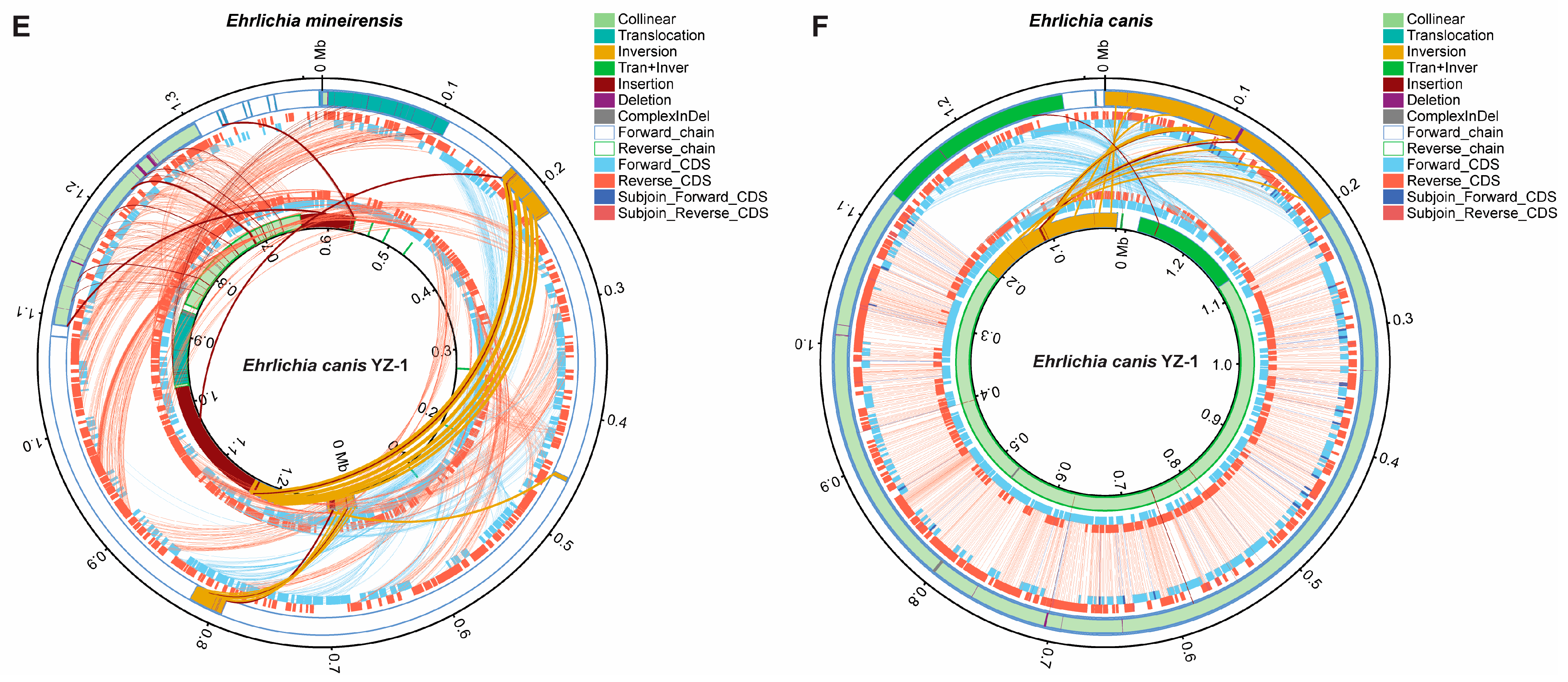
3.3.3. Structural Variation (SV) Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, X.J.; McBride, J.W.; Walker, D.H. Restriction and expansion of Ehrlichia strain diversity. Vet. Parasitol. 2007, 143, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Esemu, S.N.; Ndip, L.M.; Ndip, R.N. Ehrlichia species, probable emerging human pathogens in sub-Saharan Africa: Environmental exacerbation. Rev. Environ. Health 2011, 26, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kelly, P.; Guo, W.; Xu, C.; Wei, L.; Jongejan, F.; Loftis, A.; Wang, C. Development of a generic Ehrlichia FRET-qPCR and investigation of ehrlichioses in domestic ruminants on five Caribbean islands. Parasites Vectors 2015, 8, 506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Kelly, P.J.; Zhang, Y.; Li, M.; Li, J.; Zhang, R.; Wang, Y.; Huang, K.; You, J.; et al. Experimental infection and co-infection with Chinese strains of Ehrlichia canis and Babesia vogeli in intact and splenectomized dogs: Insights on clinical, hematologic and treatment responses. Vet. Parasitol. 2023, 323, 110032. [Google Scholar] [CrossRef] [PubMed]
- Stiles, J. Canine rickettsial infections. Vet. Clin. N. Am. Small Anim. Pract. 2000, 30, 1135–1149. [Google Scholar] [CrossRef]
- Aziz, M.U.; Hussain, S.; Song, B.; Ghauri, H.N.; Zeb, J.; Sparagano, O.A. Ehrlichiosis in Dogs: A Comprehensive Review about the Pathogen and Its Vectors with Emphasis on South and East Asian Countries. Vet. Sci. 2022, 10, 21. [Google Scholar] [CrossRef]
- Harrus, S.; Waner, T. Diagnosis of canine monocytotropic ehrlichiosis (Ehrlichia canis): An overview. Vet. J. 2011, 187, 292–296. [Google Scholar] [CrossRef]
- Conrad, M.E. Ehrlichia canis: A tick-borne rickettsial-like infection in humans living in the southeastern United States. Am. J. Med. Sci. 1989, 297, 35–37. [Google Scholar] [CrossRef]
- Forero-Becerra, E.; Patel, J.; Martínez-Díaz, H.C.; Betancourt-Ruiz, P.; Benavides, E.; Durán, S.; Olaya-Másmela, L.A.; Bolaños, E.; Hidalgo, M.; McBride, J.W. Seroprevalence and Genotypic Analysis of Ehrlichia canis Infection in Dogs and Humans in Cauca, Colombia. Am. J. Trop. Med. Hyg. 2021, 104, 1771–1776. [Google Scholar] [CrossRef]
- Miura, K.; Rikihisa, Y. Virulence potential of Ehrlichia chaffeensis strains of distinct genome sequences. Infect. Immun. 2007, 75, 3604–3613. [Google Scholar] [CrossRef][Green Version]
- Dunning Hotopp, J.C.; Lin, M.; Madupu, R.; Crabtree, J.; Angiuoli, S.V.; Eisen, J.A.; Seshadri, R.; Ren, Q.; Wu, M.; Utterback, T.R.; et al. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006, 2, e21. [Google Scholar] [CrossRef]
- Neave, M.J.; Mileto, P.; Joseph, A.; Reid, T.J.; Scott, A.; Williams, D.T.; Keyburn, A.L. Comparative genomic analysis of the first Ehrlichia canis detections in Australia. Ticks Tick-Borne Dis. 2022, 13, 101909. [Google Scholar] [CrossRef]
- Cheng, C.; Ganta, R.R. Laboratory maintenance of Ehrlichia chaffeensis and Ehrlichia canis and recovery of organisms for molecular biology and proteomics studies. Curr. Protoc. Microbiol. 2008, 9, 3A.1.1–3A.1.21. [Google Scholar] [CrossRef]
- Cabezas-Cruz, A.; Zweygarth, E.; Broniszweska, M.; Passos, L.M.; Ribeiro, M.F.; Manrique, M.; Tobes, R.; de la Fuente, J. Complete Genome Sequence of Ehrlichia mineirensis, a Novel Organism Closely Related to Ehrlichia canis with a New Host Association. Genome Announc. 2015, 3, e01450-14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Wang, C. Complete Genome Sequence of Ehrlichia canis Strain YZ-1, Isolated from a Beagle with Fever and Thrombocytopenia. Genome Announc. 2018, 6, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, G.; Kelly, P.; Zhang, Z.; Wei, L.; Yu, D.; Kayizha, S.; Wang, C. First report of Rickettsia felis in China. BMC Infect. Dis. 2014, 14, 682. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, G.; Li, J.; Kelly, P.; Li, M.; Wang, J.; Huang, K.; Qiu, H.; You, J.; Zhang, R.; et al. Molecular Detection of Rickettsia felis and Rickettsia bellii in Mosquitoes. Vector Borne Zoonotic Dis. 2019, 19, 802–809. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Saha, S.; Bridges, S.; Magbanua, Z.V.; Peterson, D.G. Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 2008, 36, 2284–2294. [Google Scholar] [CrossRef]
- Gardner, P.P.; Daub, J.; Tate, J.G.; Nawrocki, E.P.; Kolbe, D.L.; Lindgreen, S.; Wilkinson, A.C.; Finn, R.D.; Griffiths-Jones, S.; Eddy, S.R.; et al. Rfam: Updates to the RNA families database. Nucleic Acids Res. 2009, 37, D136–D140. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef]
- Magrane, M. UniProt Knowledgebase: A hub of integrated protein data. Database J. Biol. Databases Curation 2011, 2011, bar009. [Google Scholar] [CrossRef]
- Vargas, W.A.; Martín, J.M.; Rech, G.E.; Rivera, L.P.; Benito, E.P.; Díaz-Mínguez, J.M.; Thon, M.R.; Sukno, S.A. Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiol. 2012, 158, 1342–1358. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Jehl, M.A.; Arnold, R.; Rattei, T. Effective—A database of predicted secreted bacterial proteins. Nucleic Acids Res. 2011, 39, D591–D595. [Google Scholar] [CrossRef][Green Version]
- Chen, L.; Xiong, Z.; Sun, L.; Yang, J.; Jin, Q. VFDB 2012 update: Toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2012, 40, D641–D645. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Pop, M. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009, 37, D443–D447. [Google Scholar] [CrossRef]
- Carver, T.; Berriman, M.; Tivey, A.; Patel, C.; Böhme, U.; Barrell, B.G.; Parkhill, J.; Rajandream, M.A. Artemis and ACT: Viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 2008, 24, 2672–2676. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT: Iterative refinement and additional methods. Methods Mol. Biol. 2014, 1079, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009, 25, 119–120. [Google Scholar] [CrossRef]
- de Koning, A.P.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011, 7, e1002384. [Google Scholar] [CrossRef]
- Ehrlich, M.; Gama-Sosa, M.A.; Carreira, L.H.; Ljungdahl, L.G.; Kuo, K.C.; Gehrke, C.W. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 1985, 13, 1399–1412. [Google Scholar] [CrossRef] [PubMed]
- Vanyushin, B.F.; Tkacheva, S.G.; Belozersky, A.N. Rare bases in animal DNA. Nature 1970, 225, 948–949. [Google Scholar] [CrossRef] [PubMed]
- Dunn, D.B.; Smith, J.D. The occurrence of 6-methylaminopurine in deoxyribonucleic acids. Biochem. J. 1958, 68, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A genomic perspective on protein families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; Sahu, J.; Iyer, S.V.; Khamari, L.; De Silva, N.; et al. PHI-base in 2022: A multi-species phenotype database for Pathogen-Host Interactions. Nucleic Acids Res. 2022, 50, D837–D847. [Google Scholar] [CrossRef]
- Cheng, F.; Wu, J.; Fang, L.; Wang, X. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front. Plant Sci. 2012, 3, 198. [Google Scholar] [CrossRef]
- Bekebrede, H.; Lin, M.; Teymournejad, O.; Rikihisa, Y. Discovery of in vivo Virulence Genes of Obligatory Intracellular Bacteria by Random Mutagenesis. Front. Cell Infect. Microbiol. 2020, 10, 2. [Google Scholar] [CrossRef]
- Thirumalapura, N.R.; Qin, X.; Kuriakose, J.A.; Walker, D.H. Complete Genome Sequence of Ehrlichia muris Strain AS145T, a Model Monocytotropic Ehrlichia Strain. Genome Announc. 2014, 2, e01234-13. [Google Scholar] [CrossRef] [PubMed]
- Collins, N.E.; Liebenberg, J.; de Villiers, E.P.; Brayton, K.A.; Louw, E.; Pretorius, A.; Faber, F.E.; van Heerden, H.; Josemans, A.; van Kleef, M.; et al. The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proc. Natl. Acad. Sci. USA 2005, 102, 838–843. [Google Scholar] [CrossRef]
- Cabezas-Cruz, A.; Vancová, M.; Zweygarth, E.; Ribeiro, M.F.; Grubhoffer, L.; Passos, L.M. Ultrastructure of Ehrlichia mineirensis, a new member of the Ehrlichia genus. Vet. Microbiol. 2013, 167, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Mavromatis, K.; Doyle, C.K.; Lykidis, A.; Ivanova, N.; Francino, M.P.; Chain, P.; Shin, M.; Malfatti, S.; Larimer, F.; Copeland, A.; et al. The genome of the obligately intracellular bacterium Ehrlichia canis reveals themes of complex membrane structure and immune evasion strategies. J. Bacteriol. 2006, 188, 4015–4023. [Google Scholar] [CrossRef]
- Sehn, J.K. Chapter 9—Insertions and Deletions (Indels). In Clinical Genomics; Kulkarni, S., Pfeifer, J., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 129–150. [Google Scholar] [CrossRef]
- Sharp, A.J.; Locke, D.P.; McGrath, S.D.; Cheng, Z.; Bailey, J.A.; Vallente, R.U.; Pertz, L.M.; Clark, R.A.; Schwartz, S.; Segraves, R.; et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005, 77, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Freeman, J.L.; Perry, G.H.; Feuk, L.; Redon, R.; McCarroll, S.A.; Altshuler, D.M.; Aburatani, H.; Jones, K.W.; Tyler-Smith, C.; Hurles, M.E.; et al. Copy number variation: New insights in genome diversity. Genome Res. 2006, 16, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, T.T.; Kasahara, M. Visualization tools for human structural variations identified by whole-genome sequencing. J. Hum. Genet. 2020, 65, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Makovets, S.; Titheradge, A.J.; Murray, N.E. ClpX and ClpP are essential for the efficient acquisition of genes specifying type IA and IB restriction systems. Mol. Microbiol. 1998, 28, 25–35. [Google Scholar] [CrossRef]
- Lin, M.; Xiong, Q.; Chung, M.; Daugherty, S.C.; Nagaraj, S.; Sengamalay, N.; Ott, S.; Godinez, A.; Tallon, L.J.; Sadzewicz, L.; et al. Comparative Analysis of Genome of Ehrlichia sp. HF, a Model Bacterium to Study Fatal Human Ehrlichiosis. BMC Genom. 2021, 22, 11. [Google Scholar] [CrossRef]
- Nikolaev, S.; Montoya-Burgos, J.I.; Margulies, E.H.; Rougemont, J.; Nyffeler, B.; Antonarakis, S.E. Early history of mammals is elucidated with the ENCODE multiple species sequencing data. PLoS Genet. 2007, 3, e2. [Google Scholar] [CrossRef][Green Version]
- Younan, M.; Ouso, D.O.; Bodha, B.; Keitany, E.K.; Wesonga, H.O.; Sitawa, R.; Kimutai, J.; Kuria, W.; Sake, W.S.; Svitek, N.; et al. Ehrlichia spp. close to Ehrlichia ruminantium, Ehrlichia canis, and “Candidatus Ehrlichia regneryi” linked to heartwater-like disease in Kenyan camels (Camelus dromedarius). Trop. Anim. Health Prod. 2021, 53, 147. [Google Scholar] [CrossRef]




| Organism Name | GenBank # | No. of Genes | %G + C | No. of Bases | No. of Coding Bases | Coding Bases/Total Bases |
|---|---|---|---|---|---|---|
| Ehrlichia canis YZ-1 | CP025749 | 1022 | 29.00 | 1,314,789 | 956,238 | 72.73 |
| Ehrlichia canis str. Jake | NC_007354 | 985 | 28.96 | 1,315,030 | 959,246 | 72.94 |
| Ehrlichia chaffeensis str. Arkansas | NC_007799 | 1158 | 30.10 | 1,176,248 | 945,019 | 80.34 |
| Ehrlichia spp. HF | NZ_CP007474 | 988 | 29.65 | 1,148,904 | 879,236 | 76.53 |
| Ehrlichia muris AS145 | NC_023063 | 964 | 29.66 | 1,196,717 | 904,005 | 75.54 |
| Ehrlichia ruminantium Welgevonden | NC_005295 | 976 | 27.48 | 1,512,977 | 959,837 | 63.44 |
| Ehrlichia minasensis strain UFMG-EV | NZ_CDGH01000070 | 1119 | 29.89 | 1,366,818 | 982,241 | 71.86 |
| Repeat Type | Number | Total Length (bp) | Percentage in the Genome (%) |
|---|---|---|---|
| Interspersed repeats | |||
| Long-terminal repeats | 34 | 2405 | 0.1829 |
| DNA transposons | 14 | 1339 | 0.1018 |
| Long interspersed nuclear elements | 8 | 808 | 0.0615 |
| Short interspersed nuclear elements | 4 | 225 | 0.0171 |
| Rolling circle | 3 | 288 | 0.0219 |
| Unknown | 2 | 152 | 0.0116 |
| Tandem repeats | |||
| Tandem repeats | 105 | 33,841 | 2.5739 |
| Minisatellite DNA | 52 | 2500 | 0.1901 |
| Microsatellite DNA | 1 | 28 | 0.0021 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Wang, J.; Wang, C. Whole Genome Sequencing and Comparative Analysis of the First Ehrlichia canis Isolate in China. Microorganisms 2024, 12, 125. https://doi.org/10.3390/microorganisms12010125
Zhang J, Wang J, Wang C. Whole Genome Sequencing and Comparative Analysis of the First Ehrlichia canis Isolate in China. Microorganisms. 2024; 12(1):125. https://doi.org/10.3390/microorganisms12010125
Chicago/Turabian StyleZhang, Jilei, Jiawei Wang, and Chengming Wang. 2024. "Whole Genome Sequencing and Comparative Analysis of the First Ehrlichia canis Isolate in China" Microorganisms 12, no. 1: 125. https://doi.org/10.3390/microorganisms12010125
APA StyleZhang, J., Wang, J., & Wang, C. (2024). Whole Genome Sequencing and Comparative Analysis of the First Ehrlichia canis Isolate in China. Microorganisms, 12(1), 125. https://doi.org/10.3390/microorganisms12010125







