An Efficient Triplex TaqMan Quantitative PCR to Detect a Blackleg-Causing Lineage of Pectobacterium brasiliense in Potato Based on a Pangenome Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Strains
2.2. Phenotyping Pectobacterium Strains
2.3. Genome Sequencing
2.4. Pangenome Construction, Annotation, and Analysis
2.5. TaqMan Assay Design
2.6. DNA Extraction for TaqMan Assay
2.7. TaqMan Assay
3. Results
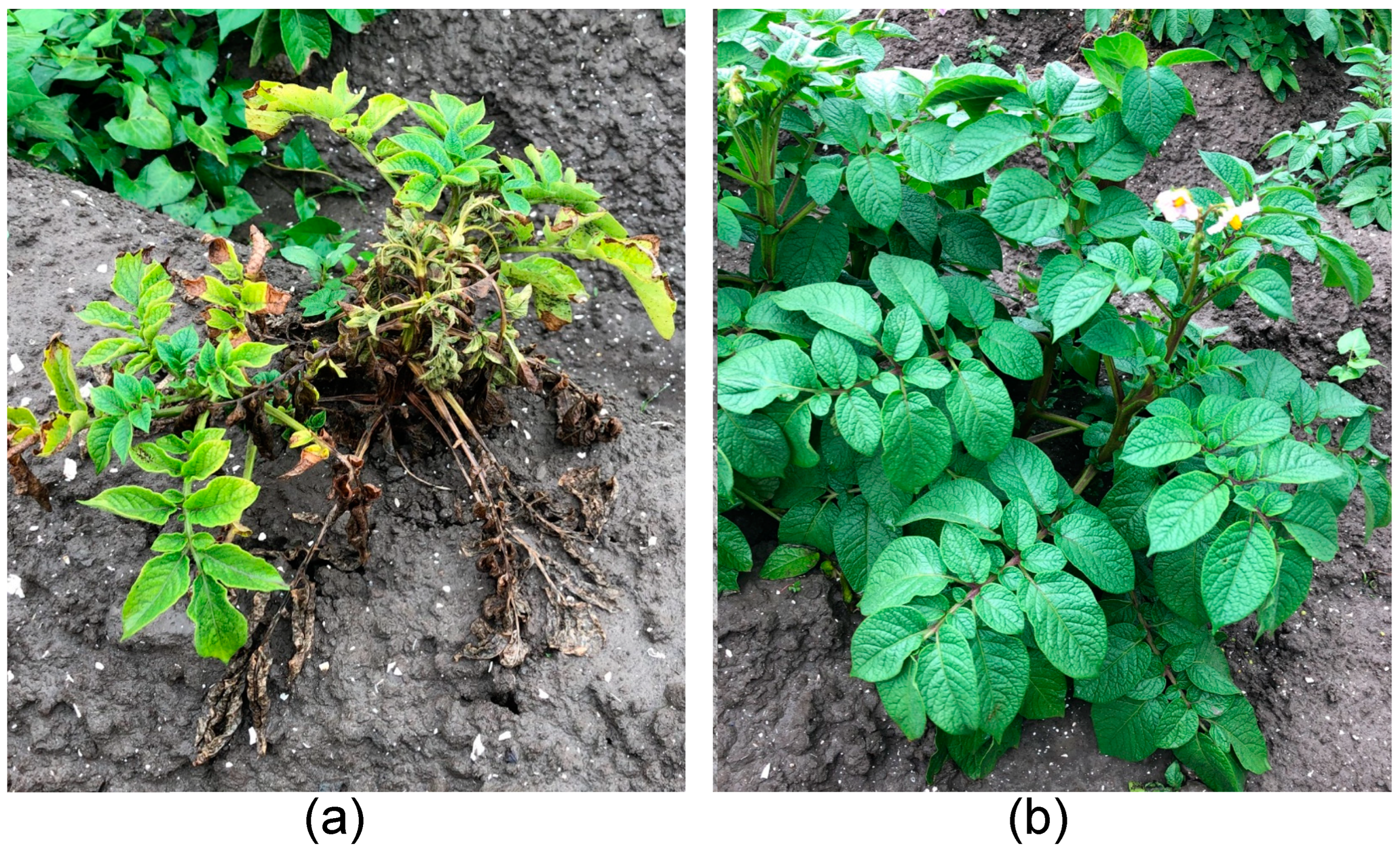
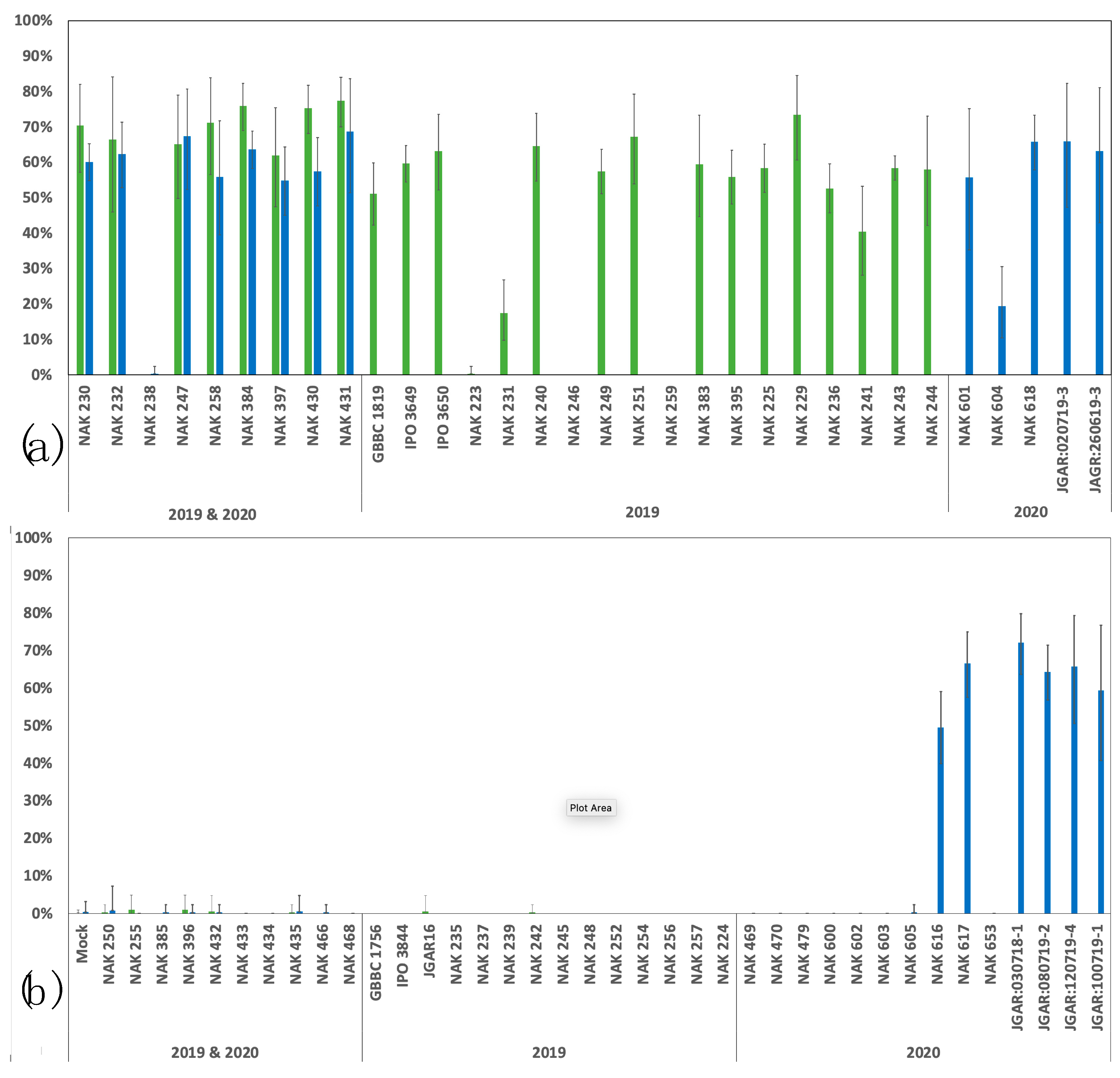
3.1. Blackleg-Phenotype Assays
3.2. Pangenome Analysis
3.3. TaqMan Results
4. Discussion
4.1. Moving from (Sub)Species Detection to Functional Detection Using a Pangenome
4.2. Diligent Gene Selection Based on Both Phylogeny and Function
4.3. Robust Detection of BL-Causing P. brasiliense
4.4. Future Diagnostics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 Plant Pathogenic Bacteria in Molecular Plant Pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Onkendi, E.M.; Moleleki, L.N. Characterization of Pectobacterium carotovorum subsp. Carotovorum and Brasiliense from Diseased Potatoes in Kenya. Eur. J. Plant Pathol. 2014, 139, 557–566. [Google Scholar] [CrossRef]
- Czajkowski, R.; Pérombelon, M.C.M.; van Veen, J.A.; van der Wolf, J.M. Control of Blackleg and Tuber Soft Rot of Potato Caused by Pectobacterium and Dickeya Species: A Review. Plant Pathol. 2011, 60, 999–1013. [Google Scholar] [CrossRef]
- Toth, I.K.; Barny, M.; Brurberg, M.B.; Condemine, G.; Czajkowski, R.; Elphinstone, J.G.; Helias, V.; Johnson, S.B.; Moleleki, L.N.; Pirhonen, M.; et al. Pectobacterium and Dickeya: Environment to Disease Development. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Springer: Cham, Switzerland, 2021. [Google Scholar]
- Pasanen, M.; Waleron, M.; Schott, T.; Cleenwerck, I.; Misztak, A.; Waleron, K.; Pritchard, L.; Bakr, R.; Degefu, Y.; van der Wolf, J.; et al. Pectobacterium parvum sp. nov., Having a Salmonella SPI-1-like Type III Secretion System and Low Virulence. Int. J. Syst. Evol. Microbiol. 2020, 70, 2440–2448. [Google Scholar] [CrossRef] [PubMed]
- Duarte, V.; de Boer, S.H.; Ward, L.J.; de Oliveira, A.M.R. Characterization of Atypical Erwinia carotovora Strains Causing Blackleg of Potato in Brazil. J. Appl. Microbiol. 2004, 96, 535–545. [Google Scholar] [CrossRef]
- Portier, P.; Pédron, J.; Taghouti, G.; Fischer-Le Saux, M.; Caullireau, E.; Bertrand, C.; Laurent, A.; Chawki, K.; Oulgazi, S.; Moumni, M.; et al. Elevation of Pectobacterium carotovorum subsp. Odoriferum to Species Level as Pectobacterium odoriferum sp. nov., Proposal of Pectobacterium brasiliense sp. nov. and Pectobacterium actinidiae sp. nov., Emended Description of Pectobacterium carotovorum and description of Pectobacterium versatile sp. nov., isolated from streams and symptoms on diverse plants. Int. J. Syst. Evol. Microbiol. 2019, 69, 3207–3216. [Google Scholar] [CrossRef]
- Leite, L.N.; Haan, E.G.; Krijger, M.; Kastelein, P.; Zouwen, P.S.; Bovenkamp, G.W.; Tebaldi, N.D.; Wolf, J.M. First Report of Potato Blackleg Caused by Pectobacterium carotovorum subsp. Brasiliensis in the Netherlands. New Dis. Rep. 2014, 29, 24. [Google Scholar] [CrossRef]
- van der Wolf, J.M.; Acuña, I.; de Boer, S.H.; Brurberg, M.B.; Cahill, G.; Charkowski, A.O.; Coutinho, T.; Davey, T.; Dees, M.W.; Degefu, Y.; et al. Diseases Caused by Pectobacterium and Dickeya Species around the World. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Springer Nature: Cham, Switzerland, 2021. [Google Scholar]
- van der Wolf, J.M.; de Haan, E.G.; Kastelein, P.; Krijger, M.; de Haas, B.H.; Velvis, H.; Mendes, O.; Kooman-Gersmann, M.; van der Zouwen, P.S. Virulence of Pectobacterium carotovorum subsp. Brasiliense on Potato Compared with That of Other Pectobacterium and Dickeya Species under Climatic Conditions Prevailing in the Netherlands. Plant Pathol. 2017, 66, 571–583. [Google Scholar] [CrossRef]
- Muzhinji, N.; Dube, J.P.; de Haan, E.G.; Woodhall, J.W.; van der Waals, J.E. Development of a TaqMan PCR Assay for Specific Detection and Quantification of Pectobacterium brasiliense in Potato Tubers and Soil. Eur. J. Plant Pathol. 2020, 158, 521–532. [Google Scholar] [CrossRef]
- Jonkheer, E.M.; Brankovics, B.; Houwers, I.M.; van der Wolf, J.M.; Bonants, P.J.M.; Vreeburg, R.A.M.; Bollema, R.; de Haan, J.R.; Berke, L.; Smit, S.; et al. The Pectobacterium Pangenome, with a Focus on Pectobacterium brasiliense, Shows a Robust Core and Extensive Exchange of Genes from a Shared Gene Pool. BMC Genom. 2021, 22, 265. [Google Scholar] [CrossRef]
- Sheikhizadeh, S.; Schranz, M.E.; Akdel, M.; de Ridder, D.; Smit, S. PanTools: Representation, Storage and Exploration of Pan-Genomic Data. Bioinformatics 2016, 32, i487–i493. [Google Scholar] [CrossRef] [PubMed]
- Jonkheer, E.M.; van Workum, D.-J.M.; Sheikhizadeh Anari, S.; Brankovics, B.; de Haan, J.R.; Berke, L.; van der Lee, T.A.J.; de Ridder, D.; Smit, S. PanTools v3: Functional Annotation, Classification and Phylogenomics. Bioinformatics 2022, 38, 4403–4405. [Google Scholar] [CrossRef] [PubMed]
- Hélias, V.; Hamon, P.; Huchet, E.; Wolf, J.v.d.; Andrivon, D. Two New Effective Semiselective Crystal Violet Pectate Media for Isolation of Pectobacterium and Dickeya. Plant Pathol. 2012, 61, 339–345. [Google Scholar] [CrossRef]
- Sheikhizadeh Anari, S.; de Ridder, D.; Schranz, M.E.; Smit, S. Efficient Inference of Homologs in Large Eukaryotic Pan-Proteomes. BMC Bioinform. 2018, 19, 340. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Sakthivel, M.; Sakthivel, M.; Karthikeyan, N.; Palani, P. Detection and Analysis of Lysozyme Activity in Some Tuberous Plants and Calotropis Procera’s Latex Antibacterial Serine Protease from Wrightia tinctoria: Purification and Characterization. J. Phytol. 2010, 2010, 65–72. [Google Scholar]
- Serrano, C.; Arce-Johnson, P.; Torres, H.; Gebauer, M.; Gutierrez, M.; Moreno, M.; Jordana, X.; Venegas, A.; Kalazich, J.; Holuigue, L. Expression of the Chicken Lysozyme Gene in Potato Enhances Resistance to Infection by Erwinia carotovora subsp. Atroseptica. Am. J. Potato Res. 2000, 77, 191–199. [Google Scholar] [CrossRef]
- Callewaert, L.; van Herreweghe, J.M.; Vanderkelen, L.; Leysen, S.; Voet, A.; Michiels, C.W. Guards of the Great Wall: Bacterial Lysozyme Inhibitors. Trends Microbiol. 2012, 20, 501–510. [Google Scholar] [CrossRef]
- Alaidarous, M.; Ve, T.; Casey, L.W.; Valkov, E.; Ericsson, D.J.; Ullah, M.O.; Schembri, M.A.; Mansell, A.; Sweet, M.J.; Kobe, B. Mechanism of Bacterial Interference with TLR4 Signaling by Brucella Toll/Interleukin-1 Receptor Domain-Containing Protein TcpB. J. Biol. Chem. 2014, 289, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Poncheewin, W.; van Diepeningen, A.D.; van der Lee, T.A.J.; Suarez-Diez, M.; Schaap, P.J. Classification of the Plant-Associated Lifestyle of Pseudomonas Strains Using Genome Properties and Machine Learning. Sci. Rep. 2022, 12, 10857. [Google Scholar] [CrossRef] [PubMed]
- Rasche, F.; Velvis, H.; Zachow, C.; Berg, G.; van Elsas, J.D.; Sessitsch, A. Impact of Transgenic Potatoes Expressing Anti-Bacterial Agents on Bacterial Endophytes Is Comparable with the Effects of Plant Genotype, Soil Type and Pathogen Infection. J. Appl. Ecol. 2006, 43, 555–566. [Google Scholar] [CrossRef]


| Target | Name | Oligo | Sequences (5′–3′) | Reporter Dye | Quencher | Size (bp) |
|---|---|---|---|---|---|---|
| LZI | LZI_F1 | Forw | CGGTAAGTTATGCCGCATCT | |||
| LZI_R1 | Rev | CACTGATCTCTTTCATTTAGCCATATC | ||||
| LZI_P1 | Probe | TGGCATTACAGAATTCATTGCCAAC | FAM | ZEN/IBFQ | ||
| gBlock-LZI | gBlock | ATGAAGAATATCATAAGTAAAAAAACGTTTATATTTTTATC CTTAATGGCATGTAGTACGGTAAGTTATGCCGCATCTTTTG ATTGTGAAAATGGAAATTCTAAGATTGAAAAAATGATTTGT TCAAATTATACTTTGAATAGACTAGATGATTTTCTCTCTGA AAACTATAAGTTGGCAATGAATTCTGTAATGCCAAATGAAG AAAAAAGTGAAATAAAAAATTCTCAGAAGATATGGCTAAAT GAAAGAGATCAGTGTAAAGATGTTAAATGCATTGGGGTATG TATTCGAGGCGTATAG | 303 | |||
| TIR | TIR-F2 | Forw | AGATAAACAAGCGAGGGTTGA | |||
| TIR-R2 | Rev | ATCTATCTCCCATTTCACCCAAG | ||||
| TIR-P2 | Probe | AAATACAGCCTCCATTAGAGTTTCCC | FAM or Yakima Yellow | ZEN/IBFQ | ||
| gBlock-TIR | gBlock | CAAGCGATTTTCTTGTTGAGTTAGAATCAAGAATATCTGAC TTAGGATATTTATACATTGACCTTTTGCATAATAACTCTGA AGATAAACAAGCGAGGGTTGAAAATGAACTTCAACAAGCAG ATATTTTTCTTCTATTAAATACAGCCTCCATTAGAGTTTCC CCTTGGGTGAAATGGGAGATAGATACAGCAAAATCGAATAA CATCTACAATATTAAAATCAACGTAAGTCCCTCAAACATTA ATACTGTCTTTAATGAAATTCGCTTGGCTATTAC | 280 | |||
| Pcb | Pcb1F a | Forw | CCTTACCAAGAAGATGTGTGTTGC | |||
| Pcb2R a | Rev | CATAAACCCGGCACGCT | ||||
| Pcb2Prb a | Probe | CAAGCGCACCTGTTGATGTCATGAGTG | FAM or Cy5 | ZEN/IBFQ or TAO/IBRQ | ||
| gBlock-Pcb_SA-NAK | gBlock | TAGAGGCCTTACCAAGAAGATGTGTGTTGCGTGAAGTGCTC ACACAGATTGTCTGATGAAAATACTGAGCAAGCGCACCTGT TGATGTCATGAGTGTAGACTCATGCTGACGCGAGCGTGCCG GGTTTATGACCTGGTGCGGAT | 144 |
| Strains | Total | Positive LZI/TIR | Negative LZI/TIR | |
|---|---|---|---|---|
| P. brasiliense | ||||
| Causing BL | 34 | 30/30 | 4/4 | |
| Not causing BL | 34 | 6/6 | 28/28 | |
| Sensitivity | 0.83 | |||
| Specificity | 0.88 |
| TaqMan Profile | Sample | Subsample | |
|---|---|---|---|
| Pb− | 30.4% | 53.0% | |
| LZI+, TIR+ | 0.0% 1 | 0.0% 1 | |
| LZI−, TIR− | 100.0% 1 | 100.0% 1 | |
| Pb+ | 69.6% | 47.0% | |
| LZI+, TIR+ | 73.4% 2 | 59.5% 2 | |
| LZI−, TIR− | 26.6% 2 | 40.5% 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van der Lee, T.A.J.; van Gent-Pelzer, M.P.E.; Jonkheer, E.M.; Brankovics, B.; Houwers, I.M.; van der Wolf, J.M.; Bonants, P.J.M.; van Duivenbode, I.; Vreeburg, R.A.M.; Nas, M.; et al. An Efficient Triplex TaqMan Quantitative PCR to Detect a Blackleg-Causing Lineage of Pectobacterium brasiliense in Potato Based on a Pangenome Analysis. Microorganisms 2023, 11, 2080. https://doi.org/10.3390/microorganisms11082080
van der Lee TAJ, van Gent-Pelzer MPE, Jonkheer EM, Brankovics B, Houwers IM, van der Wolf JM, Bonants PJM, van Duivenbode I, Vreeburg RAM, Nas M, et al. An Efficient Triplex TaqMan Quantitative PCR to Detect a Blackleg-Causing Lineage of Pectobacterium brasiliense in Potato Based on a Pangenome Analysis. Microorganisms. 2023; 11(8):2080. https://doi.org/10.3390/microorganisms11082080
Chicago/Turabian Stylevan der Lee, Theo A. J., Marga P. E. van Gent-Pelzer, Eef M. Jonkheer, Balázs Brankovics, Ilse M. Houwers, Jan M. van der Wolf, Peter J. M. Bonants, Inge van Duivenbode, Robert A. M. Vreeburg, Mathijs Nas, and et al. 2023. "An Efficient Triplex TaqMan Quantitative PCR to Detect a Blackleg-Causing Lineage of Pectobacterium brasiliense in Potato Based on a Pangenome Analysis" Microorganisms 11, no. 8: 2080. https://doi.org/10.3390/microorganisms11082080
APA Stylevan der Lee, T. A. J., van Gent-Pelzer, M. P. E., Jonkheer, E. M., Brankovics, B., Houwers, I. M., van der Wolf, J. M., Bonants, P. J. M., van Duivenbode, I., Vreeburg, R. A. M., Nas, M., & Smit, S. (2023). An Efficient Triplex TaqMan Quantitative PCR to Detect a Blackleg-Causing Lineage of Pectobacterium brasiliense in Potato Based on a Pangenome Analysis. Microorganisms, 11(8), 2080. https://doi.org/10.3390/microorganisms11082080







