Endemic High-Risk Clone ST277 Is Related to the Spread of SPM-1-Producing Pseudomonas aeruginosa during the COVID-19 Pandemic Period in Northern Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Phenotypic and Molecular Assays Associated with Antimicrobial Susceptibility and Genetic Variant Definition of blaSPM-1
2.3. Molecular and Phenotypic Detection of Virulence-Related Factors
2.4. Molecular Typing by Multilocus Sequencing Typing–MLST
2.5. Whole-Genome Sequencing (WGS) and Bioinformatics Analysis
2.6. Ethical Considerations
3. Results
3.1. Antimicrobial-Susceptibility-Related Features
3.2. Virulence-Related Features
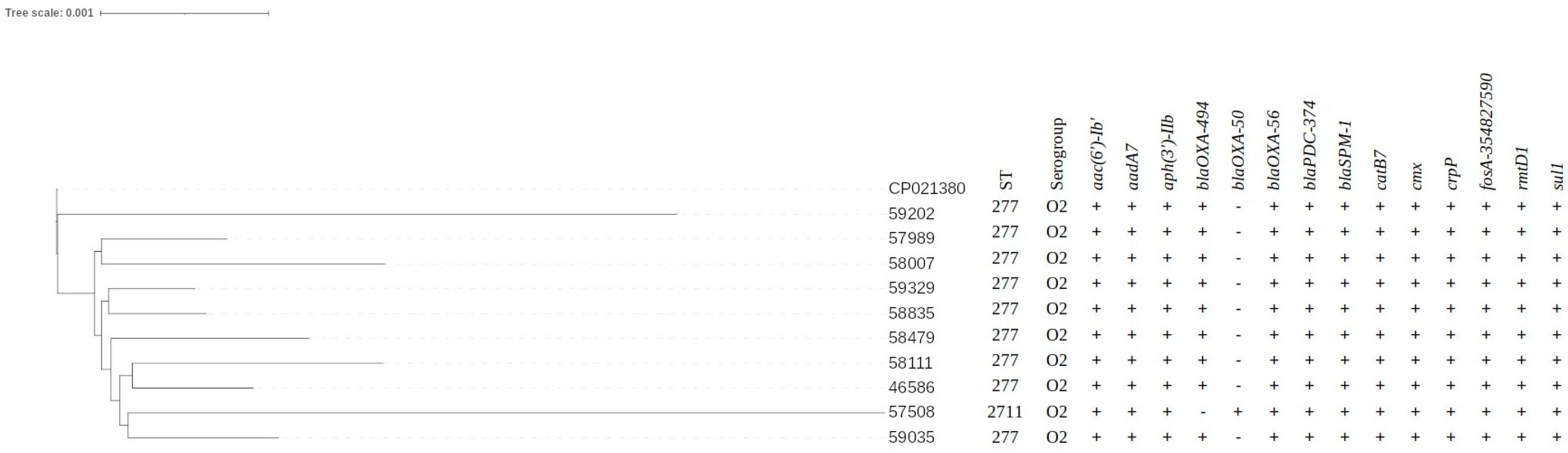
3.3. Genotyping by MLST Data
3.4. WGS Data Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Reynolds, D.; Kollef, M. The Epidemiology and Pathogenesis and Treatment of Pseudomonas aeruginosa Infections: An Update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef]
- Sarges, E.D.S.N.F.; Rodrigues, Y.C.; Furlaneto, I.P.; Melo, M.V.H.D.; Brabo, G.L.D.C.; Lopes, K.C.M.; Quaresma, A.J.P.G.; Lima, L.; Lima, K.V.B. Pseudomonas aeruginosa Type III Secretion System Virulotypes and Their Association with Clinical Features of Cystic Fibrosis Patients. Infect. Drug Resist. 2020, 13, 3771–3781. [Google Scholar] [CrossRef]
- Parkins, M.D.; Somayaji, R.; Waters, V.J. Epidemiology, Biology, and Impact of Clonal Pseudomonas aeruginosa Infections in Cystic Fibrosis. Clin. Microbiol. Rev. 2018, 31, e00019-18. [Google Scholar] [CrossRef]
- Cepas, V.; Soto, S.M. Relationship between Virulence and Resistance among Gram-Negative Bacteria. Antibiotics 2020, 9, 719. [Google Scholar] [CrossRef]
- Horna, G.; Amaro, C.; Palacios, A.; Guerra, H.; Ruiz, J. High frequency of the exoU+/exoS+ genotype associated with multidrug-resistant “high-risk clones” of Pseudomonas aeruginosa clinical isolates from Peruvian hospitals. Sci. Rep. 2019, 9, 10874. [Google Scholar] [CrossRef]
- Kadri, S.S.; Adjemian, J.; Lai, Y.L.; Spaulding, A.B.; Ricotta, E.; Prevots, D.R.; Palmore, T.N.; Rhee, C.; Klompas, M.; Dekker, J.P.; et al. Difficult-to-Treat Resistance in Gram-negative Bacteremia at 173 US Hospitals: Retrospective Cohort Analysis of Prevalence, Predictors, and Outcome of Resistance to All First-line Agents. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018, 67, 1803–1814. [Google Scholar] [CrossRef]
- Riquelme, S.A.; Liimatta, K.; Wong Fok Lung, T.; Fields, B.; Ahn, D.; Chen, D.; Lozano, C.; Sáenz, Y.; Uhlemann, A.-C.; Kahl, B.C.; et al. Pseudomonas aeruginosa Utilizes Host-Derived Itaconate to Redirect Its Metabolism to Promote Biofilm Formation. Cell Metab. 2020, 31, 1091–1106.e6. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Francis, V.I.; Stevenson, E.C.; Porter, S.L. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2017, 364, fnx104. [Google Scholar] [CrossRef]
- Lombardi, C.; Tolchard, J.; Bouillot, S.; Signor, L.; Gebus, C.; Liebl, D.; Fenel, D.; Teulon, J.-M.; Brock, J.; Habenstein, B.; et al. Structural and Functional Characterization of the Type Three Secretion System (T3SS) Needle of Pseudomonas aeruginosa. Front. Microbiol. 2019, 10, 573. [Google Scholar] [CrossRef]
- Sauvage, S.; Hardouin, J. Exoproteomics for Better Understanding Pseudomonas aeruginosa Virulence. Toxins 2020, 12, 571. [Google Scholar] [CrossRef]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Pena, C.; Cabot, G.; Gomez-Zorrilla, S.; Zamorano, L.; Ocampo-Sosa, A.; Murillas, J.; Almirante, B.; Pomar, V.; Aguilar, M.; Granados, A.; et al. Influence of Virulence Genotype and Resistance Profile in the Mortality of Pseudomonas aeruginosa Bloodstream Infections. Clin. Infect. Dis. 2015, 60, 539–548. [Google Scholar] [CrossRef]
- Sawa, T.; Momiyama, K.; Mihara, T.; Kainuma, A.; Kinoshita, M.; Moriyama, K. Molecular epidemiology of clinically high-risk Pseudomonas aeruginosa strains: Practical overview. Microbiol. Immunol. 2020, 64, 331–344. [Google Scholar] [CrossRef]
- Hasannejad-Bibalan, M.; Jafari, A.; Sabati, H.; Goswami, R.; Jafaryparvar, Z.; Sedaghat, F.; Sedigh Ebrahim-Saraie, H. Risk of type III secretion systems in burn patients with Pseudomonas aeruginosa wound infection: A systematic review and meta-analysis. Burns 2021, 47, 538–544. [Google Scholar] [CrossRef]
- Nasrin, S.; Hegerle, N.; Sen, S.; Nkeze, J.; Sen, S.; Permala-Booth, J.; Choi, M.; Sinclair, J.; Tapia, M.D.; Johnson, J.K.; et al. Distribution of serotypes and antibiotic resistance of invasive Pseudomonas aeruginosa in a multi-country collection. BMC Microbiol. 2022, 22, 13. [Google Scholar] [CrossRef]
- Al-Orphaly, M.; Hadi, H.A.; Eltayeb, F.K.; Al-Hail, H.; Samuel, B.G.; Sultan, A.A.; Skariah, S. Epidemiology of Multidrug-Resistant Pseudomonas aeruginosa in the Middle East and North Africa Region. mSphere 2021, 6, e00202-21. [Google Scholar] [CrossRef]
- Tacconelli, E. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development; Infection Control Africa Network: Cape Town, South Africa, 2017. [Google Scholar]
- World Helth Organization. Surveillance of antimicrobial resistance in Europe, 2020 data: Executive summary. In Surveillance of Antimicrobial Resistance in Europe, 2020 Data: Executive Summary; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef]
- Castanheira, M.; Fritsche, T.R.; Sader, H.S.; Jones, R.N. RmtD 16S RNA Methylase in Epidemiologically Unrelated SPM-1-Producing Pseudomonas aeruginosa Isolates from Brazil. Antimicrob. Agents Chemother. 2008, 52, 1587–1588. [Google Scholar] [CrossRef]
- Escandón-Vargas, K.; Reyes, S.; Gutiérrez, S.; Villegas, M.V. The epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev. Anti Infect. Ther. 2017, 15, 277–297. [Google Scholar] [CrossRef]
- Gales, A.C. Dissemination in distinct Brazilian regions of an epidemiccarbapenem-resistant Pseudomonas aeruginosa producing SPM metallo-β-lactamase. J. Antimicrob. Chemother. 2003, 52, 699–702. [Google Scholar] [CrossRef]
- Galetti, R.; Andrade, L.N.; Clímaco, E.C.; Pitondo-Silva, A.; Ferreira, J.C.; Darini, A.L.C. Genomic diversification and virulence features in SPM-1–producing Pseudomonas aeruginosa 13 years later. Diagn. Microbiol. Infect. Dis. 2015, 82, 179–180. [Google Scholar] [CrossRef]
- Tiri, B.; Sensi, E.; Marsiliani, V.; Cantarini, M.; Priante, G.; Vernelli, C.; Martella, L.A.; Costantini, M.; Mariottini, A.; Andreani, P.; et al. Antimicrobial Stewardship Program, COVID-19, and Infection Control: Spread of Carbapenem-Resistant Klebsiella Pneumoniae Colonization in ICU COVID-19 Patients. What Did Not Work? J. Clin. Med. 2020, 9, 2744. [Google Scholar] [CrossRef]
- Hughes, S.; Troise, O.; Donaldson, H.; Mughal, N.; Moore, L.S.P. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020, 26, 1395–1399. [Google Scholar] [CrossRef]
- Fattorini, L.; Creti, R.; Palma, C.; Pantosti, A. Bacterial coinfections in COVID-19: An underestimated adversary. Ann. Ist. Super. Sanita 2020, 56, 359–364. [Google Scholar]
- Dudoignon, E.; Caméléna, F.; Deniau, B.; Habay, A.; Coutrot, M.; Ressaire, Q.; Plaud, B.; Berçot, B.; Dépret, F. Bacterial Pneumonia in COVID-19 Critically Ill Patients: A Case Series. Clin. Infect. Dis. 2021, 72, 905–906. [Google Scholar] [CrossRef]
- Matos, E.C.O.D.; Andriolo, R.B.; Rodrigues, Y.C.; Lima, P.D.L.D.; Carneiro, I.C.D.R.S.; Lima, K.V.B. Mortality in patients with multidrug-resistant Pseudomonas aeruginosa infections: A meta-analysis. Rev. Soc. Bras. Med. Trop. 2018, 51, 415–420. [Google Scholar] [CrossRef]
- Rodrigues, Y.C.; Furlaneto, I.P.; Maciel, A.H.P.; Quaresma, A.J.P.G.; De Matos, E.C.O.; Conceição, M.L.; Vieira, M.C.D.S.; Brabo, G.L.D.C.; Sarges, E.D.S.N.F.; Lima, L.N.G.C.; et al. High prevalence of atypical virulotype and genetically diverse background among Pseudomonas aeruginosa isolates from a referral hospital in the Brazilian Amazon. PLoS ONE 2020, 15, e0238741. [Google Scholar] [CrossRef]
- Miller, R.A. Performance Standards for Antimicrobial Suscepribility Testing of Bacteria Isolated from Aquatic Animals, 3rd ed.; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2020; ISBN 978-1-68440-075-1. [Google Scholar]
- Hudzicki, J. Kirby-Bauer disk diffusion susceptibility test protocol. Am. Soc. Microbiol. 2009, 15, 55–63. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Mulet, X.; Boyd, B.J.; Drummond, C.J. Advances in drug delivery and medical imaging using colloidal lyotropic liquid crystalline dispersions. J. Colloid Interface Sci. 2013, 393, 1–20. [Google Scholar] [CrossRef]
- Mendes, R.E.; Kiyota, K.A.; Monteiro, J.; Castanheira, M.; Andrade, S.S.; Gales, A.C.; Pignatari, A.C.C.; Tufik, S. Rapid Detection and Identification of Metallo-β-Lactamase-Encoding Genes by Multiplex Real-Time PCR Assay and Melt Curve Analysis. J. Clin. Microbiol. 2007, 45, 544–547. [Google Scholar] [CrossRef]
- Curran, B.; Jonas, D.; Grundmann, H.; Pitt, T.; Dowson, C.G. Development of a Multilocus Sequence Typing Scheme for the Opportunistic Pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 2004, 42, 5644–5649. [Google Scholar] [CrossRef]
- Silveira, M.; Albano, R.; Asensi, M.; Assef, A.P.C. The draft genome sequence of multidrug-resistant Pseudomonas aeruginosa strain CCBH4851, a nosocomial isolate belonging to clone SP (ST277) that is prevalent in Brazil. Mem. Inst. Oswaldo Cruz 2014, 109, 1086–1087. [Google Scholar] [CrossRef][Green Version]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Jean, S.-S.; Harnod, D.; Hsueh, P.-R. Global Threat of Carbapenem-Resistant Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2022, 12, 823684. [Google Scholar] [CrossRef]
- Micek, S.T.; Wunderink, R.G.; Kollef, M.H.; Chen, C.; Rello, J.; Chastre, J.; Antonelli, M.; Welte, T.; Clair, B.; Ostermann, H.; et al. An international multicenter retrospective study of Pseudomonas aeruginosa nosocomial pneumonia: Impact of multidrug resistance. Crit. Care 2015, 19, 219. [Google Scholar] [CrossRef]
- Del Barrio-Tofiño, E.; Zamorano, L.; Cortes-Lara, S.; López-Causapé, C.; Sánchez-Diener, I.; Cabot, G.; Bou, G.; Martínez-Martínez, L.; Oliver, A.; GEMARA-SEIMC/REIPI Pseudomonas Study Group; et al. Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology. J. Antimicrob. Chemother. 2019, 74, 1825–1835. [Google Scholar] [CrossRef]
- Giannella, M.; Bussini, L.; Pascale, R.; Bartoletti, M.; Malagrinò, M.; Pancaldi, L.; Toschi, A.; Ferraro, G.; Marconi, L.; Ambretti, S.; et al. Prognostic Utility of the New Definition of Difficult-to-Treat Resistance Among Patients with Gram-Negative Bloodstream Infections. Open Forum Infect. Dis. 2019, 6, ofz505. [Google Scholar] [CrossRef] [PubMed]
- Rawson, T.M.; Moore, L.S.P.; Zhu, N.; Ranganathan, N.; Skolimowska, K.; Gilchrist, M.; Satta, G.; Cooke, G.; Holmes, A. Bacterial and Fungal Coinfection in Individuals with Coronavirus: A Rapid Review to Support COVID-19 Antimicrobial Prescribing. Clin. Infect. Dis. 2020, 71, 2459–2468. [Google Scholar] [CrossRef] [PubMed]
- Al-Hadidi, S.H.; Alhussain, H.; Abdel Hadi, H.; Johar, A.; Yassine, H.M.; Al Thani, A.A.; Eltai, N.O. The Spectrum of Antibiotic Prescribing During COVID-19 Pandemic: A Systematic Literature Review. Microb. Drug Resist. 2021, 27, 1705–1725. [Google Scholar] [CrossRef] [PubMed]
- Plan Nacional Para la Prevención y el Control de la Resistencia Microbiana en Los Servicios de Salud—Agência Nacional de Vigilância Sanitária—Anvisa. Available online: https://www.gov.br/anvisa/pt-br/centraisdeconteudo/publicacoes/servicosdesaude/publicacoes/plan-nacional-para-la-prevencion-y-el-control-de-la-resistencia-microbiana-en-los-servicios-de-salud/view (accessed on 9 June 2023).
- Microsoft Power BI. Available online: https://app.powerbi.com/view?r=eyJrIjoiODkzMzNiYmQtYWRkYi00NzRmLWI1ZGQtYjI5NGEzNjk1YTE0IiwidCI6ImI2N2FmMjNmLWMzZjMtNGQzNS04MGM3LWI3MDg1ZjVlZGQ4MSJ9 (accessed on 13 June 2023).
- Microsoft Power BI. Available online: https://app.powerbi.com/view?r=eyJrIjoiZjQ5ZDhjZmEtNDdhOC00MDk3LWFiNDEtNzg0MmE4MmE2MjlhIiwidCI6ImI2N2FmMjNmLWMzZjMtNGQzNS04MGM3LWI3MDg1ZjVlZGQ4MSJ9&pageName=ReportSectionac5c0437dbe709793b4b (accessed on 13 June 2023).
- Microsoft Power BI. Available online: https://app.powerbi.com/view?r=eyJrIjoiNGUxYWVjOGUtODBmYy00MzJkLWE1MDEtNWVlYTNmN2Y0ODdhIiwidCI6ImI2N2FmMjNmLWMzZjMtNGQzNS04MGM3LWI3MDg1ZjVlZGQ4MSJ9 (accessed on 13 June 2023).
- Microsoft Power BI. Available online: https://app.powerbi.com/view?r=eyJrIjoiZDIwZjYyMzUtMmYxZS00MTRjLTk0NWMtZWE2ZDUzOGRjOTVjIiwidCI6ImI2N2FmMjNmLWMzZjMtNGQzNS04MGM3LWI3MDg1ZjVlZGQ4MSJ9 (accessed on 13 June 2023).
- Mauri, C.; Maraolo, A.E.; Di Bella, S.; Luzzaro, F.; Principe, L. The Revival of Aztreonam in Combination with Avibactam against Metallo-β-Lactamase-Producing Gram-Negatives: A Systematic Review of In Vitro Studies and Clinical Cases. Antibiotics 2021, 10, 1012. [Google Scholar] [CrossRef]
- Shields, R.K.; Doi, Y. Aztreonam Combination Therapy: An Answer to Metallo-β-Lactamase–Producing Gram-Negative Bacteria? Clin. Infect. Dis. 2020, 71, 1099–1101. [Google Scholar] [CrossRef]
- Davido, B.; Fellous, L.; Lawrence, C.; Maxime, V.; Rottman, M.; Dinh, A. Ceftazidime-avibactam and aztreonam, an interesting strategy to overcome β-lactam resistance conferred by metallo-β-lactamases in Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2017, 61, e01008-17. [Google Scholar] [CrossRef] [PubMed]
- Rojo-Molinero, E.; Macià, M.D.; Rubio, R.; Moyà, B.; Cabot, G.; López-Causapé, C.; Pérez, J.L.; Cantón, R.; Oliver, A. Sequential treatment of biofilms with aztreonam and tobramycin is a novel strategy for combating Pseudomonas aeruginosa chronic respiratory infections. Antimicrob. Agents Chemother. 2016, 60, 2912–2922. [Google Scholar] [CrossRef] [PubMed]
- Jorth, P.; McLean, K.; Ratjen, A.; Secor, P.R.; Bautista, G.E.; Ravishankar, S.; Rezayat, A.; Garudathri, J.; Harrison, J.J.; Harwood, R.A.; et al. Evolved Aztreonam Resistance Is Multifactorial and Can Produce Hypervirulence in Pseudomonas aeruginosa. MBio 2017, 8, e00517-17. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Xu, C.; Zhang, X.; Wang, D.; Pan, X.; Liu, H.; Zhu, G.; Bai, F.; Cheng, Z.; Wu, W.; et al. A MexR Mutation Which Confers Aztreonam Resistance to Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 659808. [Google Scholar] [CrossRef]
- Girlich, D.; Naas, T.; Nordmann, P. Biochemical Characterization of the Naturally Occurring Oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2004, 48, 2043–2048. [Google Scholar] [CrossRef]
- Kong, K.-F.; Jayawardena, S.R.; Del Puerto, A.; Wiehlmann, L.; Laabs, U.; Tümmler, B.; Mathee, K. Characterization of poxB, a chromosomal-encoded Pseudomonas aeruginosa oxacillinase. Gene 2005, 358, 82–92. [Google Scholar] [CrossRef]
- Nicolau, C.J.; Oliver, A. Carbapenemasas en especies del género Pseudomonas. Enfermedades Infecc. Microbiol. Clin. 2010, 28, 19–28. [Google Scholar] [CrossRef]
- Streling, A.P.; Cayô, R.; Nodari, C.S.; Almeida, L.G.P.; Bronze, F.; Siqueira, A.V.; Matos, A.P.; Oliveira, V.; Vasconcelos, A.T.R.; Marcondes, M.F.M.; et al. Kinetics Analysis of β-Lactams Hydrolysis by OXA-50 Variants of Pseudomonas aeruginosa. Microb. Drug Resist. 2022, 28, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Pseudomonas aeruginosa: Resistance to the Max. Front. Microbiol. 2011, 2, 65. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Deshpande, L.M.; Woosley, L.N.; Serio, A.W.; Krause, K.M.; Flamm, R.K. Activity of plazomicin compared with other aminoglycosides against isolates from European and adjacent countries, including Enterobacteriaceae molecularly characterized for aminoglycoside-modifying enzymes and other resistance mechanisms. J. Antimicrob. Chemother. 2018, 73, 3346–3354. [Google Scholar] [CrossRef] [PubMed]
- Ghssein, G.; Ezzeddine, Z. A Review of Pseudomonas aeruginosa Metallophores: Pyoverdine, Pyochelin and Pseudopaline. Biology 2022, 11, 1711. [Google Scholar] [CrossRef]
- Fothergill, J.L.; Panagea, S.; Hart, C.A.; Walshaw, M.J.; Pitt, T.L.; Winstanley, C. Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. BMC Microbiol. 2007, 7, 45. [Google Scholar] [CrossRef]
- Sistema Nou-Rau: Biblioteca Digital da UEL. Available online: http://www.bibliotecadigital.uel.br/document/?code=vtls000151945 (accessed on 9 June 2023).
- Silva, S.T. Análise Fenotípica e Genética de Fatores Virulência de Isolados Clínicos de Pseudomonas aeruginosa Multidroga-sensível e Multidroga-Resistente de Recife-PE. Master’s Thesis, Universidade Federal de Pernambuco, Recife, Brazil, 2016. [Google Scholar]
- Adonizio, A.; Kong, K.-F.; Mathee, K. Inhibition of Quorum Sensing-Controlled Virulence Factor Production in Pseudomonas aeruginosa by South Florida Plant Extracts. Antimicrob. Agents Chemother. 2008, 52, 198–203. [Google Scholar] [CrossRef]
- Ben Haj Khalifa, A.; Moissenet, D.; Vu Thien, H.; Khedher, M. Virulence factors in Pseudomonas aeruginosa: Mechanisms and modes of regulation. Ann. Biol. Clin. 2011, 69, 393–403. [Google Scholar] [CrossRef]
- Oliver, A.; Mulet, X.; López-Causapé, C.; Juan, C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Updat. 2015, 21–22, 41–59. [Google Scholar] [CrossRef]
- Yousefi-Avarvand, A.; Khashei, R.; Sedigh Ebrahim-Saraie, H.; Emami, A.; Zomorodian, K.; Motamedifar, M. The Frequency of Exotoxin A and Exoenzymes S and U Genes Among Clinical Isolates of Pseudomonas aeruginosa in Shiraz, Iran. Int. J. Mol. Cell. Med. 2015, 4, 167–173. [Google Scholar] [PubMed]
- Sharma, P.; Faridi, F.; Mir, I.A.; Sharma, S.K. Characterization of exo-s, exo-u, and alg virulence factors and antimicrobial resistance in Pseudomonas aeruginosa isolated from migratory Egyptian vultures from India. Infect. Ecol. Epidemiol. 2014, 4, 24553. [Google Scholar] [CrossRef]
- Kiyaga, S.; Kyany’a, C.; Muraya, A.W.; Smith, H.J.; Mills, E.G.; Kibet, C.; Mboowa, G.; Musila, L. Genetic Diversity, Distribution, and Genomic Characterization of Antibiotic Resistance and Virulence of Clinical Pseudomonas aeruginosa Strains in Kenya. Front. Microbiol. 2022, 13, 835403. [Google Scholar] [CrossRef] [PubMed]
- Stanislavsky, E.S.; Lam, J.S. Pseudomonas aeruginosa antigens as potential vaccines. FEMS Microbiol. Rev. 1997, 21, 243–277. [Google Scholar] [CrossRef] [PubMed]
- Donta, S.T.; Peduzzi, P.; Cross, A.S.; Sadoff, J.; Haakenson, C.; Cryz, S.J.; Kauffman, C.; Bradley, S.; Gafford, G.; Elliston, D.; et al. Immunoprophylaxis against Klebsiella and Pseudomonas aeruginosa Infections. J. Infect. Dis. 1996, 174, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Azimi, A.; Peymani, A.; Pour, P.K. Phenotypic and molecular detection of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates from patients with burns in Tehran, Iran. Rev. Soc. Bras. Med. Trop. 2018, 51, 610–615. [Google Scholar] [CrossRef]
- Ghamgosha, M.; Shahrekizahedani, S.; Kafilzadeh, F.; Bameri, Z.; Taheri, R.A.; Farnoosh, G. Metallo-beta-Lactamase VIM-1, SPM-1, and IMP-1 Genes Among Clinical Pseudomonas aeruginosa Species Isolated in Zahedan, Iran. Jundishapur J. Microbiol. 2015, 8, e17489. [Google Scholar] [CrossRef]
- Hopkins, K.L.; Meunier, D.; Findlay, J.; Mustafa, N.; Parsons, H.; Pike, R.; Wright, L.; Woodford, N. SPM-1 metallo-β-lactamase-producing Pseudomonas aeruginosa ST277 in the UK. J. Med. Microbiol. 2016, 65, 696–697. [Google Scholar] [CrossRef]
- Wozniak, A.; Rodríguez, N.; Alcalde-Rico, M.; Castillo, C.; García, P. Primer aislado de Pseudomonas aeruginosa productora de Sao Paulo metalo-β-lactamasa (SPM-1) en un paciente chileno. Rev. Chil. Infectol. 2021, 38, 724–726. [Google Scholar] [CrossRef]
- Abdelaziz, N.A. Phenotype-genotype correlations among carbapenem-resistant Enterobacterales recovered from four Egyptian hospitals with the report of SPM carbapenemase. Antimicrob. Resist. Infect. Control 2022, 11, 13. [Google Scholar] [CrossRef]
- Fehlberg, L.C.C. Estudo Comparativo dos Mecanismos de Resistência Aos β-lactâmicos em Amostras Clínicas de Pseudomonas aeruginosa Isoladas de Infecção de Corrente Sanguínea no Brasil e Nos Estados Unidos da América. Ph.D. Thesis, Universidade Federal de São Paulo, São Paulo, Brazil, 2010. [Google Scholar]
- Almarzoky Abuhussain, S.S.; Sutherland, C.A.; Nicolau, D.P. In vitro potency of antipseudomonal β-lactams against blood and respiratory isolates of P. aeruginosa collected from US hospitals. J. Thorac. Dis. 2019, 11, 1896–1902. [Google Scholar] [CrossRef]
- Hong, D.J.; Bae, I.K.; Jang, I.-H.; Jeong, S.H.; Kang, H.-K.; Lee, K. Epidemiology and Characteristics of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa. Infect. Chemother. 2015, 47, 81. [Google Scholar] [CrossRef]
- Cacci, L.C.; Chuster, S.G.; Martins, N.; Carmo, P.R.D.; Girão, V.B.D.C.; Nouér, S.A.; Freitas, W.V.D.; Matos, J.A.D.; Magalhães, A.C.D.G.; Ferreira, A.L.P.; et al. Mechanisms of carbapenem resistance in endemic Pseudomonas aeruginosa isolates after an SPM-1 metallo-β-lactamase producing strain subsided in an intensive care unit of a teaching hospital in Brazil. Mem. Inst. Oswaldo Cruz 2016, 111, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Kalluf, K.O.; Arend, L.N.; Wuicik, T.E.; Pilonetto, M.; Tuon, F.F. Molecular epidemiology of SPM-1-producing Pseudomonas aeruginosa by rep-PCR in hospitals in Parana, Brazil. Infect. Genet. Evol. 2017, 49, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.F.; Zavascki, A.P.; Gaspareto, P.B.; Barth, A.L. Dissemination of Pseudomonas aeruginosa Producing SPM-1-like and IMP-1-like Metallo-β-lactamases in Hospitals from Southern Brazil. Infection 2007, 35, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Cezário, R.C.; Duarte De Morais, L.; Ferreira, J.C.; Costa-Pinto, R.M.; Da Costa Darini, A.L.; Gontijo-Filho, P.P. Nosocomial outbreak by imipenem-resistant metallo-β-lactamase-producing Pseudomonas aeruginosa in an adult intensive care unit in a Brazilian teaching hospital. Enfermedades Infecc. Microbiol. Clínica 2009, 27, 269–274. [Google Scholar] [CrossRef]
- Davis, M.W.; McManus, D.; Koff, A.; Jaszczur, G.R.; Malinis, M.; Dela Cruz, C.; Britto, C.J.; Price, C.; Azmy, V.; Kaman, K.; et al. Repurposing antimicrobial stewardship tools in the electronic medical record for the management of COVID-19 patients. Infect. Control Hosp. Epidemiol. 2020, 41, 1335–1337. [Google Scholar] [CrossRef]
- Daikos, G.L.; Da Cunha, C.A.; Rossolini, G.M.; Stone, G.G.; Baillon-Plot, N.; Tawadrous, M.; Irani, P. Review of Ceftazidime-Avibactam for the Treatment of Infections Caused by Pseudomonas aeruginosa. Antibiotics 2021, 10, 1126. [Google Scholar] [CrossRef]
- García-Betancur, J.C.; Appel, T.M.; Esparza, G.; Gales, A.C.; Levy-Hara, G.; Cornistein, W.; Vega, S.; Nuñez, D.; Cuellar, L.; Bavestrello, L.; et al. Update on the epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev. Anti Infect. Ther. 2021, 19, 197–213. [Google Scholar] [CrossRef]
- Sader, H.S.; Castanheira, M.; Arends, S.J.R.; Goossens, H.; Flamm, R.K. Geographical and temporal variation in the frequency and antimicrobial susceptibility of bacteria isolated from patients hospitalized with bacterial pneumonia: Results from 20 years of the SENTRY Antimicrobial Surveillance Program (1997–2016). J. Antimicrob. Chemother. 2019, 74, 1595–1606. [Google Scholar] [CrossRef]
- Cox, J.A.; Vlieghe, E.; Mendelson, M.; Wertheim, H.; Ndegwa, L.; Villegas, M.V.; Gould, I.; Levy Hara, G. Antibiotic stewardship in low- and middle-income countries: The same but different? Clin. Microbiol. Infect. 2017, 23, 812–818. [Google Scholar] [CrossRef]
- Garcia, P.J.; Alarcón, A.; Bayer, A.; Buss, P.; Guerra, G.; Ribeiro, H.; Rojas, K.; Saenz, R.; Salgado De Snyder, N.; Solimano, G.; et al. COVID-19 Response in Latin America. Am. J. Trop. Med. Hyg. 2020, 103, 1765–1772. [Google Scholar] [CrossRef]
- Mojica, M.F.; Rossi, M.-A.; Vila, A.J.; Bonomo, R.A. The urgent need for metallo-β-lactamase inhibitors: An unattended global threat. Lancet Infect. Dis. 2022, 22, e28–e34. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.M.; Carmo, M.S.; Silbert, S.; Gales, A.C. SPM-1-Producing Pseudomonas aeruginosa: Analysis of the Ancestor Relationship Using Multilocus Sequence Typing, Pulsed-Field Gel Electrophoresis, and Automated Ribotyping. Microb. Drug Resist. 2011, 17, 215–220. [Google Scholar] [CrossRef]
- De Oliveira Santos, I.C.; Pereira De Andrade, N.F.; Da Conceição Neto, O.C.; Da Costa, B.S.; De Andrade Marques, E.; Rocha-de-Souza, C.M.; Asensi, M.D.; D’Alincourt Carvalho-Assef, A.P. Epidemiology and antibiotic resistance trends in clinical isolates of Pseudomonas aeruginosa from Rio de janeiro—Brazil: Importance of mutational mechanisms over the years (1995–2015). Infect. Genet. Evol. 2019, 73, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Martins, W.M.B.S.; Narciso, A.C.; Cayô, R.; Santos, S.V.; Fehlberg, L.C.C.; Ramos, P.L.; Da Cruz, J.B.; Gales, A.C. SPM-1-producing Pseudomonas aeruginosa ST277 clone recovered from microbiota of migratory birds. Diagn. Microbiol. Infect. Dis. 2018, 90, 221–227. [Google Scholar] [CrossRef] [PubMed]
| ID | PRL | TZP | TTC | CAZ | FEP | ATM | IMP | GEN | TOB | AMK | CIP | OFX | Susceptibility Phenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 46586 | I | I | I | R | R | R | R | R | R | R | R | R | XDR # |
| 46716 | I | S | R | R | R | S | R | I | S | S | R | R | MDR |
| 54178 | S | S | I | R | R | S | R | R | R | R | R | R | MDR |
| 56158 | I | R | I | R | R | S | R | R | R | R | R | R | MDR |
| 56572 | R | S | S | S | S | R | R | S | R | S | S | R | MDR |
| 57415 | I | S | S | S | S | S | R | S | S | R | R | R | MDR |
| 57508 | R | S | I | I | S | I | R | R | S | S | S | R | XDR |
| 57564 | I | I | R | R | R | S | R | R | R | R | R | R | MDR |
| 57568 | R | R | R | R | R | I | R | R | R | R | R | R | XDR # |
| 57654 | R | I | R | R | R | S | R | R | R | R | R | R | MDR |
| 57716 | I | R | R | R | R | S | R | R | R | R | R | R | MDR |
| 57729 | R | I | R | R | R | S | R | R | R | R | R | R | MDR |
| 57863 | R | I | R | R | R | S | R | R | R | R | R | R | MDR |
| 57877 | I | S | R | R | R | I | R | R | R | R | R | R | XDR # |
| 57884 | R | I | R | R | R | S | R | R | R | R | R | R | MDR |
| 57989 | R | I | R | R | R | I | R | R | R | R | R | R | XDR # |
| 58005 | R | I | R | R | R | S | R | R | R | R | R | R | MDR |
| 58007 | I | I | R | R | R | I | R | R | R | R | R | R | XDR # |
| 58111 | R | I | R | R | R | I | R | R | R | R | R | R | XDR # |
| 58218 | I | I | R | R | R | S | R | R | R | R | R | R | MDR |
| 58276 | R | I | R | R | R | S | R | R | R | R | R | R | MDR |
| 58479 | R | S | R | R | R | R | R | R | R | R | R | R | XDR # |
| 58482 | I | I | R | R | R | S | R | R | S | S | R | R | MDR |
| 58608 | R | R | R | R | R | S | R | R | R | R | R | R | MDR |
| 58739 | S | R | R | S | R | S | R | R | S | R | S | S | MDR |
| 58798 | R | R | R | R | R | S | R | R | R | R | R | R | MDR |
| 58820 | I | I | R | R | R | S | R | R | R | R | R | R | MDR |
| 58835 | I | S | R | R | R | R | R | R | R | R | R | R | XDR # |
| 58924 | R | I | R | R | R | S | R | R | R | R | R | R | MDR |
| 59035 | R | I | R | R | R | R | R | R | R | R | R | R | XDR # |
| 59183 | R | I | R | R | R | S | R | R | R | R | R | R | MDR |
| 59202 | R | I | R | R | R | R | R | R | R | R | R | R | XDR # |
| 59233 | I | S | R | R | R | S | R | R | R | R | R | R | MDR |
| 59329 | R | I | R | R | R | R | R | R | R | R | R | R | XDR # |
| ID | Date at LabPate/IEC | Biological Source | Origin | Resistance Phenotype | ST | blaSPM−1 | blaIMP | blaVIM | blaNDM | blaKPC | blaOXA−48 | aprA | lasA | lasB | toxA | exoS | exoU | exoT | exoY | Mucoid | Pyoverdine | Pyocyanine |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 46586 | 31 July 2018 | Urine | PI/AC | XDR | 277 * | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + |
| 46716 | 8 August 2018 | Urine | PI/PA | MDR | 277 | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − |
| 54178 | 6 November 2019 | TS | PI/AC | MDR | 277 | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − |
| 56158 | 27 March 2020 | TS | PI/AC | MDR | 277 | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + |
| 56572 | 21 May 2020 | TS | PI/AC | MDR | 277 | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + |
| 57415 | 10 February 2021 | Urine | PR/PA | MDR | 277 | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − |
| 57508 | 3 March 2021 | TS | PI/PA | XDR | 2711 *§ | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + |
| 57564 | 22 March 2021 | Urine | PR/PA | MDR | 277 | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + |
| 57568 | 22 March 2021 | TS | PR/PA | XDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + | |
| 57654 | 6 April 2021 | TS | PR/PA | MDR | 277 | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − |
| 57716 | 6 May 2021 | TS | NI | MDR | 277 | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − |
| 57729 | 6 May 2021 | TS | NI | MDR | 277 | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + |
| 57863 | 24 May 2021 | Urine | NI | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − | |
| 57877 | 24 May 2021 | Blood | NI | XDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − | |
| 57884 | 24 May 2021 | TS | NI | MDR | + | − | − | − | − | − | + | + | − | + | + | − | + | + | − | − | − | |
| 57989 | 16 June 2021 | Blood | NI | XDR | 277 * | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − |
| 58005 | 16 June 2021 | CT | PI/PA | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − | |
| 58007 | 16 June 2021 | WS | NI | XDR | 277 * | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + |
| 58111 | 30 June 2021 | TS | NI | XDR | 277 * | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − |
| 58218 | 15 July 2021 | Urine | PI/PA | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − | |
| 58276 | 27 July 2021 | Liquor | PI/PA | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − | |
| 58479 | 20 August 2021 | Urine | NI | XDR | 277 * | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + |
| 58482 | 20 August 2021 | Blood | PR/PA | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − | |
| 58608 | 10 September 2021 | Urine | PI/PA | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + | |
| 58739 | 28 September 2021 | MT | PI/PA | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + | |
| 58798 | 14 October 2021 | TS | PR/PA | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − | |
| 58820 | 14 October 2021 | WS | PR/PA | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − | |
| 58835 | 14 October 2021 | IS | PR/PA | XDR | 277 * | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + |
| 58924 | 28 October 2021 | Urine | PI/PA | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + | |
| 59035 | 18 November 2021 | Blood | PI/PA | XDR | 277 * | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + |
| 59183 | 10 December 2021 | Urine | PI/PA | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − | |
| 59202 | 10 December 2021 | TS | PI/PA | XDR | 277 * | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − |
| 59233 | 16 December 2021 | TS | PI/PA | MDR | + | − | − | − | − | − | + | + | + | + | + | − | + | + | − | − | − | |
| 59329 | 5 January 2022 | Urine | PR/PA | XDR | 277 * | + | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dos Santos, P.A.S.; Rodrigues, Y.C.; Marcon, D.J.; Lobato, A.R.F.; Cazuza, T.B.; Gouveia, M.I.M.; Silva, M.J.A.; Souza, A.B.; Lima, L.N.G.C.; Quaresma, A.J.P.G.; et al. Endemic High-Risk Clone ST277 Is Related to the Spread of SPM-1-Producing Pseudomonas aeruginosa during the COVID-19 Pandemic Period in Northern Brazil. Microorganisms 2023, 11, 2069. https://doi.org/10.3390/microorganisms11082069
Dos Santos PAS, Rodrigues YC, Marcon DJ, Lobato ARF, Cazuza TB, Gouveia MIM, Silva MJA, Souza AB, Lima LNGC, Quaresma AJPG, et al. Endemic High-Risk Clone ST277 Is Related to the Spread of SPM-1-Producing Pseudomonas aeruginosa during the COVID-19 Pandemic Period in Northern Brazil. Microorganisms. 2023; 11(8):2069. https://doi.org/10.3390/microorganisms11082069
Chicago/Turabian StyleDos Santos, Pabllo Antonny Silva, Yan Corrêa Rodrigues, Davi Josué Marcon, Amália Raiana Fonseca Lobato, Thalyta Braga Cazuza, Maria Isabel Montoril Gouveia, Marcos Jessé Abrahão Silva, Alex Brito Souza, Luana Nepomuceno Gondim Costa Lima, Ana Judith Pires Garcia Quaresma, and et al. 2023. "Endemic High-Risk Clone ST277 Is Related to the Spread of SPM-1-Producing Pseudomonas aeruginosa during the COVID-19 Pandemic Period in Northern Brazil" Microorganisms 11, no. 8: 2069. https://doi.org/10.3390/microorganisms11082069
APA StyleDos Santos, P. A. S., Rodrigues, Y. C., Marcon, D. J., Lobato, A. R. F., Cazuza, T. B., Gouveia, M. I. M., Silva, M. J. A., Souza, A. B., Lima, L. N. G. C., Quaresma, A. J. P. G., Brasiliense, D. M., & Lima, K. V. B. (2023). Endemic High-Risk Clone ST277 Is Related to the Spread of SPM-1-Producing Pseudomonas aeruginosa during the COVID-19 Pandemic Period in Northern Brazil. Microorganisms, 11(8), 2069. https://doi.org/10.3390/microorganisms11082069











