Gut Microbial and Associated Metabolite Markers for Colorectal Cancer Diagnosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
2.2. Data Extraction
2.3. Methodological Quality
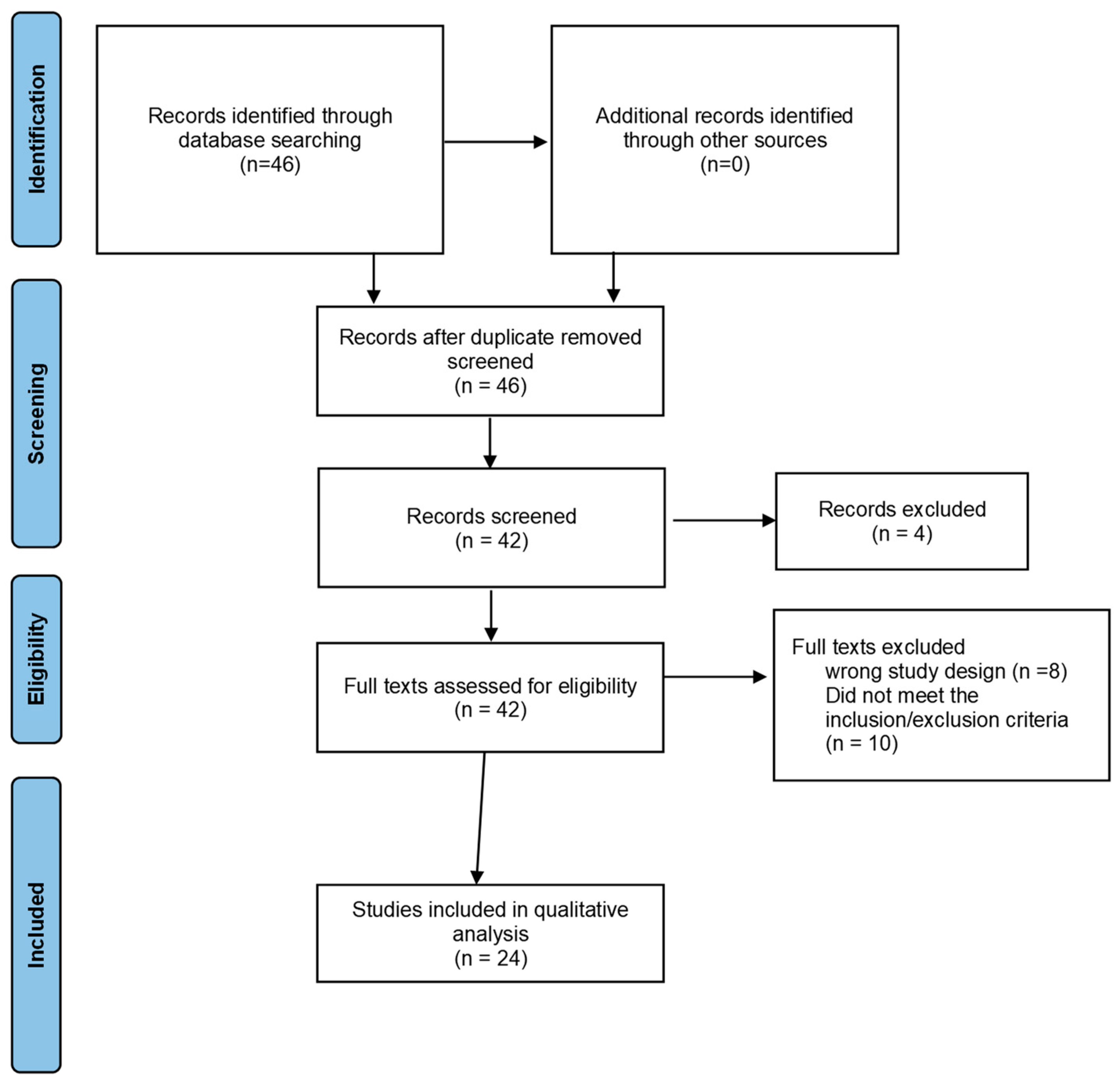
3. Results
3.1. Studies Included in the Review
3.2. Comparison Groups/Subgroups of the Studies
3.3. Interventions of the Included Studies
3.4. Methodological Quality
3.5. Measurement Outcomes
3.5.1. Primary Outcome Measures
Microbial Markers among ADA and CRC Compared to Healthy Control (HC) Using the Untargeted Microbiome Approach
Microbial Markers among ADA and CRC Compared to Healthy Control (HC) Using the Targeted Microbiome Approach
Metabolite Markers among ADA and CRC Compared to Healthy Control (HC) Using the Non-Targeted and Targeted Metabolite Approaches
3.5.2. Secondary Outcome Measures
Microbial Markers for Cancer Stages and Locations
4. Discussion
5. Study Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- McGuire, G. Switzerland: World Health Organization, International Agency for Research on Cancer; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020; Available online: https://gco.iarc.fr/today (accessed on 20 April 2023).
- Wild, C.P.; Stewart, B.W.; Wild, C. World Cancer Report 2014; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Avuthu, N.; Guda, C. Meta-Analysis of Altered Gut Microbiota Reveals Microbial and Metabolic Biomarkers for Colorectal Cancer. Microbiol. Spectr. 2022, 10, e00013-22. [Google Scholar] [CrossRef] [PubMed]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver–passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Adolph, T.E.; Gerner, R.R.; Moschen, A.R. The intestinal microbiota in colorectal cancer. Cancer Cell 2018, 33, 954–964. [Google Scholar] [CrossRef]
- Yu, J.; Feng, Q.; Wong, S.H.; Zhang, D.; yi Liang, Q.; Qin, Y.; Tang, L.; Zhao, H.; Stenvang, J.; Li, Y. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017, 66, 70–78. [Google Scholar] [CrossRef]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- Stracci, F.; Zorzi, M.; Grazzini, G. Colorectal cancer screening: Tests, strategies, and perspectives. Front. Public Health 2014, 2, 210. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.-M.; Fournier, P.-E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef]
- Boleij, A.; Hechenbleikner, E.M.; Goodwin, A.C.; Badani, R.; Stein, E.M.; Lazarev, M.G.; Ellis, B.; Carroll, K.C.; Albesiano, E.; Wick, E.C. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 2015, 60, 208–215. [Google Scholar] [CrossRef]
- Zuppi, C.; Messana, I.; Tapanainen, P.; Knip, M.; Vincenzoni, F.; Giardina, B.; Nuutinen, M. Proton nuclear magnetic resonance spectral profiles of urine from children and adolescents with type 1 diabetes. Clin. Chem. 2002, 48, 660–662. [Google Scholar] [CrossRef] [PubMed]
- Emaus, A.; Veierød, M.B.; Tretli, S.; Finstad, S.E.; Selmer, R.; Furberg, A.S.; Bernstein, L.; Schlichting, E.; Thune, I. Metabolic profile, physical activity, and mortality in breast cancer patients. Breast Cancer Res. Treat. 2010, 121, 651–660. [Google Scholar] [CrossRef]
- Parmentier-Decrucq, E.; Duhamel, A.; Ernst, O.; Fermont, C.; Louvet, A.; Vernier-Massouille, G.; Cortot, A.; Colombel, J.F.; Desreumaux, P.; Peyrin-Biroulet, L. Effects of infliximab therapy on abdominal fat and metabolic profile in patients with Crohn’s disease. Inflamm. Bowel Dis. 2009, 15, 1476–1484. [Google Scholar] [CrossRef]
- Murdoch, T.B.; Fu, H.; MacFarlane, S.; Sydora, B.C.; Fedorak, R.N.; Slupsky, C.M. Urinary metabolic profiles of inflammatory bowel disease in interleukin-10 gene-deficient mice. Anal. Chem. 2008, 80, 5524–5531. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.R.; Willsmore, J.D.; Cox, I.J.; Walker, D.G.; Cobbold, J.F.; Taylor-Robinson, S.D.; Orchard, T.R. Serum metabolic profiling in inflammatory bowel disease. Dig. Dis. Sci. 2012, 57, 2157–2165. [Google Scholar] [CrossRef] [PubMed]
- Fliss-Isakov, N.; Zelber-Sagi, S.; Webb, M.; Halpern, Z.; Shibolet, O.; Kariv, R. Distinct metabolic profiles are associated with colorectal adenomas and serrated polyps. Obesity 2017, 25, S72–S80. [Google Scholar] [CrossRef] [PubMed]
- Farshidfar, F.; Weljie, A.M.; Kopciuk, K.; Buie, W.D.; MacLean, A.; Dixon, E.; Sutherland, F.R.; Molckovsky, A.; Vogel, H.J.; Bathe, O.F. Serum metabolomic profile as a means to distinguish stage of colorectal cancer. Genome Med. 2012, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Xie, G.; Jia, W. Metabonomics of human colorectal cancer: New approaches for early diagnosis and biomarker discovery. J. Proteome Res. 2014, 13, 3857–3870. [Google Scholar] [CrossRef]
- Ohigashi, S.; Sudo, K.; Kobayashi, D.; Takahashi, O.; Takahashi, T.; Asahara, T.; Nomoto, K.; Onodera, H. Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer. Dig. Dis. Sci. 2013, 58, 1717–1726. [Google Scholar] [CrossRef] [PubMed]
- Barberini, L.; Restivo, A.; Noto, A.; Deidda, S.; Fattuoni, C.; Fanos, V.; Saba, L.; Zorcolo, L.; Mussap, M. A gas chromatography-mass spectrometry (GC-MS) metabolomic approach in human colorectal cancer (CRC): The emerging role of monosaccharides and amino acids. Ann. Transl. Med. 2019, 7, 727. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Song, X.; Khan, S.; Li, Y.; Guo, Z.; Li, C.; Wang, S.; Dong, W.; Liu, W.; Wang, B. The gut microbiota at the intersection of bile acids and intestinal carcinogenesis: An old story, yet mesmerizing. Int. J. Cancer 2020, 146, 1780–1790. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Shao, X.; Zhou, Y.; Yu, Y.; Kuai, X.; Zhou, C. Bidirectional regulation of bile acid on colorectal cancer through bile acid-gut microbiota interaction. Am. J. Transl. Res. 2021, 13, 10994. [Google Scholar] [PubMed]
- Sun, X.-Z.; Zhao, D.-Y.; Zhou, Y.-C.; Wang, Q.-Q.; Qin, G.; Yao, S.-K. Alteration of fecal tryptophan metabolism correlates with shifted microbiota and may be involved in pathogenesis of colorectal cancer. World J. Gastroenterol. 2020, 26, 7173. [Google Scholar] [CrossRef] [PubMed]
- Jalandra, R.; Dalal, N.; Yadav, A.K.; Verma, D.; Sharma, M.; Singh, R.; Khosla, A.; Kumar, A.; Solanki, P.R. Emerging role of trimethylamine-N-oxide (TMAO) in colorectal cancer. Appl. Microbiol. Biotechnol. 2021, 105, 7651–7660. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.J. Cochrane Handbook for Systematic Reviews of Interventions Version 6.3 (Updated February 2022); Welch, V.A., Ed.; Cochrane: London, UK, 2022; Available online: www.training.cochrane.org/handbook (accessed on 20 April 2023).
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. Ann. Intern. Med. 2009, 151, W65–W94. [Google Scholar] [CrossRef]
- Vandenbroucke, J.P.; Elm, E.v.; Altman, D.G.; Gøtzsche, P.C.; Mulrow, C.D.; Pocock, S.J.; Poole, C.; Schlesselman, J.J.; Egger, M.; Initiative, S. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): Explanation and elaboration. Ann. Intern. Med. 2007, 147, W163–W194. Available online: https://www.acpjournals.org/doi/10.7326/0003-4819-147-8-200710160-00010-w1 (accessed on 20 April 2023). [CrossRef]
- Cornelius, L.; Van der Klink, J.; Groothoff, J.; Brouwer, S. Prognostic factors of long term disability due to mental disorders: A systematic review. J. Occup. Rehabil. 2011, 21, 259–274. [Google Scholar] [CrossRef]
- Wang, Q.; Li, L.; Xu, R. A systems biology approach to predict and characterize human gut microbial metabolites in colorectal cancer. Sci. Rep. 2018, 8, 6225. [Google Scholar] [CrossRef]
- Konishi, Y.; Okumura, S.; Matsumoto, T.; Itatani, Y.; Nishiyama, T.; Okazaki, Y.; Shibutani, M.; Ohtani, N.; Nagahara, H.; Obama, K. Development and evaluation of a colorectal cancer screening method using machine learning-based gut microbiota analysis. Cancer Med. 2022, 11, 3194–3206. [Google Scholar] [CrossRef]
- Iwasaki, M.; Kanehara, R.; Yamaji, T.; Katagiri, R.; Mutoh, M.; Tsunematsu, Y.; Sato, M.; Watanabe, K.; Hosomi, K.; Kakugawa, Y. Association of Escherichia coli containing polyketide synthase in the gut microbiota with colorectal neoplasia in Japan. Cancer Sci. 2022, 113, 277–286. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, Y.; Zhao, L.; Zhao, F.; Feng, J.; Li, S.; Chen, H.; Sun, J.; Zhu, B.; Geng, R. Gut microbiota in patients after surgical treatment for colorectal cancer. Environ. Microbiol. 2019, 21, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Dai, X.; Zhou, C.-C.; Li, K.-x.; Zhang, Y.-j.; Lou, X.-Y.; Zhu, Y.-M.; Sun, Y.-L.; Peng, B.-X.; Cui, W. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut 2022, 71, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, N.; Li, Q.; Chu, E.S.H.; Lau, H.C.H.; Fong, W.; Liu, W.; Liang, C.; Nakatsu, G.; Su, A.C.Y.; Coker, O.O. Lactobacillus gallinarum modulates the gut microbiota and produces anti-cancer metabolites to protect against colorectal tumourigenesis. Gut 2022, 71, 2011–2021. [Google Scholar] [CrossRef]
- Sánchez-Alcoholado, L.; Laborda-Illanes, A.; Otero, A.; Ordóñez, R.; González-González, A.; Plaza-Andrades, I.; Ramos-Molina, B.; Gómez-Millán, J.; Queipo-Ortuño, M.I. Relationships of gut microbiota composition, short-chain fatty acids and polyamines with the pathological response to neoadjuvant radiochemotherapy in colorectal cancer patients. Int. J. Mol. Sci. 2021, 22, 9549. [Google Scholar] [CrossRef]
- Yu, S.Y.; Xie, Y.H.; Qiu, Y.W.; Chen, Y.X.; Fang, J.Y. Moderate alteration to gut microbiota brought by colorectal adenoma resection. J. Gastroenterol. Hepatol. 2019, 34, 1758–1765. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Zhu, Y.; Gao, Y.; Li, H.; Zhu, X.; Wei, R.; Xie, R.; Wei, Q.; Qin, H. A newly developed PCR-based method revealed distinct Fusobacterium nucleatum subspecies infection patterns in colorectal cancer. Microb. Biotechnol. 2021, 14, 2176–2186. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Carbonero, F.; Zoetendal, E.G.; DeLany, J.P.; Wang, M.; Newton, K.; Gaskins, H.R.; O’Keefe, S.J. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 2013, 98, 111–120. [Google Scholar] [CrossRef]
- Hester, C.M.; Jala, V.R.; Langille, M.G.; Umar, S.; Greiner, K.A.; Haribabu, B. Fecal microbes, short chain fatty acids, and colorectal cancer across racial/ethnic groups. World J. Gastroenterol. WJG 2015, 21, 2759. [Google Scholar] [CrossRef]
- Guo, Y.-P.; Shao, L.; Wang, L.; Chen, M.-Y.; Zhang, W.; Huang, W.-H. Bioconversion variation of ginsenoside CK mediated by human gut microbiota from healthy volunteers and colorectal cancer patients. Chin. Med. 2021, 16, 28. [Google Scholar] [CrossRef]
- Xu, R.; Wang, Q.; Li, L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genom. 2015, 16, S4. [Google Scholar] [CrossRef]
- Okumura, S.; Konishi, Y.; Narukawa, M.; Sugiura, Y.; Yoshimoto, S.; Arai, Y.; Sato, S.; Yoshida, Y.; Tsuji, S.; Uemura, K. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat. Commun. 2021, 12, 5674. [Google Scholar] [CrossRef]
- Tang, Q.; Cang, S.; Jiao, J.; Rong, W.; Xu, H.; Bi, K.; Li, Q.; Liu, R. Integrated study of metabolomics and gut metabolic activity from ulcerative colitis to colorectal cancer: The combined action of disordered gut microbiota and linoleic acid metabolic pathway might fuel cancer. J. Chromatogr. A 2020, 1629, 461503. [Google Scholar] [CrossRef]
- Katsidzira, L.; Ocvirk, S.; Wilson, A.; Li, J.; Mahachi, C.; Soni, D.; DeLany, J.; Nicholson, J.; Zoetendal, E.; O’keefe, S. Differences in fecal gut microbiota, short-chain fatty acids and bile acids link colorectal cancer risk to dietary changes associated with urbanization among Zimbabweans. Nutr. Cancer 2019, 71, 1313–1324. [Google Scholar] [CrossRef]
- Ocvirk, S.; Wilson, A.S.; Posma, J.M.; Li, J.V.; Koller, K.R.; Day, G.M.; Flanagan, C.A.; Otto, J.E.; Sacco, P.E.; Sacco, F.D. A prospective cohort analysis of gut microbial co-metabolism in Alaska Native and rural African people at high and low risk of colorectal cancer. Am. J. Clin. Nutr. 2020, 111, 406–419. [Google Scholar] [CrossRef]
- Ai, D.; Pan, H.; Li, X.; Wu, M.; Xia, L.C. Association network analysis identifies enzymatic components of gut microbiota that significantly differ between colorectal cancer patients and healthy controls. PeerJ 2019, 7, e7315. [Google Scholar] [CrossRef]
- Kim, M.; Vogtmann, E.; Ahlquist, D.A.; Devens, M.E.; Kisiel, J.B.; Taylor, W.R.; White, B.A.; Hale, V.L.; Sung, J.; Chia, N. Fecal metabolomic signatures in colorectal adenoma patients are associated with gut microbiota and early events of colorectal cancer pathogenesis. mBio 2020, 11, e03186-19. [Google Scholar] [CrossRef]
- Ahlquist, D.A.; Sargent, D.J.; Loprinzi, C.L.; Levin, T.R.; Rex, D.K.; Ahnen, D.J.; Knigge, K.; Lance, M.P.; Burgart, L.J.; Hamilton, S.R. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann. Intern. Med. 2008, 149, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Nugent, J.L.; McCoy, A.N.; Addamo, C.J.; Jia, W.; Sandler, R.S.; Keku, T.O. Altered tissue metabolites correlate with microbial dysbiosis in colorectal adenomas. J. Proteome Res. 2014, 13, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Mishra, R.; Cen, C.; Tang, Y.; Ma, C.; Wasti, S.; Wang, Y.; Ou, Q.; Chen, K.; Zhang, J. Metagenomic analyses expand bacterial and functional profiling biomarkers for colorectal cancer in a Hainan cohort, China. Curr. Microbiol. 2021, 78, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Guertin, K.A.; Li, X.S.; Graubard, B.I.; Albanes, D.; Weinstein, S.J.; Goedert, J.J.; Wang, Z.; Hazen, S.L.; Sinha, R. Serum Trimethylamine N-oxide, Carnitine, Choline, and Betaine in Relation to Colorectal Cancer Risk in the Alpha Tocopherol, Beta Carotene Cancer Prevention StudySerum TMAO, Related Metabolites, and Colorectal Cancer Risk. Cancer Epidemiol. Biomark. Prev. 2017, 26, 945–952. [Google Scholar] [CrossRef]
- Group ACPS. The alpha-tocopherol, beta-carotene lung cancer prevention study: Design, methods, participant characteristics, and compliance. Ann. Epidemiol. 1994, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Yang, J.; Seo, H.; Lee, W.H.; Ho Lee, D.; Kym, S.; Park, Y.S.; Kim, J.G.; Jang, I.-J.; Kim, Y.-K. Colorectal cancer diagnostic model utilizing metagenomic and metabolomic data of stool microbial extracellular vesicles. Sci. Rep. 2020, 10, 2860. [Google Scholar] [CrossRef] [PubMed]
- Song, E.M.; Byeon, J.-S.; Lee, S.M.; Yoo, H.J.; Kim, S.J.; Lee, S.-H.; Chang, K.; Hwang, S.W.; Yang, D.-H.; Jeong, J.-Y. Fecal fatty acid profiling as a potential new screening biomarker in patients with colorectal cancer. Dig. Dis. Sci. 2018, 63, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Genua, F.; Mirković, B.; Mullee, A.; Levy, M.; Gallagher, W.M.; Vodicka, P.; Hughes, D.J. Association of circulating short chain fatty acid levels with colorectal adenomas and colorectal cancer. Clin. Nutr. ESPEN 2021, 46, 297–304. [Google Scholar] [CrossRef]
- D’asheesh, T.i.A.; Hussen, B.M.; Al-Marzoqi, A.H.; Ghasemian, A. Assessment of oncogenic role of intestinal microbiota in colorectal cancer patients. J. Gastrointest. Cancer 2021, 52, 1016–1021. [Google Scholar] [CrossRef]
- Coker, O.O.; Liu, C.; Wu, W.K.K.; Wong, S.H.; Jia, W.; Sung, J.J.; Yu, J. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome 2022, 10, 35. [Google Scholar] [CrossRef]
- Goedert, J.J.; Sampson, J.N.; Moore, S.C.; Xiao, Q.; Xiong, X.; Hayes, R.B.; Ahn, J.; Shi, J.; Sinha, R. Fecal metabolomics: Assay performance and association with colorectal cancer. Carcinogenesis 2014, 35, 2089–2096. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Ahn, J.; Sampson, J.N.; Shi, J.; Yu, G.; Xiong, X.; Hayes, R.B.; Goedert, J.J. Fecal microbiota, fecal metabolome, and colorectal cancer interrelations. PLoS ONE 2016, 11, e0152126. [Google Scholar] [CrossRef] [PubMed]
- Clos-Garcia, M.; Garcia, K.; Alonso, C.; Iruarrizaga-Lejarreta, M.; D’Amato, M.; Crespo, A.; Iglesias, A.; Cubiella, J.; Bujanda, L.; Falcón-Pérez, J.M. Integrative analysis of fecal metagenomics and metabolomics in colorectal cancer. Cancers 2020, 12, 1142. [Google Scholar] [CrossRef]
- Cubiella, J.; Clos-Garcia, M.; Alonso, C.; Martinez-Arranz, I.; Perez-Cormenzana, M.; Barrenetxea, Z.; Berganza, J.; Rodríguez-Llopis, I.; D’amato, M.; Bujanda, L. Targeted UPLC-MS metabolic analysis of human faeces reveals novel low-invasive candidate markers for colorectal cancer. Cancers 2018, 10, 300. [Google Scholar] [CrossRef]
- Cubiella, J.; Vega, P.; Salve, M.; Díaz-Ondina, M.; Alves, M.T.; Quintero, E.; Álvarez-Sánchez, V.; Fernández-Bañares, F.; Boadas, J.; Campo, R. Development and external validation of a faecal immunochemical test-based prediction model for colorectal cancer detection in symptomatic patients. BMC Med. 2016, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Qiu, Y.; Zou, X.; Chen, T.; Xie, G.; Cheng, Y.; Dong, T.; Zhao, L.; Feng, B.; Hu, X. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 2013, 12, 3000–3009. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef] [PubMed]
- Zackular, J.P.; Rogers, M.A.; Ruffin IV, M.T.; Schloss, P.D. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. 2014, 7, 1112–1121. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Niu, M.; Pan, J.; Du, N.; Liu, S.; Li, H.; He, Q.; Mao, J.; Duan, Y.; Du, Y. Bacteroides, butyric acid and t10, c12-CLA changes in colorectal adenomatous polyp patients. Gut Pathog. 2021, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, V.; Löfgren-Burström, A.; Zingmark, C.; Edin, S.; Larsson, P.; Karling, P.; Alexeyev, O.; Rutegård, J.; Wikberg, M.L.; Palmqvist, R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 2017, 141, 2528–2536. [Google Scholar] [CrossRef]
- Gao, R.; Wang, Z.; Li, H.; Cao, Z.; Gao, Z.; Chen, H.; Zhang, X.; Pan, D.; Yang, R.; Zhong, H. Gut microbiota dysbiosis signature is associated with the colorectal carcinogenesis sequence and improves the diagnosis of colorectal lesions. J. Gastroenterol. Hepatol. 2020, 35, 2109–2121. [Google Scholar] [CrossRef]
- Yusuf, F.; Adewiah, S.; Fatchiyah, F. The level short chain fatty acids and HSP 70 in colorectal cancer and non-colorectal cancer. Acta Inform. Med. 2018, 26, 160. [Google Scholar] [CrossRef]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE 2013, 8, e70803. [Google Scholar] [CrossRef]
- Yang, Y.; Misra, B.B.; Liang, L.; Bi, D.; Weng, W.; Wu, W.; Cai, S.; Qin, H.; Goel, A.; Li, X. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics 2019, 9, 4101. [Google Scholar] [CrossRef] [PubMed]
- Russ, C.A.; Zertalis, N.A.; Nanton, V. Gut Bacterial Microbiome Profiles Associated with Colorectal Cancer Risk: A Systematic Review and Meta-Analysis. medRxiv 2021, 21258404. [Google Scholar] [CrossRef]
- Yu, L.; Zhao, G.; Wang, L.; Zhou, X.; Sun, J.; Li, X.; Zhu, Y.; He, Y.; Kofonikolas, K.; Bogaert, D. A systematic review of microbial markers for risk prediction of colorectal neoplasia. Br. J. Cancer 2022, 126, 1318–1328. [Google Scholar] [CrossRef] [PubMed]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef] [PubMed]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Mancabelli, L.; Milani, C.; Lugli, G.A.; Turroni, F.; Ferrario, C.; van Sinderen, D.; Ventura, M. Meta-analysis of the human gut microbiome from urbanized and pre-agricultural populations. Environ. Microbiol. 2017, 19, 1379–1390. [Google Scholar] [CrossRef]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A review of methods and databases for metagenomic classification and assembly. Brief. Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef]
| Level | Description |
|---|---|
| Strong | Consistent results (≥70%) from at least 2 high-quality studies |
| Moderate | 1 high-quality study and consistent findings (≥70%) in 1 or more low-quality studies |
| Limited | Findings in 1 high-quality * study or consistent results (≥70%) among low-quality studies |
| NO | No study identified |
| Conflicting | Inconsistent results, irrespective of study quality |
| Author | Study Type | Recruitment Strategy and Selection Criteria | Number of Subjects and Groups | Location and Time Frame | |||
|---|---|---|---|---|---|---|---|
| Sun et al. [26] | Case-control study for untargeted microbiome and targeted metabolites identification, specifically Tryptophan and its metabolites in CRC patients | Male and female Aged 18–80 yrs ADA, CRC, HC | Healthy control = 38 24 24  14 1456.85 yrs ± 10.99 | ADA = 33 23 23  10 1061.18 yrs ± 8.53 | CRC = 46 32 32  14 1463.63 yrs ± 11.39 | The China–Japan Friendship Hospital, China March 2019 and December 2019 | |
| Kim et al. [50] | Case-control study for untargeted metabolites and microbiome identification in CRC patients Ps. The samples were obtained from cross sectional study, which gives this study a cross-sectional nature | All samples selected here have been enrolled in previous study [51] Male and female Aged 50–80 yrs ADA, CRC, and HC. | Healthy control = 102 62 62  40 4050–59 yrs = 18 60–69 yrs = 49 >70 yrs = 35 | ADA = 102 62 62  40 4050–59 yrs = 17 60–69 yrs = 50 >70 yrs = 35 | CRC = 6 20 20  16 1650–59 yrs = 6 60–69 yrs = 19 >70 yrs = 11 | ND 2001 to 2007 | |
| Nugent et al. [52] | Case-control study for targeted microbiota (Lactobacillus sp., Escherichia coli, Bifidobacterium sp., Clostridium sp., Bacteroides sp., and Eubacteria) and untargeted metabolites identification in CRC patients | Male and female Aged > 30 yrs ADA and HC | Healthy control = 15 4 4  11 1155.0 yrs ± 1.1 | ADA = 15 6 6  9 954.3 yrs ± 1.1 | University of North Carolina Hospitals, USA ND | ||
| Chang et al. [53] | Case-control study for untargeted microbiome in CRC patients | Only Male Aged 38–77 yrs CRC and HC | Healthy control = 12 12 12 | CRC = 6 6 6 | Haikou people’s Hospital, Hainan, China ND | ||
| Metagenomics sequences of 59 patients with CRC were obtained from the NCBI database (ref_CRC, Metagenomics sequencing data: PRJEB7774). | |||||||
| Guertin et al. [54] | Case-control study for targeted metabolites, trimethylamine N-oxide, Carnitine, Choline, and Betaine in CRC patients “Nested case-control study within the Alpha Tocopherol and Beta Carotene Cancer Prevention (ATBC) Study, described in detail elsewhere [55] | Gender ND Aged 50–69 yrs CRC and HC | Healthy control = 644 | CRC = 644 | USA ATBC study (1985–1988)–(1993) [55] | ||
| Tumor location Proximal colon = 169 Distal colon = 153 Rectum ICD-9 = 282 | |||||||
| Kim et al. [56] | Case-control study for untargeted microbiome and untargeted metabolites in CRC patients | Male and female Aged 45–80 yrs CRC and HC | Healthy control = 40 22 22  18 1849–78 yrs | CRC = 32 20 20  16 1645–80 yrs | CRC patients from Seoul National University Bundang Hospital and Chung-Ang University Hospital, South Korea HC individuals from Haewoondae Baek Hospital, South Korea April 2016–April 2018. | ||
| Tumor Stage 0 = 1 I = 7 II = 12 III = 9 IV = 3 | |||||||
| Tumor location Cesum = 2 Ascending = 6 Transverse = 1 Sigmoid = 12 Rectal = 7 | |||||||
| Song et al. [57] | Pilot, case-control study for targeted metabolites, long and short fatty acid in CRC patients | Male and female Aged 45–70 yrs ADA, CRC, and HC | Healthy control = 28 22 22  6 651.1 yrs ± 6.0 | ADA = 27 25 25  1 153.6 yrs ± 7.2 | CRC = 26 16 16  10 1059.7 yrs ± 12.2 | Asan Institute for Life Sciences, University of Ulsan College of Medicine, South Korea July 2014 and August 2014 | |
| Tumor stage I = 3 IIa = 5 IIc = 1 IIIb = 11 IIIc = 3 IVa = 3 | |||||||
| Presence of lymph node metastasis = 16 Presence of colonoscopic obstruction = 5 Tumor location Proximal cancer (above splenic flexure) = 3 Distal cancer (below splenic flexure) = 23 | |||||||
| Genua et al. [58] | Case-control study for targeted metabolites, Acetic Acid, Propionic Acid, i-Butyric Acid, Butyric Acid, 2-MethylButyric Acid, i-Valeric Acid, Valeric Acid from serum in CRC patients | Male and female Cohort Irish and Czech Aged 45–70 yrs Tubular tubulovillous adenoma (TA/TVA), High-grade dysplasia (HGD), CRC, and HC | Irish cohort 128 | The Adelaide & Meath Hospital in Dublin, Ireland Thomayer Hospital in Prague, Czech Republic. | |||
Healthy control = 36 17 17  19 1958 yrs ± 7 | TA/TVA = 48 30 30  18 1861.5 yrs ± 11 | HGD = 18 11 11  7 759 yrs ± 7 | CRC = 26 13 13  13 1356 yrs ± 23 | ||||
| Czech cohort 85 | |||||||
Healthy control = 27 12 12  15 1556 yrs ± 10 | CRC = 58 40 40  18 1864 yrs ± 15 | ||||||
| D’asheesh et al. [59] | Case-control study for targeted microbiota Lactobaccilus acidophilus, Lactobacillus Plantarum, and Enterococcus faecalis | Aged 20–76 yrs Gender ND CRC and HC | Healthy control = 300 45.3 ± 2.5 | CRC = 30055.34 ± 3.66 | Iran March 2014 to October 2019 | ||
| Coker et al. [60] | Case-control study for untargeted microbiome and targeted metabolites | Male and female Aged 58–83 yrs ADA, CRC, and HC | Healthy control 128 59 59  69 6964.03 yrs ± 6.84 | ADA 140 64 64  54 5465.84 yrs ± 5.53 | CRC 118 64 64  54 5473.21 yrs ± 10.37 | Prince of Wales Hospital, the Chinese University of Hong Kong ND | |
| Goedert et al. [61] | Case-control study for untargeted metabolites | Male and female Aged 46–75 yrs CRC and HC | Healthy control 102 55.9% 55.9%  44.1% 44.1%58.3 yrs ± 12.9 | CRC 48 64.6% 64.6%  35.4% 35.4%62.9 yrs ± 13.7 | 1985–1989 Washington DC area hospitals, USA | ||
| Tumor stage Non-invasive = 20.8% Invasive, no known metastases = 41.7% Known metastases = 35.4% Missing = 2.1% | |||||||
| Tumor location Right colon = 29.1% Left colon = 33.3% Rectal = 27.1% Missing = 10.4% | |||||||
| Sinha et al. [62] | Case-control study for untargeted microbiome and untargetd metabolites | Male and female Aged 45–76 yrs CRC and HC | Healthy control = 89 55.5% 55.5%  40.5% 40.5%58.4 yrs ± 13 | CRC = 42 59.5% 59.5%  40.5% 40.5%63.4 yrs ± 13.1 | ND 1985–1987 | ||
| Tumor stage Non-invasive = 21.4% Invasive, no known metastases = 42.9% Known metastases = 33.3% Missing = 2.1% | |||||||
| Clos-Garcia et al. [63] | Case-control study for targeted metabolites as in [64] and untargeted microbiome identification in CRC patients | Male and female Aged >18 yrs ADA, CRC, and HC | Healthy control = 77 35 35  48 4864.62 yrs | ADA = 69 41 41  41 4167.99 yrs | CRC = 99 60 60  39 3970.16 yrs | Samples batch 1 and 2 from COLONPREDICT study [65] Batch 3 from Instituto de Investigación Sanitario Galicia Sur, Spain ND | |
| Tan et al. [66] | Case-control study for untargeted metabolites in CRC patients | CRC and HC Aged 24–82 yrs | Healthy control = 102 31–76 yrs | CRC = 101 24–82 yrs | The Ruijin Hospital affiliated with Shanghai Jiao Tong University School of Medicine, China ND | ||
| Tumor stage I = 26 II = 43 III = 26 IV = 6 | |||||||
| Tumor location Ascending = 21 Descending = 9 Sigmoid colon = 7 Rectum = 63 | |||||||
| Flemer et al. [67] | Case-control study for untargeted microbiome from stool and mucosa in CRC patients | Female and male Aged 27–82 yrs CRC, ADA, and HC | Healthy control = 56 | Polyps ADA = 21 | CRC = 59 | Mercy University Hospital, Ireland ND | |
| Zeller et al. [68] | Case-control study for untargeted microbiome from stool and mucosa in CRC patients | Female and male Aged 34–69 yrs Adenoma (small < 1 cm and large > 1 cm) HC from different cohorts from France and Germany | Healthy control = 358 Cohort France = 61 Cohort Germany = 297 | ADA = 42 Cohort France ADA small = 27 ADA large = 15 | CRC = 91 | F group Assistance Publique-Hôpitaux de Paris (academic hospitals) G population the Department of Surgery at the University Hospital Heidelberg and the affiliated Hospital Salem H population From my microbe project http://my.microbes.eu/ (accessed on 12 June 2023) ND | |
| Cohort France = 61 Tumor stage 0 = 0 I = 15 II = 7 III = 10 IV = 21 | Cohort Germany = 38 Tumor stage 0 = 25 I = 0 II = 0 III = 13 IV = 0 | ||||||
| Zackular et al. [69] | Case-control study for untargeted microbiome from stool in CRC patients | Male and female Aged >18 yrs ADA, CRC, and HC | Healthy control = 30 11 11  19 1955.3 yrs (±9.2) | ADA = 30 18 18  12 1261.3 yrs (±11.1) | CRC = 30 21 21  9 959.4 yrs (±11) | Toronto (Canada), Boston (USA), Houston (USA), and Ann Arbor (USA) ND | |
| Ohigashi et al. [22] | Case-control study for targeted metabolites and microbiome from stool in CRC patients | Male and female Aged 52–81 yrs ADA, CRC, and HC | Healthy control = 27 16 16  11 1165.6 yrs ± 13.5 | ADA = 22 11 11  11 1166.6 yrs ± 9.2 | CRC = 93 49 49  44 4468.9 yrs ± 12.1 | ND November 2007–October 2010 | |
| Tumor stage Dukes A (36 patients) Dukes B (19 patients) Dukes C (24 patients) Dukes D (14 patients) | |||||||
| Chen et al. [70] | Case-control study for untargeted metabolites and microbiome, followed by targeted microbiota using functional genes from stool in CRC patients | Male and female Aged 40–63 yrs ADA and HC | Healthy control = 30 13 13  17 1750.33 yrs ± 10.87 | ADA = 30 20 20  10 1053.23 yrs ± 10.14 | The First Affiliated Hospital of Kunming Medical University, China November 2017 to April 2018 | ||
| Eklöf et al. [71] | Case-control study for targeted microbiome in CRC patients | Male and female Aged > 34 yrs CRC, ADA, HC | Healthy control = 65 35 35  30 3034–80 yrs | Dysplasia ADA = 134 80 80  54 5434–80 yrs | CRC = 39 20 20  19 1934–80 yrs | The University Hospital in Umeå, Sweden September 2008 to March 2013 | |
| Tumor stage I = 2 II = 21 III = 8 IV = 7 | |||||||
| Tumor location | |||||||
| Total | Dysplasia | CRC | |||||
| Right | 37 | 12 | 49 | ||||
| Left | 59 | 17 | 76 | ||||
| Rectum | 38 | 10 | 40 | ||||
| Gao et al. [72] | Case-control study for untargeted microbiome in CRC patients | Male and female Aged ND CRC, precancer (ADA), HC | Healthy control = 442 60.65% 60.65% 39.35% 39.35%65.79 yrs ± 12.73 | Precancer (ADA) = 195 (31) 62.5% 62.5% 37. 5% 37. 5%63.07 yrs ± 12.84 | CRC = 155 29.48% 29.48% 70.52% 70.52%64.96 yrs ± 10.44 | The Shanghai Tenth People’s Hospital, Tongji University School of Medicine and Changzheng Hospital affiliated with the Naval Medical University, China The discovery cohort from January 2014–November 2015 The validation cohort from March 2016–December 2017 | |
| Tumor stage 0 = 25 (16.13%) I = 51 (32.9%) II = 56 (36.13%) III = 11.7 (10%) IV = 12 (7.74%) | |||||||
| Tumor location Ascending colon = 25 (16.13%) Transverse colon = 7 (4.52%) Descending colon = 10 (6.45%) Sigmoid colon = 33 (21.29%) Rectum = 70 (45.16%) Undefined = 5 (2.3%) | |||||||
| Yusuf et al. [73] | Case-control study for targeted metabolites, short-chain fatty acids, acetate, propionate and butyrate acids in CRC patients | Male and female Aged >18 yrs CRC and HC | Healthy control = 14 9 9  5 550 yrs ± 17.6 | CRC = 14 10 10  4 4 53.8 yrs ± 13.3 | General Teaching Hospital Banda Aceh, Indonesia ND | ||
| Weir et al. [74] | Case-control study for untargeted microbiome and untargeted metabolites followed by targeted for short chain fatty acids in CRC patients | Male and female Aged >18 yrs CRC and HC | Healthy control = 11 7 7  3 350 yrs ± 17.6 | CRC = 10 8 8  2 253.8 yrs ± 13.3 | The University of Colorado Health-Poudre Valley Hospital in Fort Collins, CO, USA ND | ||
| Tumor stage * T1 = 2 T2 = 3 T3 = 4 Tis = 1 * Tis: Carcinoma in situ: intraepithelial or invasion of lamina propria; T1: Tumor invades submucosa; T2: Tumor invades muscularis propria; T3:Tumor invades through muscularis propria into the subserosa or into nonperitonealized pericolic or perirectal tissue. | |||||||
| Tumor location Ascending 3 Rectum 3 Sigmoid 4 | |||||||
| Yang et al. [75] | Case-control study for untargeted microbiome and metabolites in CRC patients | Male and female Aged >60 and <60 yrs CRC and HC | Healthy control = 50 17 17  33 33>60 yrs = 33 <60 yrs = 17 | CRC = 50 26 26  24 24>60 yrs = 24 <60 yrs = 26 | Ongji University Affiliated Tenth People’s Hospital (Shanghai, China) January 2014 to September 2014 | ||
| Author | Group | Intervention | Sample Type | Metric | |
|---|---|---|---|---|---|
| Sun et al. [26] | Experimental group AD and CRC Control group | Targeted metabolites identification | Untargeted microbiome identification | Fecal specimen | +/− of Trp and its metabolites Indole/Trap ratio Distribution (abundance) at bacterial genera level |
| Tryptophan (Trap) and its metabolites, such as L-Trp, L-Kynurenine (KYN), indole, skatole, indole-3-carboxylic acid (I3CA), Indole-3-aldehyde (IALD), Indole-3-acetate (IAA), Indolepropionic acid (IPA), indoxyl-3-sulfate (I3S), and Indole-3-acetadehyde (IAALD) using Ultraperformance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) analysis | 16S geneRNA gene sequencing using an Illumina NovaSeq PE250 | ||||
| Kim et al. [50] | Experimental group AD and CRC Control group | Untargeted metabolites identification | Untargeted microbiome identification | Fecal specimen | Distribution (abundances) of metabolites Distribution (abundance) bacterial genera |
| UPLC-MS/MS platform | 16S gene RNA gene sequencing using the Illumina MiSeq system | ||||
| Nugent et al. [52] | Experimental group AD Control group | Untargeted metabolites identification | Targeted microbiome identification | Rectal mucosal biopsy | +/− of metabolites Distribution (abundance) of bacterial genera/species |
| Liquid chromatography and gas chromatography time of flight mass spectrometry | For Lactobacillus sp., Escherichia coli, Bifidobacterium sp., Clostridium sp., Bacteroides sp., and Eubacteria using qPCR with primers that amplify 16S rDNA | ||||
| Chang et al. [53] | Experimental group CRC Control group | Untargeted microbiome identification | Fecal specimen | Distribution (abundance) of bacterial species | |
| Whole-genome shotgun sequencing Illumina HiSeq | |||||
| Guertin et al. [54] | Experimental group CRC Control group | Targeted metabolites identification | Serum specimen | +/− of serum metabolites, trimethylamine N-oxide, Carnitine, Choline, and Betaine Odds ratio of serum metabolites, trimethylamine N-oxide, Carnitine, Choline, and Betaine | |
| Trimethylamine N-oxide, Carnitine, Choline, and Betaine in CRC patients using liquid chromatography (LC) tend mass spectrometry (MS) | |||||
| Kim et al. [56] | Experimental group CRC Control group | Untargeted metabolites identification | Untargeted microbiome identification | Stool to extract bacterial extra vesicles (EV) | Distribution (Abundance) of metabolites Fold change difference of the means Distribution of bacterial genera |
| Gas chromatography-time-of-flight mass spectrometry | 16S gene RNA gene sequencing by MiSeq Illumina. | ||||
| Song et al. [57] | Experimental group CRC Control group | Targeted metabolites identification | Fecal specimen | Distribution (Abundance) of metabolites Mean ± SD | |
| Long and short fatty acids using gas chromatography—mass spectrometry | |||||
| Genua et al. [58] | Experimental group TA/TVA HGD CRC Control group | Targeted metabolites identification | Plasma specimen | +/− of the following metabolites, Acetic Acid, Propionic Acid, i-Butyric Acid, Butyric Acid, 2-MethylButyric Acid, i-Valeric Acid, Valeric Acid Mean/IQ | |
| Acetic Acid, Propionic Acid, i-Butyric Acid, Butyric Acid, 2-MethylButyric Acid, i-Valeric Acid, Valeric Acid using gas chromatography | |||||
| D’asheesh et al. [59] | Experimental group CRC Control group | Targeted microbiome identification | Fecal specimen | Fold change and CFU/ml | |
| Lactobacillus acidophilus, Lactobacillus palntarom and Enterococcus faecalis By real-time PCR | |||||
| Coker et al. [60] | Experimental group ADA and CRC Control group | Targeted metabolites identification | Untargeted microbiome identification | Fecal specimen | Distribution (Abundance) of metabolites Fold change Distribution (Abundance) of bacterial species |
| Methyl and ethyl chloroformate (MCF and ECF) derivatized compounds identified previously using gas chromatography coupled to time-of-flight mass spectrometer (GC-TOFMS) analysis | Whole-genome shotgun sequencing of all samples was carried out on an Illumina HiSeq. | ||||
| Goedert et al. [61] | Experimental group CRC Control group | Untargeted metabolites identification | Fecal specimen | Distribution (Abundance) of metabolites | |
| High-performance liquid chromatography/tandem mass spectrometry | |||||
| Sinha et al. [62] | Experimental group CRC Control group | Untargeted metabolites identification | Untargeted microbiome identification | Fecal specimen | Distribution (Abundance) of metabolites Distribution of bacterial genera Odds ratio for both microbiota and metabolites |
| HPLC-GC/MS-MS | 16S rRNA gene sequencing | ||||
| Clos-Garcia et al. [63] | Experimental group ADA, CRC Control group | Targeted metabolites identification | Untargeted microbiome identification | Fecal specimen | Distribution (Abundance) of metabolites Distribution of bacterial genera |
| UHPLC-MS | 16S rRNA gene sequencing | ||||
| Tan et al. [66] | Experimental group CRC Control group | Untargeted metabolites identification | Serum specimen | Distribution (Abundance) of metabolites % | |
| Gas chromatography time-of-flight mass spectrometry (GC−TOFMS)UPLC−QTOFMS | |||||
| Flemer et al. [67] | Experimental group ADA CRC Control group | Untargeted microbiome identification | Fecal specimen and mucosa biopsy | Distribution of bacterial species | |
| 16S rRNA gene sequencing | |||||
| Zeller et al. [68] | Experimental group ADA CRC Control group | Untargeted microbiome identification | Fecal specimen and mucosa biopsy | Distribution (Abundance) of bacterial genera | |
| Whole-genome shotgun sequencing of fecal samples) 16S rRNA gene sequencing (DNA from 48 tissue sample pairs (tumor and healthy mucosa) and 129 fecal samples | |||||
| Zackular et al. [69] | Experimental group ADA CRC Control group | Untargeted microbiome identification | Fecal specimen | Distribution (Abundance) of bacterial genera | |
| 16S rRNA gene sequencing analysis | |||||
| Ohigashi et al. [22] | Experimental group ADA CRC Control group | Targeted metabolites identification | Targeted microbiome identification | Fecal specimen | Distribution (Abundance) of metabolite. Bacterial counts |
| Organic acids, identification from stools using high-performance liquid chromatography system. | Clostridium leptum, Bacteroides fragilis, Bifidobacterium, Atopobium, Prevotella, Clostridium difficile, Clostridium perfringens, Lactobacillus casei, Lactobacillus gasseri, Lactobacillus plantarum, Lactobacillus reuteri, Lactobacillus ruminis, Lactobacillus sakei, Lactobacillus brevis, Lactobacillus fermentum, Lactobacillus fructiborans Enterobacteriaceae, Enterococcus, Staphylococcus, Pseudomonas using real-time PCR | ||||
| Chen et al. [70] | Experimental group ADA Control group | Untargeted metabolites identification | Untargeted microbiome identification | Fecal specimen | Abundance/distribution and concentration of metabolite. Bacterial species distribution/abundance Fold-change in gene expression of bacterial species producing specific metabolites. |
| Ion chromatography and ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). | 16S rRNA gene sequencing analysis followed by real-time PCR to identify bacteria that produced specific metabolites | ||||
| Targeted microbiome identification | |||||
| Real-time PCR analysis, butyrate-producing bacteria, determined by the presence of the butyryl-coenzyme-A-CoA transferase (bcoA) gene, secondary bile acid-producing bacteria, determined by the presence of the Bile acid 7α-dehydroxylation (baiCD) gene, conjugated linoleic acid-producing bacteria, determined by the presence of the plasminogen activator inhibitor 1(pai-1) gene, plasmid-encoded cfr gene (clbA) gene and the polypeptide outer membrane usher protein (afaC) gene of the afa-1 operon were used to detect Putative inactive phenolphthiocerol synthesis polyketide synthase type I (pks1) bacteria and afa-1 adhesin-expressing diffusely adhering Escherichia coli (DAEC), respectively For F. nucleatum 16S rRNA gene | |||||
| Eklöf et al. [71] | Experimental group ADA/dysplasia CRC Control group | Targeted microbiome identification | Fecal specimen | +/− of clbA and afaC +, F. nucleatum bacteria | |
| qPCR clbA gene colibactin-producing bacteria, diffusely adherent Escherichia coli harboring the afa-1 operon, and F. nucleatum | |||||
| Gao et al. [72] | Experimental group ADA CRC Control group | Untargeted microbiome identification | Fecal specimen | Distribution (Abundance) of bacterial species | |
| 16S rRNA gene sequencing analysis | |||||
| Yusuf et al. [73] | Experimental group CRC Control group | Targeted metabolites identification | Fecal specimen | +/− absence of acetate, propionate and butyrate acids | |
| Acetate, propionate and butyrate acids by gas chromatography | |||||
| Weir et al. [74] | Experimental group CRC Control group | Untargeted metabolites identification | Untargeted microbiome identification | Fecal specimen | Distribution (Abundance) of bacterial species, % abundant, fold change Distribution (abundance) |
| Gas chromatography-mass spectrometry (GC-MS) | 16S rRNA gene sequencing analysis | ||||
| Targeted metabolites identification | |||||
| Gas chromatography-mass spectrometry (GC-MS) | |||||
| Yang et al. [75] | Experimental group CRC Control group | Untargeted metabolites identification | Untargeted microbiome identification | Fecal specimen | Distribution (Abundance) of bacterial species, |
| Gas chromatography-mass spectrometry (GC-MS) | 16S rRNA gene sequencing analysis | ||||
| Author | Comparison Group | Bacterial or Metabolite Markers | Performance to Detect ADA or CRC | Identification Technique | |
|---|---|---|---|---|---|
| AUC (CI 95%) | Sen/Spec | ||||
| Sun et al. [26] | ADA vs. HC | 3 metabolites IPA IALD Indole/Trap ratio | ND | ND | 16S rRNA gene sequencing. Ultraperformance liquid chromatography coupled to tandem mass spectrometry. |
| ADA vs. HC | 4 metabolites Skatole IALD I3CA Indoles | ND | ND | ||
| CRC vs. HC | 10 Bacteria Bacteroides Bacilli Clostidales_Incertae_Sedis XI Clostridia Fusobacteria Verrucomicrobia Corynebacteriacea Enterobacteriacea 5 metabolites KYN IPA IALD I3CA Indole/Trap ratio | ND | ND | ||
| Kim et al. [50] | AD vs HC | 24 metabolites Endocannabinoid N acetyl-cadverine Bilirubin ZZ Lionleoyl ethanolamide Oleoyl ethanolamide Palmitoyl ethanolamide 3-Hydroxy-palmitate Myristoleate Palmitoleate 1-Linoleoyl-GPE 1-Palmitioyl -GPE Secondary bile acid 3b-Hydroxy-5-cholenoic acid Deoxycholate Polyunsaturated fatty acid Docosahexaenoate Docosapentaenoate Hexadecadienoate Sphingolipid N-palmitoyl-saphinganine Hexadecasphinganine Sphinganine Piperine 3,7-Dimethyl-urate | ND | ND | UPLC-MS/MS platform |
| CRC vs. HC | 8 metabolites Polyunsaturated fatty acid Docosahexaenoate Docosapentaenoate Hexadecadienoate Sphingolipid N-palmitoyl-saphinganine Hexadecasphinganine Sphinganine Piperine 3,7-Dimethyl-urate | ND | ND | ||
| Nugent et al. [52] | ADA vs. HC | 23 metabolites Galactose, 13,14-dihydro-15-keto-PGE2, 5-oxoproline, 2,4-diaminobutyric acid, Pentadecanoic acid, 5-hydroxyindoleacetic acid, Phosphoric acid, 2-aminoethanol, Dihydroceramide, Ornithine, linoleic acid, Petroselinic acid, LysoPC (18:2(9Z,12Z)), Myo-inositol, Diketogulonic acid, Prostaglandin E2, Methionine, 2-aminobutyric acid, Oleamide, Glycine, Maltitol, 2-phenylglycine, 2-phenylacetamide, N6-acetyl-L-lysine | ND | ND | Liquid chromatography and gas chromatography time of flight mass spectrometry |
| Chang et al. [53] | CRC vs. HC | 18 bacteria Parvimonas micra Fusobacterium nucleatum Clostridium saccharoperbutylacetonicum Clostridium beijerinckii Eubacterium celluloslvens Lachnoclostridium phytofermentans Clostridium butyricum Herbiirix luporum Balcillus cereus Blautia sp. SCOSB48 Anaerobutyrucium hallii Lachnospiraceae bacterium Choco86 Eubacterium eligens Blautia hansenii Longibaculum SPKGMB06250 Clostridum sporogenes Faecalibacterium prausnitizi Anaerostipes hardus | ND | ND | Whole-genome shotgun sequencing |
| Guertin et al. [54] | CRC vs. HC | 1 metabolite Serum choline | ND | ND | Liquid chromatography (LC) tandem mass spectrometry (MS) |
| Kim et al. [56] | CRC vs. HC | 2 Bacteria Solanum melongena, Collinsella | 95% | ND | 16S rRNA gene sequencing Gas chromatography-time-of-flight mass spectrometry |
| 2 metabolites Leucine and Oxalic acid | 92% | ND | |||
| Both bacteria+ metabolites Solanum melongena, Collinsella, Leucine and Oxalic acid | 100% | ND | |||
| Song et al. [57] | CRC vs. HC | 4 metabolites Monounsaturated fatty acid (MUFAs), Oleic acid, ω-6-polyunsaturated fatty acids (ω-6 PUFAs), and Linoleic acid | ND | ND | Gas chromatography-mass Spectrometry |
| Genua et al. [58] | ADA vs. CRC | 1 metabolite 2-MethylButyric acid | Gas chromatography | ||
| CRC vs. HC | 4 metabolites Acetic acid, Propnic acid, i-Valeric, and Valeric acid | ND | ND | ||
| D’asheesh et al. [59] | CRC vs. HC | 3 Bacteria Lactobacillus acidophilus, Lactobacillus palntarom, and Enterococcus faecalis | ND | ND | Real-time PCR |
| Coker et al. [60] | ADA vs. CRC | 6 bacteria Roseburia inulinivorans Xanthmonas perforans Fusobacterium nucleatum Eiknella corrodens Parvimonas micra Peptostreptococcus anaerobius 11 metabolites 2-Hydroxy butyric acid Gamma Aminobutyric acid L-alanine L-Aspartic acid Norvaline Orinthine Oxoadipic acid Oxoglutaric acid Palmitoleic acid Pimelic acid | Only bacteria 94.17% (91.5–96.83) | ND | Whole-genome shotgun sequencing Gas chromatography coupled to time-of-flight mass Spectrometer (GC-TOFMS) |
| ADA vs. HC | 14 bacteria Roseburia inulinivorans Xanthmonas gardneri Fusobacterium nucleatum Prevotella intermedia Peptostreptococcus stomatis Sutterella parviruba 4 metabolites Alpha-Linoleici acid L-Homoserine Phenylacetic acid Phenyllactic ac | Only bacteria 87.59% (83.58, 91.6%) | ND | ||
| CRC vs. HC | 14 bacteria Eubacteria cellulosolvens Lachinospiraceae_bacterium-3-1-57FAA-CT1 Clostridium bolteae Streptococcus tigurinus Xanthmonas gardneri Eikenella corrodens Oscillibacter valericigens Actinomyces viscosus Synergistes_sp_1_syn1 Clostridium symbiosum Prevotella intermedia Slackia exigua Prevotella nigrescens Porphymonas gingivalis 2 metabolites L-Asparagine Phenyllactic acid | Both 14 bacteria and 2 metabolites 93.7% (91.07, 96.42%) | ND | ||
| Goedert et al. [61] | CRC vs. HC | 10 metabolites 3-Dehydrocarnitine, p aminobenzoate (PABA) α-Tocopherol, γ-Tocopherol, Pterin, N-2-Furoyl-glycine, p-Hydroxybenzaldehyde, Sitostanol, Conjugated linoleate-18-2N7, Palmitoyl-sphingomyelin, Mandelate | 77% | ND | High-performance liquid chromatography/tandem mass spectrometry |
| Sinha et al. [62] | CRC vs. HC | 4 Bacteria Fusobacterium, g-Porphyromonas, Clostridia, Lachnospiraceae 5 metabolites p-hydroxy-benzaldehyde, Palmitoyl-sphin-gomyelin p-aminobenzoate, Conjugated linoleate, and Mandelate | ND | ND | 16S rRNA gene sequencing HPLC-GC/MS-MS |
| Clos-Garcia et al. [63] | ADA vs. H | 1 metabolite Triacylglycerol | ND | ND | 16S rRNA gene sequencing UHPLC-MS |
| ADA vs. CRC | 4 Bacteria Streptococcus Parvvimonas Coriobacteriaceae Adlercreutzia 3 metabolites cholesteryl esters, sphingolipids, Glycerophospatidylcholine | ND | ND | ||
| CRC vs. HC | 7 Bacteria Fusobacterium, Streptococcus, Parvimonas, Coprococcus, Blatia, Clostridum, Staphylococcus 3 metabolites Cholesteryl esters, sphingolipids, Glycerophospatidylcholine | ND | ND | ||
| Tan et al. [66] | CRC vs. HC | 72 metabolites This involved the following categories: Tricarboxylic acid (TCA) cycle, urea cycle, glutamine, fatty acids, and gut flora metabolism Tan et al. [66] | ND | ND | Gas chromatography time-of-flight mass spectrometry (GC−TOFMS) UPLC−QTOFMS |
| Flemer et al. [67] | CRC vs. HC | 6 Bacteria Bacteroides Roseburia Ruminococcus Oscillibacter Lachinospiraceae incertae Coporoccus | 87% | ND | 16S rRNA gene sequencing |
| Zeller et al. [68] | CRC vs. HC | 2 Bacteria Fusobacterium nucleatum subsp. vincentii and Fusobacterium nucleatum subsp. animalis | 85% (84–87%) | ND | Whole-genome shotgun sequencing/16S rRNA gene sequencing |
| Zackular et al. [69] | ADA vs. HC | 6 Bacteria Fusobacterium, Porphyromonas, Lachnospiraceae, Enterobacteriaceae, Bacteroides, Lachnospiraceae Clostridiales | 79.8% (68.7–90.8%) | ND | 16S rRNA gene sequencing |
| ADA vs. CRC | 4 Bacteria Fusobacterium, Porphyromonas, Parasutterella Pacscolarctobacterium | 82.3% (72.2–92.3%) | ND | ||
| CRC vs. HC | 6 Bacteria Fusobacterium, Porphyromonas, Lachnospiraceae, Enterobacteriaceae, Bacteroides, Lachnospiraceae and Clostridiales | 83.9% (74–93.8%) | ND | ||
| Ohigashi et al. [22] | ADA vs. CRC | 3 Bacteria Clostridium leptum, Bacteroides fragilis, Staphylococc | ND | ND | Real-time PCR Liquid chromatography system |
| CRC vs. HC | 7 Bacteria C. coccoides, C. leptum, B. fragilis, Bifidobacterium, Atopobium, Enterobacteriaceae, Staphylococcu 4 Metabolites Acetic acid, Propionic acid, Butyric acid, and Valeric acid | ND | ND | ||
| Chen et al. [70] | ADA vs. HC | 1 Bacterium Bacteroidete 3 Metabolites Acetic acid, butyric acid, and t10, c12-CLA | Both 90% (70–90%) | ND | 16S rRNA gene sequencing analysis followed by real-time PCR. Ion chromatography and ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). |
| Eklöf et al. [71] | ADA/dysplasia vs. CRC | 1 Bacterium F. nucleatum | 73.7% | 84.6% and 63.1% | Real-time PCR |
| Gao et al. [72] | ADA vs. HC and CRC vs. HC | 18 Bacteria Rhodococcus, Anaerostipes, Escherichia_Shigella, Akkermansia, Gemella, Clostridium_XVIII, Alkaliphilus Paenibacillus, Enterococcus, Fusobacterium, Fusicatenibacter, Blautia Porphyromonas, Faecalibacterium, Parvimonas, Peptostreptococcus, Clostridium_IV Bacillus | ADA vs. HC 61.6% (52–71%) CRC vs. HC 85.8% (78–93%) | ADA vs. HC 83.6% and 39% CRC vs. HC 66.7% and 98% | 16S rRNA gene sequencing |
| Yusuf et al. [73] | CRC vs. HC | 3 Metabolites Acetate, propionate and butyrate acids | ND | ND | Gas Chromatography |
| Weir et al. [74] | CRC vs. HC | 18 Bacteria Bacteroides finegoldii, Bacteroides intestinalis, Prevotella copri, Prevotella oris, Ruminococcus obeum, Dorea formicigenerans, Lachnobacterium bovis, Lachnospira pectinoschiza, Pseudobutyrivibrio ruminis, Bacteroides capillosus, Ruminococcus albus, Dialister invisus, Dialister pneumosintes, Megamonas hypermegale, Acidaminobacter unclassified, Phascolarctobacterium unclassified, Citrobacter farmer, Akkermansia muciniphila, | ND | ND | 16S rRNA gene sequencing analysis Gas chromatography—mass spectrometry (GC-MS) |
| 20 Metabolites Alanine, Glutamate, Glycine, Aspartic acid, Leucine, Lysine, Proline, Threonine, valine, Phenylalanine, Benzeneacetic acid, Propionic acid, pantothenic acid, Cholesterol derivatives, Oleic acid, Linoleic acid, Elaidic acid, Glycerol, Monooleoylglycerol, Ursodeoxycholic acid | ND | ND | |||
| Yang et al. [75] | CRC vs. HC | 13 Bacteria Escherichia-Shigella, Parvimonas, Fusobacterium, CFT112H7_norank, Porphyromonas. Firmicutes, Clostridiales, Clostridia, Lachnospiraceae, Ruminococcaceae, Selenomonadales, Negativicutes, and Faecalibacterium | ND | ND | Gas chromatography—mass spectrometry (GC-MS) 16S rRNA gene sequencing analysis |
| 2 metabolites Cadaverine putrescine | Only metabolites, each one alone: 74% 67.2 | ND | |||
| Author | Recruitment/5 | Examiner/2 | Methodology/5 | Outcomes/2 | Missing Data/7 | Statistical Analysis/5 | Results/2 | Overall Score/28 | Overall Score 100% |
|---|---|---|---|---|---|---|---|---|---|
| Zhen Sun et al. [26] | 4 | 0 | 3 | 2 | 7 | 3 | 2 | 21 | 77.7 |
| Kim et al. [50] | 4 | 0 | 5 | 2 | 7 | 5 | 2 | 25 | 92.5 |
| Nugent et al. [52] | 4 | 0 | 2 | 2 | 7 | 2 | 2 | 19 | 70.3 |
| Chang et al. [53] | 0 | 0 | 1 | 2 | 7 | 3 | 1 | 14 | 51.8 |
| Guertin et al. [54] | 1 | 2 | 5 | 2 | 7 | 5 | 2 | 24 | 88.8 |
| Kim et al. [56] | 4 | 0 | 4 | 2 | 7 | 5 | 2 | 24 | 88.8 |
| Song et al. [57] | 4 | 0 | 3 | 2 | 7 | 3 | 1 | 20 | 74.1 |
| Genua et al. [58] | 2 | 0 | 5 | 2 | 6 | 5 | 1 | 20 | 74.1 |
| D’asheesh et al. [59] | 3 | 0 | 3 | 2 | 4 | 2 | 0 | 14 | 51.8 |
| Coker et al. [60] | 4 | 0 | 5 | 2 | 7 | 5 | 2 | 25 | 92.5 |
| Goedert et al. [61] | 2 | 1 | 2 | 2 | 6 | 2 | 1 | 16 | 59.3 |
| Sinha et al. [62] | 2 | 0 | 5 | 2 | 7 | 5 | 2 | 23 | 85.2 |
| Clos-Garcia et al. [63] | 1 | 0 | 5 | 2 | 7 | 5 | 2 | 23 | 81.1 |
| Tan et al. [66] | 4 | 0 | 5 | 2 | 7 | 3 | 1 | 22 | 81.1 |
| Flemer et al. [67] | 4 | 0 | 5 | 2 | 7 | 5 | 2 | 25 | 92.6 |
| Zeller et al. [68] | 4 | 0 | 5 | 2 | 7 | 5 | 2 | 25 | 92.6 |
| Zackular et al. [69] | 4 | 0 | 5 | 1 | 6 | 3 | 2 | 21 | 77.8 |
| Ohigashi et al. [22] | 4 | 0 | 3 | 2 | 6 | 1 | 1 | 17 | 62.9 |
| Chen et al. [70] | 4 | 0 | 3 | 2 | 6 | 4 | 1 | 20 | 74.1 |
| Eklöf et al. [71] | 2 | 0 | 3 | 2 | 6 | 3 | 1 | 17 | 62.9 |
| Gao et al. [72] | 3 | 0 | 2 | 2 | 7 | 2 | 1 | 17 | 62.9 |
| Yusuf et al. [73] | 3 | 0 | 1 | 2 | 6 | 2 | 1 | 15 | 55.5 |
| Weir et al. [74] | 4 | 0 | 2 | 2 | 7 | 2 | 1 | 18 | 66.7 |
| Yang et al. [75] | 4 | 0 | 5 | 2 | 7 | 3 | 2 | 23 | 85.2 |
| a. Untargeted Microbiome Identification | ||||||||||
| Study (Appraisal Quality) | Increased in ADA vs. HC | Increased in CRC vs. ADA | Increased in CRC vs. HC | |||||||
| Nugent et al. [52] 66.6% (L) | Bifidobacterium sp. Eubacteria | |||||||||
| Chang et al. [53] 51.8% (L) | Streptococcus gallolyticus, Haemophillus parainfluenza, Dialister sp. Marseille-P5638, Ruthenibacterium lactatiformans | |||||||||
| Kim et al. [56] 88.8% (H) | Bifidobacterium, Collinsella, Blautia, Lachnoclostridium Lachnospiraceae, Dorea Eubacterium coprostanoligenes group Ruminococcaceae-Ruminococcus Faecalibacterium, Subdoligranulum Catenibacterium, Parvimonas Ruminiclostridium, Enterobacter Diaphorobacter | |||||||||
| Sinha et al. [62] 85.2% (H) | Fusobacterium, Porphyromonas Clostridia, Lachnospiraceae | |||||||||
| Flemer et al. [67] 92.6% (H) | Bacteroides, Roseburia Ruminococcus, Oscillibacter Porphyromonas, Peptostreptococcus, Parvimonas, Fusobacterium | |||||||||
| Zeller et al. [68] 92.6% (H) | Fusobacterium nucleatum subsp. vincentii Fusobacterium nucleatum subsp. Animalis Fusobacterium nucleatum subsp. nucleatum Fusobacterium nucleatum subsp. polymorphum Porphyromonas asaccharolytica Prevotella nigrescens Peptostreptococcus stomatis Parvimonas sp. Parvimonas micra Olsenella uli Parvimonas sp. Streptococcus anginosus | Fusobacterium nucleatum subsp. vincentii Fusobacterium nucleatum subsp. Animalis Fusobacterium nucleatum subsp. nucleatum Pseudoflavonifractor capillosus Fusobacterium nucleatum subsp. polymorphum Porphyromonas asaccharolytica Ruminococcaceae bacterium Prevotella nigrescens Peptostreptococcus stomatis Leptotrichia hofstadii Parvimonas sp. Parvimonas micra Bacteroides fragilis Bilophila wadsworthia Neisseria sp. Campylobacter rectus Selenomonas sputigena Leptotrichia buccalis Clostridium hylemonae Clostridium symbiosum | ||||||||
| Zackular et al. [69] 77.8% (H) | Ruminococcaceae Clostridium Pseudomonas Porphyromonadaceae | Fusobacterium Bacteroides Phascolarctobacterium Porphyromonas | Fusobacterium Porphyromonas Lachnospiraceae Enterobacteriaceae | |||||||
| Chen et al. [70] 74.1 (H) | Bacteroides Escherichia Faecalibacterium Citrobacter | |||||||||
| Gao et al. [72] 62.9% (L) | Bacillus cereus Bacillus thuringiensis Bacillus amyloliquefaciens Cronobacter sakazakii | Alcanivorax hongdengensis Burkholderia mallei Clostridium ramosum Coprobacillus sp. Fusobacterium sp. | Streptococcus intermedius Peptostreptococcus stomatis Parvimonas micra F. nucleatum | |||||||
| Weir et al. [74] 66.7% (L) | Acidaminobacter Citrobacter farmer Akkermansia muciniphila | |||||||||
| Yang et al. [75] 85.2% (H) | Enterobacteriaceae Fusobacterium | |||||||||
| Increased in ADA vs. HC | ||||||||||
| Overlapping microbial markers | No common microbial markers 4 studies [52,69,70,72] | |||||||||
| Level of evidence | Conflicting | |||||||||
| Increased in CRC vs. ADA | ||||||||||
| Overlapping microbial markers | Fusobacterium sp. 3 studies [68,69,72] | Porphyromonas 2 studies [68,69] | ||||||||
| Level of evidence | Strong | Strong | ||||||||
| Increased in CRC vs. HC | ||||||||||
| Overlapping microbial markers | Lachnospiraceae-Lachnoclostridium 3 studies [56,62,69] | Ruminococcaceae-Ruminococcus 4 studies [56,62,67,68] | Parvimonas Parvimonas micra 4 studies [56,67,68,72] | Enterobacteriaceae 2 studies [69,75] | Fusobacterium sp. 5 studies [62,67,68,69,75] | Bacteroides 2 studies [67,68] | Peptostreptococcus sp. 2 studies [67,72] | Clostridia sp. C. hylemonae C. symbiosum 2 studies [62,68] | Porphyromonas 4 studies [62,67,68,69] | Streptococcus sp. S. gallolyticus, S. intermedius 2 studies [53,72] |
| Level of evidence | Strong | Strong | Strong | Strong | Strong | Strong | Strong | Strong | Strong | Limited |
| b. Targeted microbiome identification | ||||||||||
| Study (Appraisal quality) | Increased in ADA vs. HC | Increased in CRC vs. ADA | Increased in CRC vs. HC | |||||||
| D’asheesh et al. [59] 51.8 (L) | Bifidobacterium sp. Eubacteria | Enterococcus faecalis | ||||||||
| Clos-Garcia et al. [63] 81.1% (H) | Staphylococcus and Parvimonas | Fusobacterium, Staphylococcus and Parvimonas | ||||||||
| Ohigashi et al. [22] 62.9% (L) | C. difficile C. perfringens, Pseudomonas *,1 | |||||||||
| Eklöf et al. [71] 62.92% (L) | F. nucleatum | |||||||||
| Increased in ADA vs. HC | ||||||||||
| Overlapping microbial markers | Only one study was reported. [12] | |||||||||
| Level of evidence | NO | |||||||||
| Increased in CRC vs. ADA | ||||||||||
| Overlapping microbial markers | Only one study was reported. [63] | |||||||||
| Level of evidence | NO | |||||||||
| Increased in CRC vs. HC | ||||||||||
| Overlapping microbial markers | Fusobacterium sp. 2 studies [63,71] | |||||||||
| Level of evidence | Moderate | |||||||||
| c. Untargeted Metabolites Identification | ||||||||||
| Study (Appraisal quality) | Increased in ADA vs. HC | Increased in CRC vs. HC | ||||||||
| Kim et al. [56] 92.5% (H) | Endocannabinoid N acetyl-cadverine Bilirubin ZZ Lionleoyl ethanolamide Oleoyl ethanolamide Palmitoyl ethanolamide 3-Hydroxy-palmitate Myristoleate Palmitoleate 1-Linoleoyl-GPE 1-Palmitioyl -GPE | Polyunsaturated fatty acid Docosahexaenoate Docosapentaenoate Hexadecadienoate | ||||||||
| Secondary bile acid 3b-Hydroxy-5-cholenoic acid Deoxycholate | Sphingolipid N-palmitoyl-saphinganine Hexadecasphinganine Sphinganine Piperine 3,7-Dimethyl-urate | |||||||||
| Nugent et al. [52] 66.7% (L) | The inflammatory metabolite prostaglandin E2 | |||||||||
| Kim et al. [50] 88.8% (H) | Aminoacids Leucine Isoleucine Alanine Lysine Tyramine Aminoisobutyric acid | Amino alcohol Ethanolamine Aromatic alcohol Phenol | ||||||||
| Carboxylic acid Furoic acid Succinic acid Oxalic acid | Fatty acid Butanoic acid Hexanoic acid Palmitic acid Oleic acid | |||||||||
| Godert et al. [61] 59.3% (L) | Heme-related molecules Heme Z-18565 X_19549 | Cofactors. and vitamin α-Tocopherol γ-Tocopherol Pterin | ||||||||
| Xenobiotics 4-Acetamidophenol 2-Hydroxyacetaminophen sulfate 3-Cystein-S-YL-acetaminophen p-Acetamidophenylglucuronide Para-aminobenzoic acid (PABA) N-2-Furoyl-glycine Sitostanol p-Hydroxybenzaldehyde Mandelate | Peptides/Aminoacids Histidine Cis-Urocanate Tryptophyl-glycine Leucyl-tryptophan Alanyl-histidine Histidyl-glycine Tyrosylglutamine Histidyl-alanine Valyl-aspartate Pyro-glutamyl-glycine Alanyl-leucine Alanyl-tryptophan Histidylphenylalanine Leucyl-glutamate Leucyl-serine α-Glutamyl-valine Prolyl-alanine Valyl-histidine | |||||||||
| Lipids Palmitoyl-sphingomyelin Conjugated linoleate-18-2N7 3-Dehydrocarnitine | ||||||||||
| Shina et al. [62] 85.5% (H) | Palmitoyl_Sphingomyelin p_Hydroxybenzaldhyde | |||||||||
| Tan et al. [66] 81.1% (H) | Fatty acid metabolism β-hydroxybutyrate betaine Glycerol Oleamide Oleic acid Erythrotetrofuranose Carnitine (18:1) Linolic acid Acetyl carnitine Elaidic acid 3-oxodecanoic acid Palmitic acid | valine, leucine, and isoleucine degradation Allisoleucine | Arginine and proline metabolism Creatinine | |||||||
| Purine nucleotide synthetics Xanthosine | Cystine & methionine metabolism Cystine | Carbohydrate metabolism Threitol | ||||||||
| Phospholipid metabolism Sphinganine CPA(18:0/0:0) | Glutathione metabolism 2-hydroxybutyric acid 2-aminobutanoic acid TCA cycle Pyruvate Vitamin B6 metabolism Glycolaldehyde | Others Tetrahydrogestrinone Allyl isothiocyanate Proline | ||||||||
| Weir et al. [74] 66.7% (L) | Aminoacids Alanine Glutmate Glycine Aspartic acid Leucine Lysine Proline Serine Threonine Valine Phenylalanine | Carboxylic acids Beneneacetic acid Propionic acid Mysteric acid Pantothenic acid | ||||||||
| Steroids Cholesterol derivative | ||||||||||
| Yang et al. [75] 85.2% (H) | 4-Methylvaleric acid 9-(2-Carboxyethyl)-2,2,4,4-tetramethyl-1,2,3,4-tetrahydro-gamma-carboline Adenosine Butanoic acid d-2-Aminobutyric acid DL-Ornithine D-Proline, n-propoxycarbonyl-, hexadecyl ester Heptanedioic acid Heptanoic acid Hexane, 2,5-dimethyl L-5-Hydroxytryptophan L-Lysine L-Tryptophan L-Norleucine L-Norvaline Pentanoic acid N-Acetyl-D-glucosamine Cadaverine | |||||||||
| Increased in ADA vs. HC | ||||||||||
| Overlapping metabolite markers | No common metabolites 5 studies [50,52,56,74,75] | |||||||||
| Level of evidence | Conflicting | |||||||||
| Increased in CRC vs. HC | ||||||||||
| Overlapping metabolite markers | Palmitoyl-sphingomyelin 2 studies [61,62] | Proline 2 studies [66,74] | ||||||||
| Level of evidence | Moderate | Moderate | ||||||||
| d. Targeted metabolites identification | ||||||||||
| Study (Appraisal Quality) | Increased in ADA vs. HC | Increased in CRC vs. ADA | Increased in CRC vs. HC | |||||||
| Zhen Sun et al. [26] 77.7% (H) | Kynurenin(KYN) Indole-3-aldehyde (IALD) and Indole-3-carboxylic acid (I3CA) The ratio of KYN to Trp (KYN/Trp ratio) | Kynurenin(KYN) Indole-3-aldehyde (IALD) and Indole-3-carboxylic acid (I3CA) The ratio of KYN to Trp (KYN/Trp ratio) | ||||||||
| Guertin et al. [54] 88.8% (H) | Serum choline | |||||||||
| Song et al. [57] 74.1% (L) | Monounsaturated fatty acids (MUFAs) C18:1ω-9 Oleic acid ω-6 polyunsaturated fatty acids (PUFAs) C18:2ω-6 Linoleic acid | |||||||||
| Genua et al. [58] 74.1% (L) | 2-MethylButyric Acid Acetic Acid Propionic acids | |||||||||
| Coker et al. [60] 92.5% (H) | Phenyllactic acid, Phenylacetic acid, L-Phenylalanine, L-Valine, L-Alpha-aminobutyric acid, L-Proline, L-Alanine Oxoglutaric acid, L-Isoleucine, Gamma-Aminobutyric acid, L-Leucine, Glycine, L-Methionine, L-Tyrosine, L-Aspartic acid, Butyric acid, Glutathione, Succinic acid, 2-Hydroxybutyric acid, Malic acid, 3-Aminoisobutanoic acid, Ornithine, Beta-Alanine, Myristic acid, Oxoadipic acid, Alpha-Linolenic acid, L-Serine, Nicotinic acid, Linoleic acid, Pelargonic acid, Pyroglutamic acid, Glutaric acid, Hexanoic acid, L-Homoserine, 5-Dodecenoic acid, Pimelic acid | L-alanine, glycine L-proline L-aspartic acid L-valine L-leucine L-serine myristic acid phenyl lactic acid oxoglutaric acid L-phenylalanine L-alpha-aminobutyric acid phenylacetic acid palmitoleic acid 3-aminoisobutanoic acid norvaline | ||||||||
| Ohigashi et al. [22] 62.9% (M) | Succinic acid | |||||||||
| Yusuf et al. [73] 55.5% (M) | The opposite decrease in Acetate Propionate butyrate acids | |||||||||
| Increased in ADA vs. HC | ||||||||||
| Overlapping microbial markers | Only one study [19] | |||||||||
| Level of evidence | NO | |||||||||
| Increased in CRC vs. ADA | ||||||||||
| Overlapping microbial markers | Only one study [60] | |||||||||
| Level of evidence | NO | |||||||||
| Increased in CRC vs. HC | ||||||||||
| Overlapping microbial markers | No common metabolites 6 studies [26,54,57,60,73,75] | |||||||||
| Level of evidence | Conflicting | |||||||||
| e. Untargeted microbial markers for tumor stages and locations | ||||||||||
| Study (Appraisal Quality) | Microbial Markers in CRC Early Stage I | Microbial Markers in CRC III Stage | Microbial Markers in CRC IV, Late Stage | Microbial Markers in Distal Cancers vs. Proximal Cancers | Microbial Markers in Rectal vs. Proximal Cancers | Microbial Markers in Proximal Cancer | ||||
| Flemer et al. [67] 92.6% (H) | Alistipes Akkermansia Halomonas Shewanella | Alistipes Akkermansia Halomonas Shewanella | Faecalibacterium Blautia Clostridium | |||||||
| Gao et al. [72] 62.9% (M) | Escherichia/Shigella | Bacteroides | Saccharibacteria incertaesedis | Escherichia/Shigella | ||||||
| Microbial markers in CRC early stage I | ||||||||||
| Overlapping microbial markers | Only one study reported. [72] | |||||||||
| Level of evidence | NO | |||||||||
| Microbial markers in CRC III stage | ||||||||||
| Overlapping microbial markers | Only one study reported. [72] | |||||||||
| Level of evidence | NO | |||||||||
| Microbial markers in CRC IV, late-stage | ||||||||||
| Overlapping microbial markers | Only one study reported. [72] | |||||||||
| Level of evidence | NO | |||||||||
| Microbial markers in distal cancers vs. proximal cancers | ||||||||||
| Overlapping microbial markers | No common metabolites Two studies [67,72] | |||||||||
| Level of evidence | Conflicting | |||||||||
| Microbial markers in rectal vs. proximal cancers | ||||||||||
| Overlapping microbial markers | Only one study reported. [67] | |||||||||
| Level of evidence | NO | |||||||||
| Microbial markers in proximal cancer | ||||||||||
| Overlapping microbial markers | Only one study reported. [67] | |||||||||
| Level of evidence | NO | |||||||||
| f. Targeted microbial markers for tumor stages and locations | ||||||||||
| Study (Appraisal Quality) | Microbial Markers in CRC IV, Late Stage | Microbial Markers on Right Side | ||||||||
| Clos-Garcia et al. [63] 81.1% (H) | Bulleidia Fusobacterium Butyrivibrio Peptostreptococcus Staphylococcus Parvimonas Selenomonas | |||||||||
| Ohigashi et al. [22] 62.9% (M) | Clostridium perfringens | |||||||||
| Microbial markers in CRC IV, late-stage | ||||||||||
| Overlapping microbial markers | Only one study reported. [63] | |||||||||
| Level of evidence | NO | |||||||||
| Microbial markers on right side | ||||||||||
| Overlapping microbial markers | Only one study reported. [22] | |||||||||
| Level of evidence | NO | |||||||||
| g. Untargeted metabolite markers for tumor stage and location | ||||||||||
| Study (Appraisal Quality) | Microbial Markers in CRC Late Stage IV vs. Early Stage I | |||||||||
| Tan et al. [66] 81.1% (H) | Beta hydroxybuturate | |||||||||
| Microbial markers in CRC late stage IV vs. early stage I | ||||||||||
| Overlapping microbial markers | Only one study reported. [66] | |||||||||
| Level of evidence | NO | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhhazmi, A.A.; Alhamawi, R.M.; Almisned, R.M.; Almutairi, H.A.; Jan, A.A.; Kurdi, S.M.; Almutawif, Y.A.; Mohammed-Saeid, W. Gut Microbial and Associated Metabolite Markers for Colorectal Cancer Diagnosis. Microorganisms 2023, 11, 2037. https://doi.org/10.3390/microorganisms11082037
Alhhazmi AA, Alhamawi RM, Almisned RM, Almutairi HA, Jan AA, Kurdi SM, Almutawif YA, Mohammed-Saeid W. Gut Microbial and Associated Metabolite Markers for Colorectal Cancer Diagnosis. Microorganisms. 2023; 11(8):2037. https://doi.org/10.3390/microorganisms11082037
Chicago/Turabian StyleAlhhazmi, Areej A., Renad M. Alhamawi, Reema M. Almisned, Hanouf A. Almutairi, Ahdab A. Jan, Shahad M. Kurdi, Yahya A. Almutawif, and Waleed Mohammed-Saeid. 2023. "Gut Microbial and Associated Metabolite Markers for Colorectal Cancer Diagnosis" Microorganisms 11, no. 8: 2037. https://doi.org/10.3390/microorganisms11082037
APA StyleAlhhazmi, A. A., Alhamawi, R. M., Almisned, R. M., Almutairi, H. A., Jan, A. A., Kurdi, S. M., Almutawif, Y. A., & Mohammed-Saeid, W. (2023). Gut Microbial and Associated Metabolite Markers for Colorectal Cancer Diagnosis. Microorganisms, 11(8), 2037. https://doi.org/10.3390/microorganisms11082037





