Genome-Wide DNA Changes Acquired by Candida albicans Caspofungin-Adapted Mutants
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
2.2. DNA-Sequencing and Analysis
3. Results
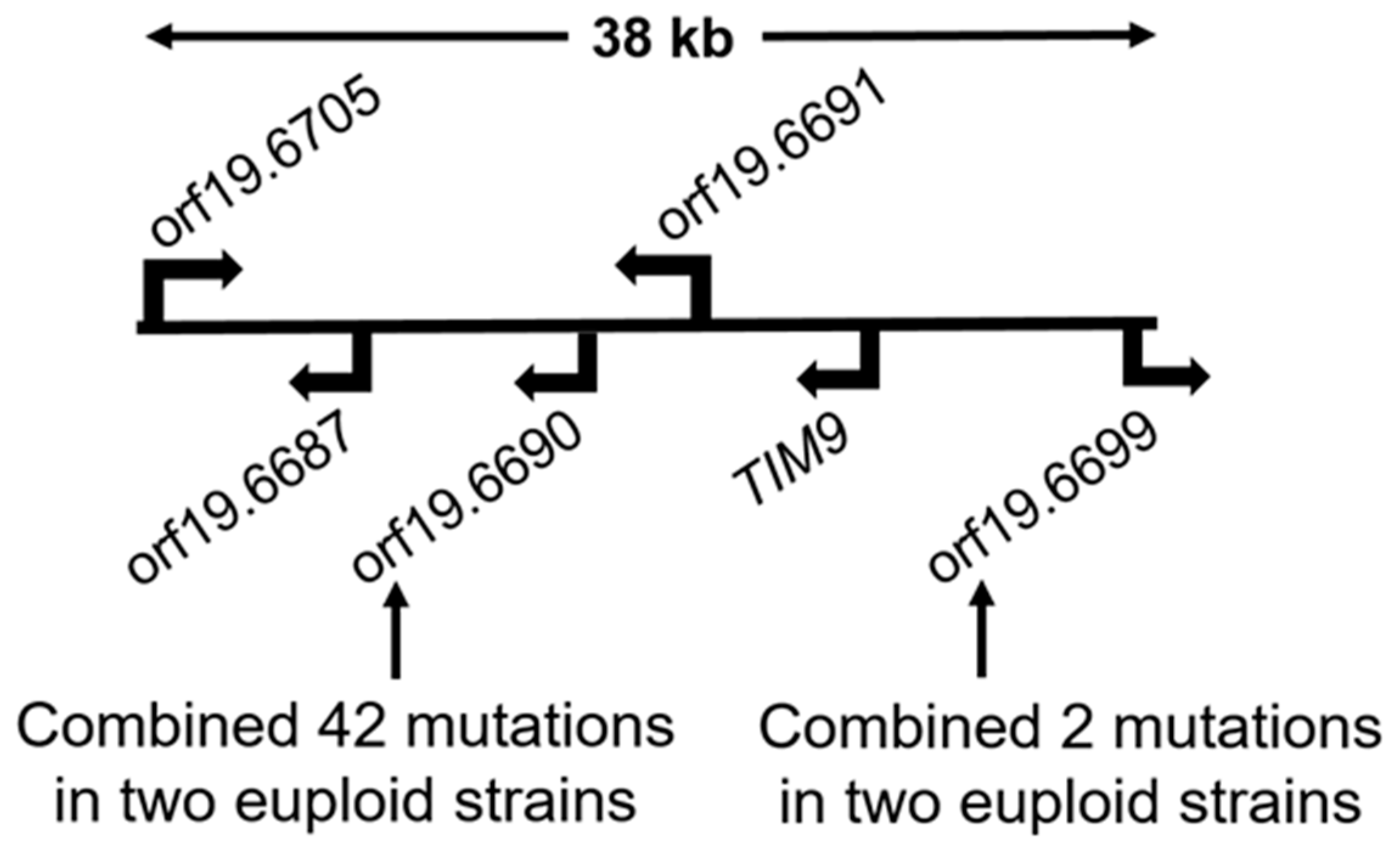
3.1. DNA Changes
| Strain | Description | Phenotype ** | Source |
|---|---|---|---|
| SC5314 | Parental strain, normal diploid | Caspofungin susceptible | A.D. Johnson laboratory [23] |
| SMC60-2-5 | Same as SC5314, but Ch5 monosomy, MTLα. | Caspofungin tolerant | [9] |
| JRCT1 | Parental strain, normal diploid | Caspofungin susceptible | Same as above |
| JMC200-3-4 | Same as JRCT1, but Ch5 monosomy, MTLα. | Caspofungin tolerant | Same as above |
| JMC160-2-5 | Same as JRCT1, no ploidy change. | Same as above | Same as above |
| JMC200-2-5 | Same as JRCT1, no ploidy change. | Same as above | Same as above |
3.2. Comparison of DNA Changes to Previously Reported Expression Changes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arendrup, M.C.; Perlin, D.S. Echinocandin resistance: An emerging clinical problem? Curr. Opin. Infect. Dis. 2014, 27, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.; Cowen, L.E. Genomic approaches to antifungal drug target identification and validation. Annu. Rev. Microbiol. 2022, 76, 369–388. [Google Scholar] [CrossRef] [PubMed]
- Butts, A.; Palmer, G.E.; Rogers, P.D. Antifungal adjuvants: Preserving and extending the antifungal arsenal. Virulence 2017, 8, 198–210. [Google Scholar] [CrossRef]
- Matsumoto, E.; Boyken, L.; Tendolkar, S.H.; McDanel, J.; Castanheira, M.; Pfaller, M.; Diekema, D. Candidemia surveillance in Iowa: Emergence of echinocandin resistance. Diagn. Microbiol. Infect. Dis. 2014, 79, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Coste, A.T.; Kritikos, A.; Li, J.; Khanna, N.; Goldenberger, D.; Garzoni, C.; Zehnder, C.; Boggian, K.; Neofytos, D.; Riat, A.; et al. Fungal infection network of Switzerland (FUNGINOS). Emerging echinocandin-resistant Candida albicans and glabrata in Switzerland. Infection 2020, 48, 761–766. [Google Scholar] [CrossRef]
- Park, S.; Kelly, R.; Kahn, J.N.; Robles, J.; Hsu, M.-J.; Register, E.; Li, W.; Vyas, V.; Fan, H.; Abruzzo, G.; et al. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 2005, 49, 3264–3273. [Google Scholar] [CrossRef]
- Sah, S.K.; Bhattacharya, S.; Yadav, A.; Husain, F.; Ndiaye, A.B.K.T.; Kruppa, M.D.; Hayes, J.J.; Rustchenko, E. Multiple genes of Candida albicans influencing echinocandin susceptibility in caspofungin-adapted mutants. Antimicrob. Agents Chemother. 2022, 66, e00977-22. [Google Scholar] [CrossRef]
- Sah, S.K.; Hayes, J.J.; Rustchenko, E. The role of aneuploidy in the emergence of echinocandin resistance in human fungal pathogen Candida albicans. PLoS Pathog. 2021, 17, e1009564. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, L.; Wakabayashi, H.; Myers, J.; Jiang, Y.; Cao, Y.; Jimenez-Ortigosa, C.; Perlin, D.S.; Rustchenko, E. Tolerance to caspofungin in Candida albicans is associated with at least three distinctive mechanisms that govern expression of FKS genes and cell wall remodeling. Antimicrob. Agents Chemother. 2017, 61, e00071-17. [Google Scholar] [CrossRef]
- Sah, S.K.; Yadav, A.; Rustchenko, E. At least 10 genes on chromosome 5 of Candida albicans are downregulated in concert to control cell wall and to confer adaptation to caspofungin. bioRxiv 2023. [Google Scholar] [CrossRef]
- Ford, C.B.; Funt, J.M.; Abbey, D.; Issi, L.; Guiducci, C.; Martinez, D.A.; Delorey, T.; Li, B.Y.; White, T.C.; Cuomo, C.; et al. The evolution of drug resistance in clinical isolates of Candida albicans. eLife 2015, 4, e00662. [Google Scholar] [CrossRef]
- Ene, J.V.; Bennett, R.J.; Anderson, M.Z. Mechanisms of genome evolution in Candida albicans. Curr. Opin. Microbiol. 2019, 52, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Janbon, G.; Sherman, F.; Rustchenko, E. Appearance and properties of L-sorbose-utilizing mutants of Candida albicans obtained on a selective plate. Genetics 1999, 153, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Rustchenko-Bulgac, E. Variations of Candida albicans electrophoretic karyotypes. J. Bacteriol. 1991, 20, 6586–6596. [Google Scholar] [CrossRef] [PubMed]
- Rustchenko, E. Chromosome instability in Candida albicans. FEMS Yeast Res. 2007, 1, 2–11. [Google Scholar] [CrossRef]
- Sherman, F. Getting started with yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar]
- Kravets, A.; Yang, F.; Bethlendy, G.; Cao, Y.; Sherman, F.; Rustchenko, E. Adaptation of Candida albicans to growth on sorbose via monosomy of chromosome 5 accompanied by duplication of another chromosome carrying a gene responsible for sorbose utilization. FEMS Yeast Res. 2014, 14, 708–713. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Muzzey, D.; Schwartz, K.; Weissman, J.S.; Sherlock, G. Assembly of a phased diploid Candida albicans genome facilitates allele-specific measurements and provides a simple model for repeat and indel structure. Genome Biol. 2013, 14, R97. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Rescheneder, P.; von Haeseler, A. NextGenMap: Fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 2013, 29, 2790–2791. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Vesztrocy, A.W.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef]
- Ahmad, A.; Kabir, M.A.; Kravets, A.; Andaluz, E.; Larriba, G.; Rustchenko, E. Chromosome instability and unusual features of some widely used strains of Candida albicans. Yeast 2008, 25, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Husain, F.; Yadav, A.; Sah, S.K.; Hayes, J.J.; Rustchenko, E. Candida albicans strains adapted to caspofungin due to aneuploidy become highly tolerant under continued drug pressure. Microorganisms 2022, 11, 23. [Google Scholar] [CrossRef]
- Thewes, S. Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryot. Cell 2014, 13, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.E. Hsp90 orchestrates stress response signaling governing fungal drug resistance. PLoS Pathog. 2009, 5, e1000471. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.L.; Qin, L.; Miao, Z.; Grys, B.T.; Diaz, J.D.L.C.; Ting, K.; Krieger, J.R.; Tong, J.; Tan, K.; Leach, M.D.; et al. The Candida albicans transcription factor Cas5 couples stress responses, drug resistance and cell cycle regulation. Nat. Commun. 2017, 8, 499. [Google Scholar] [CrossRef]
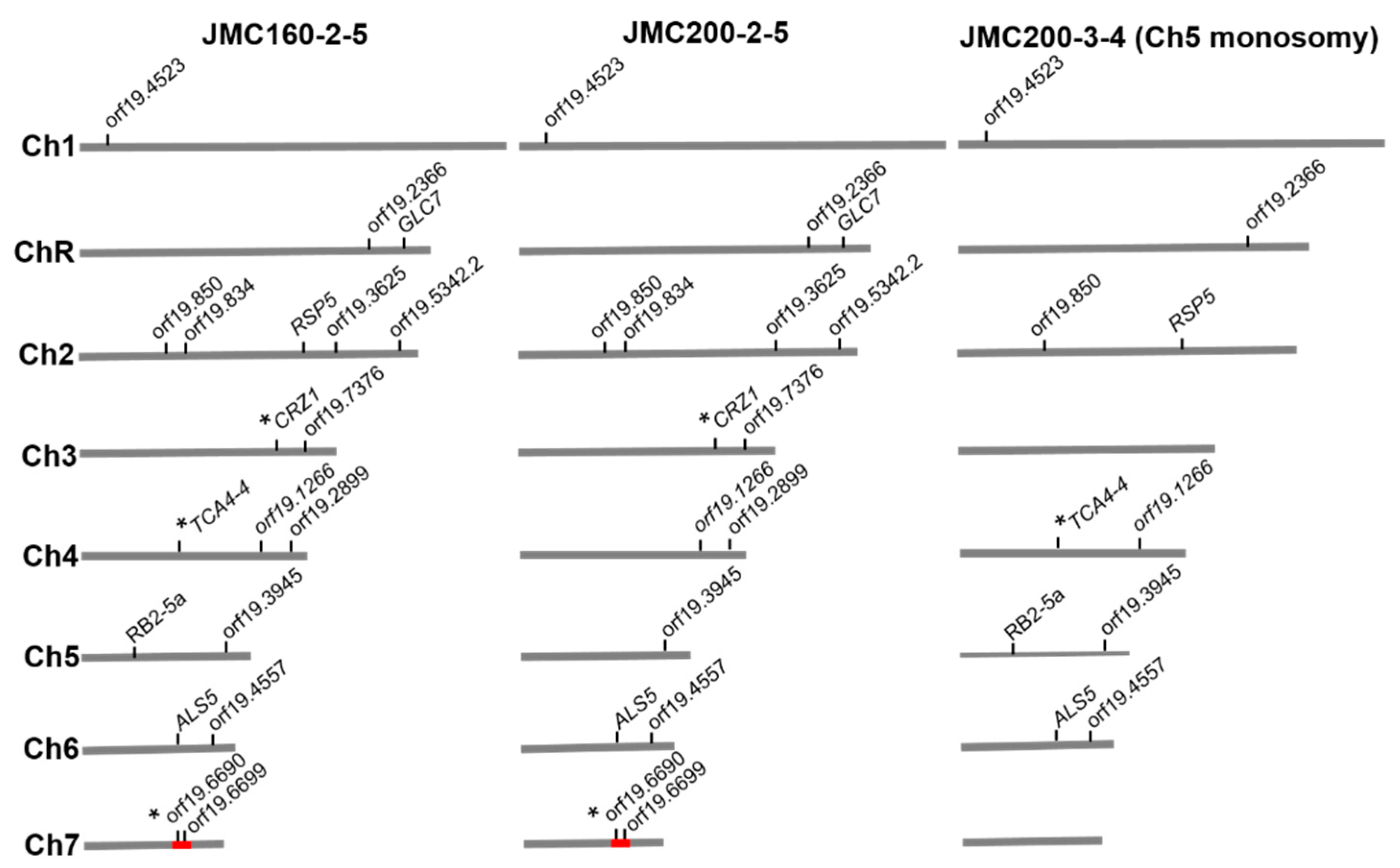
| Euploid | Ch5 Mono | ||||
|---|---|---|---|---|---|
| Gene Name | Gene Identifier | Systematic Name Chromosome | JMC160- 2-5 | JMC200- 2-5 | JMC200- 3-4 |
| orf19.2366 | CR_06990W R | 1 | 1 | 1 | |
| GLC7 | orf19.6285 | CR_07650W R | 1 | 1 | 0 |
| orf19.4523 | C1_02020W 1 | 1 | 1 | 1 | |
| orf19.834 | C2_03910C 2 | 1 | 1 | 0 | |
| orf19.850 | C2_03700W 2 | 1 | 1 | 1 | |
| orf19.3625 | C2_08540C 2 | 1 | 1 | 0 | |
| RSP5 | orf19.3628 | C2_08500W 2 | 2 | 0 | 1 |
| orf19.5342.2 | C2_10650W 2 | 1 | 1 | 0 | |
| CRZ1 | orf19.7359 | C3_05780C 3 | 4 | 3 | 0 |
| orf19.7376 | C3_05950W 3 | 2 | 1 | 0 | |
| Tca4-4 | C4_03210C 4 | 6 | 0 | 2 | |
| orf19.1266 | C4_05800C 4 | 1 | 1 | 1 | |
| orf19.2899 | C4_06360C 4 | 2 | 1 | 0 | |
| orf19.3945 | C5_04610W 5 | 1 | 1 | 1 | |
| RB2-5a | C5_01660C 5 | 1 | 0 | 2 | |
| orf19.4557 | C6_04120C 6 | 0 | 1 | 1 | |
| ALS5 | orf19.5736 | C6_03690W 6 | 1 | 1 | 1 |
| orf19.6699 | C7_03650W 7 | 1 | 1 | 0 | |
| orf19.6690 | C7_03580C 7 | 25 | 17 | 0 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuber, J.; Sah, S.K.; Mathews, D.H.; Rustchenko, E. Genome-Wide DNA Changes Acquired by Candida albicans Caspofungin-Adapted Mutants. Microorganisms 2023, 11, 1870. https://doi.org/10.3390/microorganisms11081870
Zuber J, Sah SK, Mathews DH, Rustchenko E. Genome-Wide DNA Changes Acquired by Candida albicans Caspofungin-Adapted Mutants. Microorganisms. 2023; 11(8):1870. https://doi.org/10.3390/microorganisms11081870
Chicago/Turabian StyleZuber, Jeffrey, Sudisht K. Sah, David H. Mathews, and Elena Rustchenko. 2023. "Genome-Wide DNA Changes Acquired by Candida albicans Caspofungin-Adapted Mutants" Microorganisms 11, no. 8: 1870. https://doi.org/10.3390/microorganisms11081870
APA StyleZuber, J., Sah, S. K., Mathews, D. H., & Rustchenko, E. (2023). Genome-Wide DNA Changes Acquired by Candida albicans Caspofungin-Adapted Mutants. Microorganisms, 11(8), 1870. https://doi.org/10.3390/microorganisms11081870






