Abstract
Molybdenum (Mo) is vital for the activity of a small but essential group of enzymes called molybdoenzymes. So far, specifically five molybdoenzymes have been discovered in eukaryotes: nitrate reductase, sulfite oxidase, xanthine dehydrogenase, aldehyde oxidase, and mARC. In order to become biologically active, Mo must be chelated to a pterin, forming the so-called Mo cofactor (Moco). Deficiency or mutation in any of the genes involved in Moco biosynthesis results in the simultaneous loss of activity of all molybdoenzymes, fully or partially preventing the normal development of the affected organism. To prevent this, the different mechanisms involved in Mo homeostasis must be finely regulated. Chlamydomonas reinhardtii is a unicellular, photosynthetic, eukaryotic microalga that has produced fundamental advances in key steps of Mo homeostasis over the last 30 years, which have been extrapolated to higher organisms, both plants and animals. These advances include the identification of the first two molybdate transporters in eukaryotes (MOT1 and MOT2), the characterization of key genes in Moco biosynthesis, the identification of the first enzyme that protects and transfers Moco (MCP1), the first characterization of mARC in plants, and the discovery of the crucial role of the nitrate reductase–mARC complex in plant nitric oxide production. This review aims to provide a comprehensive summary of the progress achieved in using C. reinhardtii as a model organism in Mo homeostasis and to propose how this microalga can continue improving with the advancements in this field in the future.
1. Chlamydomonas reinhardtii as a Model in Mo Homeostasis
Microalgae are unicellular photosynthetic microorganisms that are considered one of the primary producers on our planet. Although microalgae are predominantly found in aquatic ecosystems, they can also thrive in various habitats, including soil as part of the rhizosphere [1]. Furthermore, microalgae can also be present within organisms, such as in coral reefs [2], and are a fundamental constituent of lichens [3]. Conversely, microalgae have been found to inhabit regions with extreme climates, such as polar regions [4]. Microalgae are responsible for a significant proportion of total carbon fixation [5], making them fundamental to sustaining different ecosystems [6]. However, they can also have adverse effects, such as algae blooms, which cause serious environmental, economic, and health damage [7]. Microalgae display a wide variety of cell sizes, morphologies, and architectures, and possess a diverse metabolic capacity that provides several distinctive features for scientific investigation [8]. The biotechnological use of microalgae has experienced exponential growth in recent years in various applications [9]. Notable examples of these applications include wastewater treatment, biofuel production, hydrogen production, the production of high-value substances, the use of microalgae as biofertilizers in agriculture, as well as their use as food additives and in cosmetics [10]. In recent years, the symbiotic relationship between microalgae and bacteria has gained significant traction in biotechnology [11].
The green microalgae C. reinhardtii (hereafter Chlamydomonas) was first isolated in a potato field in Massachusetts (USA) in 1945 [12], and since then, it has become an important model organism [13] because it possesses numerous attributes that have contributed to its importance. Chlamydomonas exhibits a fast growth rate, with a duplication time of approximately 8 h. It can also be easily cultivated under laboratory conditions either autotrophically, heterotrophically, or mixotrophically. This allows for the selection of mutants to study the mechanisms responsible for growth under different environmental conditions. Chlamydomonas shares basic metabolic characteristics with higher plants and also with animals [14,15]. It has both an asexual life cycle as a unicellular haploid alga, as well as a sexual life cycle with the formation of diploid cells. This allows for genetic crosses and the transfer of genes/mutations to the resulting offspring. The Chlamydomonas genome has been sequenced [15], and there is a large collection of mutants, many of which have their mutation molecularly labeled [16]. In addition, a multitude of molecular tools have been developed and optimized for the study of Chlamydomonas, including techniques for transforming its three genomes (nuclear, chloroplastic, and mitochondrial). All of this has allowed for the development of a number of biochemical, metabolic, and physiological studies that have contributed to the understanding of the structure, function, and regulation of several biological processes [17]. Among others, the research topics that have used Chlamydomonas as a reference organism include the metabolism of nitrogen [18], sulfur [19], phosphorus [20], amino acid [21], and lipids [22], the biosynthesis of carotenoids [23], starch [24], heme groups [25], Fe-S clusters [26], chlorophyll [27], and glycerolipids [28], as well as the function of chaperones [29], proteases [30], thioredoxins [31], and flagella [32] and the response to different types of stresses [33].
Chlamydomonas has also emerged as an organism with great biotechnological potential [34]. It is worth noting its use for the production of vaccines [35], antibodies [36], or in human clinical trials aimed to improve gastrointestinal health [37]. Furthermore, Chlamydomonas is also an excellent model for studying the basis of mutualistic interactions with bacteria, based on carbon–nitrogen exchange [38]. Certain mutualistic interactions have biotechnological potential applicable to bioremediation and hydrogen production [39,40]. The recent development of gene editing techniques such as CRISPR-Cas9 in Chlamydomonas could be a decisive step in increasing the biotechnological potential of this organism [41].
Regarding molybdenum (Mo) homeostasis, Chlamydomonas has been successfully used to uncover different steps of this process such as Mo uptake by identifying the first eukaryotic Mo transporters [42,43] and Moco storage, protection, and transfer by means of the identification and characterization of the Moco carrier protein (MCP1) [44,45]. Furthermore, Chlamydomonas has been used to obtain mutants affected in Mo metabolism [46] and to characterize Mo-using proteins like nitrate reductase [47] and mARC [48]. Therefore, an important part of the knowledge currently available about plant Mo homeostasis has been generated directly from Chlamydomonas, demonstrating its crucial role in this field of research over the last 30 years. In this review, we focus on the advancements in Mo homeostasis achieved through the use of Chlamydomonas and how the knowledge acquired has been instrumental in the progress made in other model organisms.
2. Molybdenum Uptake
Organisms require a sufficient cellular supply of Mo to ensure the activity of important enzymes such as nitrate reductase, sulfite oxidase, xanthine dehydrogenase, aldehyde oxidase, and mARC. Eukaryotic Mo homeostasis is a complex process that involves strategies for Mo uptake from the medium. These processes include Mo storage, which can occur through binding to other molecules in the cell or compartmentalization into organelles. Additionally, there are mechanisms for long-distae transport and regulatory mechanisms to coordinate intracellular Mo content, and cellular responses to these changes. The different stages of Mo homeostasis, including its transport into the cell, the formation of active Mo cofactor, cellular accumulation, and the activity and regulation of different molybdoenzymes, will be discussed below, with special attention given to those aspects where the use of Chlamydomonas has produced significant advances in this field.
With an average of 1.5 parts per million, Mo ranks as the 54th most abundant element in the Earth’s crust, while in oceans, it ranks as the 25th most abundant element with an average of 10 parts per billion [49]. In the soil, the bioavailability of Mo depends on the abundance of the oxyanion molybdate (MoO42−), which is the predominant form of Mo at a pH greater than 4.2. All organisms take up Mo from the outside in the form of MoO42−. A direct relationship between Mo availability in soils and plant growth has been reported, showing a drastic decrease in growth under low Mo conditions [50]. High concentrations of molybdate also have drastic effects; for example, in Chlamydomonas, molybdate concentrations greater than 50 mM have been shown to be toxic to wild-type strains. However, Chlamydomonas mutants deficient in Mo transport are resistant to these high concentrations [51]. Therefore, a precise Mo homeostatic mechanism is required to ensure proper cellular Mo availability and to prevent toxicities in environments with high molybdate concentrations.
For quite some time it was unknown whether eukaryotes had a specific molybdate transporter or if molybdate entered non-specifically through sulfate or phosphate transporters, anions sharing structural similarity with molybdate [52]. However, experiments using different mutants of Chlamydomonas have shown that there should be at least two molybdate transporters, one mediating high affinity transport and the other one mediating low affinity transport [51] (Figure 1). Low-affinity molybdate transport activity was blocked by similar concentrations of sulfate, whereas the high-affinity transport activity was not blocked by sulfate. These transport activities were genetically linked to the loci NIT5 and NIT6 in Chlamydomonas. Although the identity of the NIT5 locus is still unknown, the NIT6 locus has been found to be the CNX1E gene [53]. As will be discussed later, CNX1E catalyzes the last reaction for the biosynthesis of Moco. These results indicate that there is a close relationship between the protein involved in the last step of Moco biosynthesis (CNX1E) and the molybdate transporters, which could potentially be attributed to a physical connection between both proteins [54]. This hypothesis is reinforced by the fact that CNX1E is associated with the cytoskeleton, favoring localization close to the plasma membrane where MOT1 could be localized. However, this interesting possibility has not been proven so far and requires further experimentation in order to be confirmed. Two families of eukaryotic molybdate transporters (MOT1 and MOT2) have been identified for the first time using Chlamydomonas. The MOT1 transporter family was mistakenly annotated in databases as belonging to the sulfate transporter SULTR family due to some structural homology with them. However, this homology is low, and MOT1 transporters lack the fundamental STAS domain required for the ability to transport sulfate in the SULTR transporter family [42]. The MOT1 transporter family is characterized by having two conserved domains (FGXMPXCHG(S/A)GGLAXQ(Y/H)XFG(A/G)RXG and PXPVQPMKX(I/L)(A/G)AXA) that serve to molecularly label members belonging to this family. The Chlamydomonas MOT1 transporter shows high affinity for molybdate (Km 7 nM). Its transport activity is not inhibited by sulfate but is blocked by tungstate (the structural analog of molybdate). Interestingly, the expression of MOT1 does not depend on the presence of molybdate in the medium but on that of nitrate, which increases its expression. This form of gene regulation may be due to the fact that the molybdoenzyme nitrate reductase is essential for growth on nitrate and suggests that there must be a close relationship and coordination between Mo homeostasis and nitrate assimilation. This discovery in Chlamydomonas has enabled the identification of putative MOT1 member proteins in bacteria, fungi, and plants. However, MOT1 appears not to be present in animal genomes. In Arabidopsis thaliana, MOT1 is also a high-affinity transporter involved in transporting molybdate from the soil. Impaired plant growth occurs when the MOT1 transporter is absent [55,56]. In the legume plants Lotus japonicus and Medicago truncatula, MOT1 proteins have been described to facilitate molybdate transport, whereas in L. japonicus, the MOT1 reported member seems to be involved in Mo uptake by the roots [57]. In M. truncatula, both MOT1.2 and MOT1.3 mediate Mo supply to the nodules during nitrogen fixation. M. truncatula mutants lacking these transporter show deficient N-fixation activity although nitrogenase was present within the nodules [58,59], probably because nitrogenase is a Mo-dependent enzyme that requires an adequate supply of Mo. Furthermore, it has been shown that polymorphisms in the A. thaliana MOT1.1 gene within a plant population can be used as markers of leaf Mo content [60].
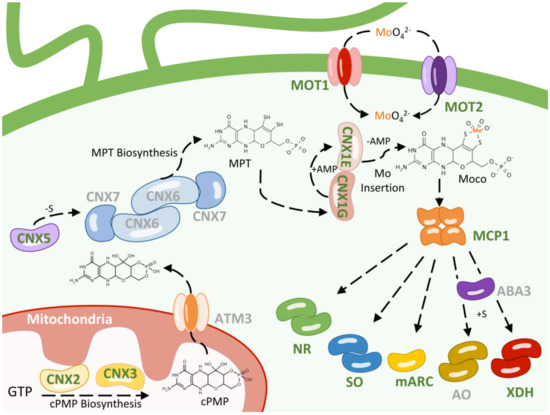
Figure 1.
Molybdenum homeostasis in Chlamydomonas. The biosynthetic machinery, maturation, and distribution of Moco in a Chlamydomonas cell are shown. The basic steps of Moco biosynthesis, from GTP to Moco, including enzymes that contain Moco, are depicted. Proteins catalyzing the individual steps are shown in different colors. The names of proteins that have been notably studied in Chlamydomonas are shown in green, and the others are shown in gray. All known intermediates of the pathway are presented sequentially in the three steps in which Moco is synthesized. The MOT1 and MOT2 proteins transport the molybdate-anion to the cytosol. The CNX2 and CNX3 proteins catalyze the conversion of GTP to cPMP. The mitochondrial ABC transporter ATM3 is involved in transporting cPMP from the mitochondria to the cytosol. MPT-synthase, consisting of CNX6 and CNX7, converts cPMP to MPT, which is then sulfurated by CNX5. CNX1G activates MPT by converting it to MPT-AMP, which is then transferred to CNX1E. CNX1E deadenylates MPT-AMP and incorporates Mo into MPT to produce Moco. ABA3 catalyzes the addition of a sulfur atom coordinated to Mo to the XDH and AO families of molybdoenzymes. Moco can bind to the Moco carrier protein MCP1, where it is accumulated or transferred to form the different Mo enzymes nitrate reductase (NR), sulfite oxidase (SO), xanthine dehydrogenase (XDH), aldehyde oxidase (AO), and mARC. AMP (adenosine monophosphate); for a more detailed explanation, refer to the text.
The MOT2 transporter family was also first molecularly identified in Chlamydomonas [43]. The members of this family do not have the typical conserved domain of MOT1 transporters and have low structural homology with MOT1. MOT2 being of high affinity shows lower affinity to transport molybdate (Km 550 nM) compared to MOT1 and is blocked by tungstate. Interestingly, the expression of the MOT2 gene is activated by molybdate deficiency, and does not respond to the availability of nitrate, which represents opposite regulation compared to MOT1. Proteins homologs to MOT2 are present in plants and animals. It has been shown that the human MOT2 is capable of transporting molybdate when expressed heterologously in yeast, suggesting a similar role in Mo homeostasis in mammals [43].
Apart from these two identified transporters, other transporters related to Mo homeostasis might be present in eukaryotes, mediating uptake, export, or re-direction to organelles for storage. In this sense, the Chlamydomonas mutant db6 has been characterized as having a lower intracellular concentration of molybdate and reduced activity of the molybdoenzymes xanthine dehydrogenase and aldehyde oxidase. These effects can be restored by increasing the molybdate concentration in the medium to 10 mM. Additionally, the Chlamydomonas mutant db6 shows high resistance to elevated concentrations of tungstate in the medium [61]. These characteristics suggest that db6 is a mutant in molybdate transport, although the molecular identity of the gene involved has not yet been resolved.
3. Moco Biosynthesis
As mentioned, in eukaryotic organisms, for Mo to be biologically active, it must complex with a pterin-derive molecule called MPT to form Moco. The different steps of this biosynthetic pathway will be shown below, focusing on those aspects where Chlamydomonas has made significant contributions. The proteins involved in the Moco biosynthetic pathway have a high degree of homology across all studied organisms. According to the different intermediates formed, this pathway can be divided into three main steps: cyclic pyranopterin monophosphate (cPMP) biosynthesis, MPT biosynthesis, and Mo insertion [62]. In Chlamydomonas, seven genes are essential for Moco biosynthesis, within these three steps. These genes follow the CNX (cofactor of nitrate reductase and xanthine dehydrogenase) nomenclature, as it occurs in plants and fungi [63].
3.1. cPMP Biosynthesis
In plants, the first step of the Moco biosynthesis pathway is catalyzed in the mitochondria by the enzymes CNX2 and CNX3 and involves the conversion of a molecule of GTP into cyclic pyranopterin monophosphate (cPMP) [64] (Figure 1). In Chlamydomonas, individual mutants affected either in CNX2 or CNX3 genes have been identified. Characterization of such mutants indicates that they are unable to grow on nitrate and have elevated expression of the nitrate reductase gene due to a putative persistent nitrate positive signal [65]. Chlamydomonas CNX2 and CNX3 mutants are also resistant to high concentrations of mutagenic base analogs due to the involvement of the molybdoenzyme mARC in their detoxification [66]. Once cPMP is synthesized, it must be transported to the cytosol, which is carried out in Arabidopsis by the mitochondrial transporter ATM3 [67]. The ATM3 genes have not yet been characterized in Chlamydomonas. However, a mutant of the ATM3 gene (LMJ.RY0402.180694) is available in the molecularly tagged mutant library of Chlamydomonas [16].
3.2. MPT Biosynthesis
The second step consists of the insertion of two sulfur atoms into cPMP, which converts it to MPT. This reaction is catalyzed by MPT synthase, a tetrameric protein consisting of two large subunits (CNX6) and two small subunits (CNX7) [68] (Figure 1). To date, Chlamydomonas mutants have not been characterized in the CNX6 and CNX7 genes. However, in the library of molecularly labeled Chlamydomonas mutants [16], a mutant affected in the CNX6 gene (LMJ.RY0402.047904) is present but not in the CNX7 gene, likely due to its small size (252 pb of the predicted coding sequence). After the transfer of the sulfur atoms, MPT synthase must be resulfurated in a reaction that is mediated by MPT synthase sulfurase, which is encoded by the CNX5 gene [69]. A Chlamydomonas mutant affected in the CNX5 gene is also available [65]. Similarly to CNX2 and CNX3, the Chlamydomonas CNX5 mutant is also resistant to high concentrations of mutagenic base analogs [66].
cPMP purified from Escherichia coli overexpressing strains have been used for the treatment of human Moco deficiency [70]. However, in 2021, a chemically synthesized form of cPMP, called ‘Fosdenopterin’, was granted FDA approval and received marketing authorization as a Nulibry product [71]. It can be assumed that the accumulation of cPMP could also occur in the Chlamydomonas CNX5 mutant. Therefore, it would be interesting to evaluate how this accumulation compares to that of E. coli or the chemically synthesized ‘Fosdenopterin’, to determine if the purification of cPMP from this Chlamydomonas mutant is more feasible from an economical point of view.
3.3. Mo Insertion
After MPT formation, a Mo atom coordinates to the sulfur atoms of MPT to form a biologically active Moco, a process that is mediated by adenylation [72,73] (Figure 1). In plants, the two-domain enzyme CNX1 catalyzes this insertion. The CNX1G domain of CNX1 catalyzes, in an Mg2+ and ATP-dependent reaction, the adenylation of MPT, forming transient MPT-AMP [74]. Then, the CNX1E domain of CNX1 catalyzes the deadenylation of MPT-AMP, a reaction that triggers Mo insertion [75]. In Chlamydomonas, unlike most other eukaryotic organisms, the two domains CNX1G and CNX1E are encoded by two separate genes [46]. Therefore, in this aspect, Chlamydomonas more closely resembles prokaryotic organisms such as E. coli where these two proteins are also separated and encoded in different genes [76]. In agreement with that, the Chlamydomonas CNX1G and CNX1E genes are able to complement mutations in the E. coli homolog mutants [46]. The mutations Nit4 and Nit6 in Chlamydomonas arise from a single alteration in CNX1E (V171A and G183D, respectively), which results in its loss of function. Additionally, it has been shown that the Chlamydomonas CNX1E gene exhibits intragenic complementation, suggesting that in Chlamydomonas, CNX1E forms multimeric complexes [53]. The Moco of xanthine dehydrogenase and aldehyde oxidase family enzymes requires an additional step to become biologically active. This step involves the addition of an additional sulfur atom coordinated to Mo, a reaction catalyzed by the Moco sulfurase enzyme, encoded by the ABA3 gene [77] (Figure 1). In the other molybdoenzymes, instead of this sulfur atom, Mo is coordinated to the thiol group of a cysteine from the active center [78]. So far, Chlamydomonas mutant strains affected in ABA3 have not been reported. However, an ABA3 mutant (LMJ.RY0402.059375) seems to also be available in the labeled mutant library of Chlamydomonas [16].
4. Moco Storage
Active Moco is highly susceptible to inactivation in the presence of oxygen. Therefore, photosynthetic organisms should have some mechanism to protect this cofactor against oxidation [79]. The Chlamydomonas Moco Carrier Protein 1 (MCP1) was the first protein identified in eukaryotes to be involved in this process of Moco protection [80,81]. The main function of MCP1 seems to be Moco’s protection from oxidation, but additionally, this protein has also been reported to have a function in Moco storage and transference to the corresponding molybdoenzyme [45] (Figure 1). The three-dimensional structure of MCP1 has been resolved, with it being a tetramer with a typical Rossmann fold, and a putative Moco binding pocket has been proposed [44]. Proteins showing similar characteristics to Chlamydomonas MCP1 have also been identified and characterized in the cyanobacterium Rippkaea orientalis [82] and the microalga Volvox carteri [83]. Based on the three-dimensional structure of MCP1, orthologous proteins have also been identified in A. thaliana, which have been named MoBPs (molybdenum binding proteins) [84,85]; however, their function in Mo homeostasis still needs to be clarified.
In prokaryotic organisms, proteins involved in Moco binding/storage have not been identified yet. However, in certain bacteria such as Azotobacter vinelandii, the existence of proteins that bind, accumulate, and buffer intracellular concentrations of molybdate, called molybdate-binding proteins, has been described [86,87]. These Mo-binding proteins seem to not be present in eukaryotic genomes, and Mo storage may occur using different proteins or alternative strategies. In this sense, it has been proposed that in plants like the legume Medicago sativa, excess intracellular Mo might be chelated by organic acids such as malate or citrate [88].
5. Chlamydomonas Moco Enzymes
As previously mentioned, five molybdoenzymes have been identified in Chlamydomonas. In the next section, we will present the main advances made in the study of these enzymes, with special attention given to those in which the results obtained in Chlamydomonas have led to significant progress.
5.1. Xanthine Dehydrogenase
Plant xanthine dehydrogenase (XDH) catalyzes the oxidation of hypoxanthine to xanthine and xanthine to uric acid with NAD+ as the physiological electron acceptor (Figure 2). Structural and functional studies of Xanthine Dehydrogenase have recently been updated [89]. Chlamydomonas is capable of using the purines xanthine and hypoxanthine as a nitrogen source [90], which indicates that it must have an active XDH enzyme to search for additional nitrogen sources when ammonia or nitrate are not present. In this regard, it has been observed that the Chlamydomonas XDH gene is strongly expressed under nitrogen-depleted conditions in culture medium [91]. Chlamydomonas was the first microalga in which XDH was purified and characterized. Chlamydomonas XDH is a homodimeric enzyme with high molecular weight (330 kDa) that contains not only Moco but also Fe-S and FAD as cofactors [92].
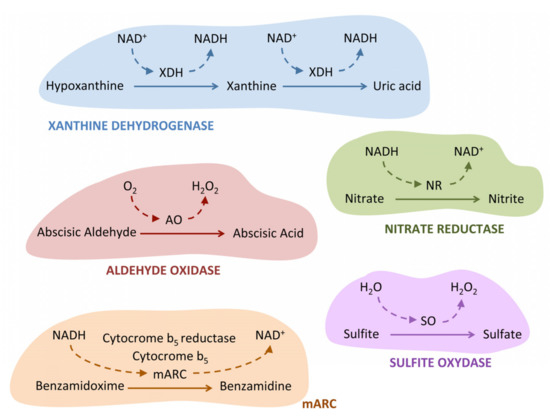
Figure 2.
Schematic representation of the enzymatic activity catalyzed by the five molybdoenzymes of Chlamydomonas. Not all substrates for each of the enzymes are shown. A characteristic example has been chosen as the substrate for each of the molybdoenzymes. The electron donors (NADH and O2) or acceptors (NAD+ and H2O) involved in each of the reactions are indicated for each of the enzymes. To simplify the figure, it has not been shown that NR can also act as an electron donor for mARC. For more details, refer to the text.
5.2. Aldehyde Oxidase
Plant aldehyde oxidase (AO) catalyzes the oxidation of abscisic aldehyde to the phytohormone abscisic acid (ABA) (Figure 2) [93]. AO and XDH share great structural homology and have the same cofactor distribution. One reason for the high similarity in sequence and structure is that, in the eukaryotes, the AO gene evolved from a duplication of the XDH gene [94]. So far, the AO protein has not been characterized in Chlamydomonas, but it is presumed to play a critical role in protecting against various types of stresses. In this regard, it has been observed that ABA protects Chlamydomonas cells against the increase in reactive oxygen species that occur in response to salt stress [95], enhances the transport of bicarbonate, and is responsible for regulating the diurnal cycle of upward and downward Chlamydomonas movement in response to gravity in the water column [96]. The Chlamydomonas AO mutant (LMJ.RY0402.043929) is available in the labeled mutant library of Chlamydomonas [16], and its study could fundamentally contribute to elucidating the role of ABA in the response to different types of stresses.
5.3. Sulfite Oxidase
The plant sulfite oxidase (SO) enzyme functions as a homodimeric protein and plays a key role in sulfur catabolism by facilitating the two-electron oxidation of sulfite to sulfate using H2O as an electron acceptor and releasing hydrogen peroxide (Figure 2) [97]. This recent review covers the main structural and functional aspects of sulfite oxidase [98]. SO seems to protect plants against the toxicity of sulfite in an environment with high levels of SO2 [99]. There are some differences between animal and plant SO. In vertebrates, the SO enzyme is localized in the mitochondrial intermembrane space and contains heme, in addition Moco, as a cofactor [100]. In contrast, plant SO is localized in the peroxisome [101] and only contains Moco as a cofactor [102]. Interestingly, the Chlamydomonas enzyme resembles the SO of vertebrates more than that of plants since it contains a heme group and has been found in the mitochondrial proteome [103]. Furthermore, Chlamydomonas SO is overexpressed in the presence of nitrate as a nitrogen source. The reason behind this overexpression is not fully clear, but it could be related to the need of Chlamydomonas to regulate the balance of sulfur and nitrogen within its cellular metabolism. In the presence of nitrate, cells can use sulfur to produce other compounds necessary for their growth, such as cysteine, which may increase the need for SO to maintain the sulfur balance in the cell. The Chlamydomonas SO mutant (LMJ.RY0402.221179) is available in the labeled mutant library of Chlamydomonas [16], although it has not yet been characterized.
5.4. Nitrate Reductase
The nitrate reductase (NR) enzyme is involved in two key processes: the critical step in nitrate assimilation, which is its reduction to nitrite (Figure 2), and the reduction of nitrite to nitric oxide (NO). Structural and mechanistic insights on nitrate reductases can be seen in the review [104]. The Chlamydomonas NR gene (NIA1) encodes a homodimeric protein. Each monomer has three prosthetic groups: FAD, heme b557, and Moco [105]. In NR, the flow of electrons goes from NAD(P)H to FAD, then to the heme b557, and finally ends in the Moco domain, where nitrate is reduced to nitrite [106]. Interestingly, the NR Moco domain is also able to use nitrite as a substrate, when it is present in high concentrations, reducing it to NO [107]. NO is an important and ubiquitous signaling molecule that is involved in many different biological processes, fundamental for cell survival under different types of stresses [108]. Two mechanisms exist for the production of NO, oxidative and reductive. The oxidative mechanism is catalyzed by the enzyme NO synthase (NOS) and uses L-arginine as a substrate [109]. However, in plants, the presence of NOS has only been determined in some microalgae such as Ostreococcus tauri [110], but not in Chlamydomonas. Therefore, in plants, the reduction of nitrite to NO through different mechanisms appears to be the most important way for NO synthesis [111]. Using Chlamydomonas, it has been shown, for the first time in a photosynthetic organism, that NR can not only reduce nitrite to NO by itself but also with the assistance of another molybdoenzyme, the mARC protein [48]. This aspect will be explained below.
5.5. mARC
mARC is the latest Moco enzyme discovered in eukaryotic organisms. It was firstly identified in pig liver mitochondria, and its name (mitochondrial amidoxime reducing component) is derived from the first substrate characterized for this enzyme, amidoximes [112]. mARC is able to reduce the amidoxime N-hydroxylate pro-drug benzamidoxime to benzamidine (Figure 2) and is therefore supposed to be involved in xenobiotic detoxification. However, since then, a wide variety of other mARC substrates have been identified. The first characterization of a mARC protein in a photosynthetic was carried out in Chlamydomonas [66]. The mARC enzyme in Chlamydomonas has Moco showing zinc-dependent activity. In agreement with that, proteomic studies have shown that in Chlamydomonas, zinc deficiency increases the cellular content of mARC by more than 30 times [113]. Moreover, the characterization of the Chlamydomonas mARC enzyme revealed that it belongs to the SO protein family, as it has a completely conserved cysteine within the active center that is used to chelate Moco [66]. In contrast to humans, where mARC appears to be a monomeric enzyme, Chlamydomonas mARC seems to form high molecular weight oligomeric complexes consisting of 10–12 monomers. These complexes could be involved in protecting the oxygen-labile Moco [114].
In addition to amidoximes, the human mARC is also involved in the reduction of sulfamethoxazole hydroxylamine to sulfamethoxazole [115], N4-hydroxy-L-arginine (NOHA) to arginine [116], and nitrite to NO [117]. It has been characterized that Chlamydomonas mARC can reduce the base analogue 6-hydroxylaminopurine (HAP) to adenine [66]. Therefore, it could be involved in the detoxification of mutagenic components. An interesting aspect about mARC proteins is that they can bind to different electron donors able to transfer electrons from NADH to reduce the corresponding substrates. Chlamydomonas mARC uses a cytochrome b5 and a cytochrome b5 reductase as electron donors for HAP reduction [66].
Fascinatingly, it has been reported that Chlamydomonas mARC is also capable of reducing nitrite to the second messenger NO. However, for this action, it does not use cytochrome b5 as an electron donor but instead uses the molybdoenzyme NR [48]. In agreement with that, in Chlamydomonas, NR mutants overexpress mARC, and vice versa, and furthermore, Chlamydomonas mARC does not appear to be located in the mitochondria as in humans but instead seems to be in the cytoplasm where its partner NR is located [48]. It is known that plant NR itself is capable of reducing nitrite to NO [118]; however, this ability is blocked in the presence of high millimolar concentrations of nitrate [119], which does not occur when NR acts as an electron donor in Chlamydomonas to produce NO from nitrite reduction together with mARC [48]. Whereas C. reinhardtii has only one mARC enzyme, in the higher plant A. thaliana, two mARC proteins (mARC1 and mARC2) have been identified, both of which are capable of reducing N-hydroxylated compounds, but only mARC2 seems to be involved in nitrite reduction to NO using NR as an electron donor [120]. These data suggest that the NR–mARC pair is a very efficient machinery for synthesizing NO under physiological conditions, in the presence of both nitrate and nitrite, and may have a very important role in modulating cellular NO levels. Apart from NO, the Chlamydomonas complex NR–mARC has been shown to regulate N2O production, a greenhouse gas, through NO synthesis [121]. The mARC proteins have been proposed as moonlighting proteins [122]. Moonlighting proteins are proteins able to perform more than one biological function within the cell, that is, they have the ability to perform different molecular tasks or participate in different cellular processes [123]. Therefore, the large number of different substrates and partners discovered for mARC support its classification as a moonlighting protein.
6. Concluding Remarks and Future Outlook
In this review, we have shown how the green microalga C. reinhardtii has been used to identify various molecular components of Mo homeostasis, ranging from molybdate transport to molybdoenzyme activities. These advances have subsequently been applied to identify the corresponding molecular components in other model organisms. However, there are several issues that remain unresolved in which Chlamydomonas could also play a crucial role. For example, one of them is the mechanism mediating Mo export from the cell, with it being important to avoid Mo toxicity or for long-distance transport in multicellular organisms. The transporters mediating this process are still unknown. The isolation and characterization of Chlamydomonas strains resistant to high Mo concentrations could clarify this issue. Another point that remains unanswered is whether there are regulatory interconnections between the maintenance of the intracellular concentration of Mo and the activity of Moco biosynthesis. In this regulation, the relationship between the molybdate transporters and the proteins that biosynthesize Moco may play a fundamental role. Therefore, it would be interesting to clarify whether there is a relationship between the molybdate transporters and the final protein involved in the biosynthesis of Moco (CNX1E). Regarding molybdoenzymes, the role of the NR–mARC complex in the production of NO is well defined, but the role of the NO produced in the production of N2O has just begun to be studied. Surely in the future, very interesting results will be obtained through studying this process in Chlamydomonas.
Author Contributions
A.L.: conceptualization and original draft preparation; A.L. and M.T.-J. wrote the paper; E.L.-M.: review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Gobierno de España, Ministerio de Ciencia e Innovacion (Grant PID2020-118398GB-I00), Junta de Andalucía (Grant ProyExcel_00483), the “Plan Propio” from University of Cordoba, and a grant awarded by the Torres-Gutierrez Foundation.
Data Availability Statement
All data required to evaluate the conclusions of this paper are included in the main text.
Acknowledgments
This paper is dedicated to Emilio Fernandez Reyes for his more than 30 years of studying Mo homeostasis using Chlamydomonas reinhardtii as a reference organism. He was the driving force behind the research on Mo homeostasis in Chlamydomonas, the pillar that has allowed for its advancement and our great teacher whom we will never be able to repay for all the learnings received.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Quintas-Nunes, F.; Brandão, P.R.; Barreto Crespo, M.T.; Glick, B.R.; Nascimento, F.X. Plant Growth Promotion, Phytohormone Production and Genomics of the Rhizosphere-Associated Microalga, Micractinium rhizosphaerae sp. nov. Plants 2023, 12, 651. [Google Scholar] [CrossRef] [PubMed]
- Maire, J.; Buerger, P.; Chan, W.Y.; Deore, P.; Dungan, A.M.; Nitschke, M.R.; van Oppen, M.J.H. Effects of Ocean Warming on the Underexplored Members of the Coral Microbiome. Integr. Comp. Biol. 2022, 62, 1700–1709. [Google Scholar] [CrossRef] [PubMed]
- Grimm, M.; Grube, M.; Schiefelbein, U.; Zühlke, D.; Bernhardt, J.; Riedel, K. The Lichens’ Microbiota, Still a Mystery? Front. Microbiol. 2021, 12, 623839. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovska, M.; Vakulenko, G.; Smith, D.R.; Zhang, X.; Hüner, N.P.A. Temperature Stress in Psychrophilic Green Microalgae: Minireview. Physiol. Plant. 2022, 174, e13811. [Google Scholar] [CrossRef]
- Falkowski, P.G. The Role of Phytoplankton Photosynthesis in Global Biogeochemical Cycles. Photosynth. Res. 1994, 39, 235–258. [Google Scholar] [CrossRef]
- Tarafdar, A.; Sowmya, G.; Yogeshwari, K.; Rattu, G.; Negi, T.; Awasthi, M.K.; Hoang, A.T.; Sirohi, R. Environmental Pollution Mitigation through Utilization of Carbon Dioxide by Microalgae. Environ. Pollut. 2023, 328, 121623. [Google Scholar] [CrossRef]
- Kazmi, S.S.U.H.; Yapa, N.; Karunarathna, S.C.; Suwannarach, N. Perceived Intensification in Harmful Algal Blooms Is a Wave of Cumulative Threat to the Aquatic Ecosystems. Biology 2022, 11, 852. [Google Scholar] [CrossRef]
- Gerotto, C.; Norici, A.; Giordano, M. Toward Enhanced Fixation of CO2 in Aquatic Biomass: Focus on Microalgae. Front. Energy Res. 2020, 8, 213. [Google Scholar] [CrossRef]
- Parmar, P.; Kumar, R.; Neha, Y.; Srivatsan, V. Microalgae as next Generation Plant Growth Additives: Functions, Applications, Challenges and Circular Bioeconomy Based Solutions. Front. Plant Sci. 2023, 14, 1073546. [Google Scholar] [CrossRef]
- Fabris, M.; Abbriano, R.M.; Pernice, M.; Sutherland, D.L.; Commault, A.S.; Hall, C.C.; Labeeuw, L.; McCauley, J.I.; Kuzhiuparambil, U.; Ray, P.; et al. Emerging Technologies in Algal Biotechnology: Toward the Establishment of a Sustainable, Algae-Based Bioeconomy. Front. Plant Sci. 2020, 11, 279. [Google Scholar] [CrossRef]
- Zhang, B.; Li, W.; Guo, Y.; Zhang, Z.; Shi, W.; Cui, F.; Lens, P.N.L.; Tay, J.H. Microalgal-Bacterial Consortia: From Interspecies Interactions to Biotechnological Applications. Renew. Sustain. Energy Rev. 2020, 118, 109563. [Google Scholar] [CrossRef]
- Harris, E.H. Introduction into Chlamydomonas and Its Laboratory Use. Chlamydomonas Sourcebook; Oxford Academic Press: Oxford, UK, 2009. [Google Scholar]
- Salomé, P.A.; Merchant, S.S. A Series of Fortunate Events: Introducing Chlamydomonas as a Reference Organism. Plant Cell 2019, 31, 1682–1707. [Google Scholar] [CrossRef]
- Llamas, A.; Tejada-Jiménez, M.; Fernández, E.; Galván, A. Molybdenum Metabolism in the Alga Chlamydomonas Stands at the Crossroad of Those in Arabidopsis and Humans. Metallomics 2011, 3, 578–590. [Google Scholar] [CrossRef]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef]
- Fauser, F.; Vilarrasa-Blasi, J.; Onishi, M.; Ramundo, S.; Patena, W.; Millican, M.; Osaki, J.; Philp, C.; Nemeth, M.; Salomamp, P.A.; et al. Systematic Characterization of Gene Function in the Photosynthetic Alga Chlamydomonas reinhardtii. Nat. Genet. 2022, 54, 705–714. [Google Scholar] [CrossRef]
- Goodenough, U. The Chlamydomonas Sourcebook. Volume 1: Introduction to Chlamydomonas and Its Laboratory Use; Elsevier Academic Press: Cambridge, MA, USA, 2023. [Google Scholar] [CrossRef]
- Bellido-Pedraza, C.M.; Calatrava, V.; Sanz-Luque, E.; Tejada-Jiménez, M.; Llamas, A.; Plouviez, M.; Guieysse, B.; Fernández, E.; Galván, A. Chlamydomonas reinhardtii, an Algal Model in the Nitrogen Cycle. Plants 2020, 9, 903. [Google Scholar] [CrossRef]
- Saroussi, S.; Sanz-Luque, E.; Kim, R.G.; Grossman, A.R. Nutrient Scavenging and Energy Management: Acclimation Responses in Nitrogen and Sulfur Deprived Chlamydomonas. Curr. Opin. Plant Biol. 2017, 39, 114–122. [Google Scholar] [CrossRef]
- Irihimovitch, V.; Yehudai-Resheff, S. Phosphate and Sulfur Limitation Responses in the Chloroplast of Chlamydomonas reinhardtii. FEMS Microbiol. Lett. 2008, 283, 1–8. [Google Scholar] [CrossRef]
- Calatrava, V.; Hom, E.F.Y.; Llamas, A.; Fernández, E.; Galvan, A. Nitrogen Scavenging from Amino Acids and Peptides in the Model Alga Chlamydomonas reinhardtii. The Role of Extracellular L-Amino Oxidase. Algal Res. 2019, 38, 101395. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Kong, F.; Wang, P.; Lee, Y.; Kang, B.-H. The Disassembly of Lipid Droplets in Chlamydomonas. New Phytol. 2021, 231, 1359–1364. [Google Scholar] [CrossRef]
- Rengel, R.; Giraldez, I.; Díaz, M.J.; García, T.; Vigara, J.; Leon, R. Simultaneous Production of Carotenoids and Chemical Building Blocks Precursors from Chlorophyta Microalgae. Bioresour. Technol. 2022, 351, 127035. [Google Scholar] [CrossRef] [PubMed]
- Sirohi, R.; Joun, J.; Choi, H.I.; Gaur, V.K.; Sim, S.J. Algal Glycobiotechnology: Omics Approaches for Strain Improvement. Microb. Cell Fact. 2021, 20, 163. [Google Scholar] [CrossRef] [PubMed]
- Calderon, R.H.; de Vitry, C.; Wollman, F.A.; Niyogi, K.K. Rubredoxin 1 Promotes the Proper Folding of D1 and Is Not Required for Heme B559 Assembly in Chlamydomonas Photosystem II. J. Biol. Chem. 2023, 299, 102968. [Google Scholar] [CrossRef] [PubMed]
- Przybyla-Toscano, J.; Couturier, J.; Remacle, C.; Rouhier, N. Occurrence, Evolution and Specificities of Iron-Sulfur Proteins and Maturation Factors in Chloroplasts from Algae. Int. J. Mol. Sci. 2021, 22, 3175. [Google Scholar] [CrossRef] [PubMed]
- Roach, T.; Baur, T.; Kranner, I. β-Cyclocitral Does Not Contribute to Singlet Oxygen-Signalling in Algae, but May Down-Regulate Chlorophyll Synthesis. Plants 2022, 11, 2155. [Google Scholar] [CrossRef]
- Yang, M.; Xie, X.; Kong, F.T.; Xie, K.P.; Yu, S.H.; Ma, J.Y.; Xue, S.; Gong, Z. Differences in Glycerolipid Response of Chlamydomonas reinhardtii Starchless Mutant to High Light and Nitrogen Deprivation Stress under Three Carbon Supply Regimes. Front. Plant Sci. 2022, 13, 860966. [Google Scholar] [CrossRef]
- Kreis, E.; Niemeyer, J.; Merz, M.; Scheuring, D.; Schroda, M. CLPB3 Is Required for the Removal of Chloroplast Protein Aggregates and Thermotolerance in Chlamydomonas. J. Exp. Bot. 2023, erad109. [Google Scholar] [CrossRef]
- Zou, Y.; Bozhkov, P.V. Chlamydomonas Proteases: Classification, Phylogeny, and Molecular Mechanisms. J. Exp. Bot. 2021, 72, 7680–7693. [Google Scholar] [CrossRef]
- Marchetti, G.M.; Füsser, F.; Singh, R.K.; Brummel, M.; Koch, O.; Kümmel, D.; Hippler, M. Structural Analysis Revealed a Novel Conformation of the NTRC Reductase Domain from Chlamydomonas reinhardtii. J. Struct. Biol. 2022, 214, 107829. [Google Scholar] [CrossRef]
- Marshall, W.F. The Flagellar Length Control System: Exploring the Physical Biology of Organelle Size. Phys. Biol. 2023, 20, 021001. [Google Scholar] [CrossRef]
- Sasso, S.; Stibor, H.; Mittag, M.; Grossman, A.R. From Molecular Manipulation of Domesticated Chlamydomonas reinhardtii to Survival in Nature. Elife 2018, 7, e39233. [Google Scholar] [CrossRef]
- Masi, A.; Leonelli, F.; Scognamiglio, V.; Gasperuzzo, G.; Antonacci, A.; Terzidis, M.A. Chlamydomonas reinhardtii: A Factory of Nutraceutical and Food Supplements for Human Health. Molecules 2023, 28, 1185. [Google Scholar] [CrossRef]
- Dehghani, J.; Movafeghi, A.; Mathieu-Rivet, E.; Mati-Baouche, N.; Calbo, S.; Lerouge, P.; Bardor, M. Microalgae as an Efficient Vehicle for the Production and Targeted Delivery of Therapeutic Glycoproteins against SARS-CoV-2 Variants. Mar. Drugs 2022, 20, 657. [Google Scholar] [CrossRef]
- Rosales-Mendoza, S.; García-Silva, I.; González-Ortega, O.; Sandoval-Vargas, J.M.; Malla, A.; Vimolmangkang, S. The Potential of Algal Biotechnology to Produce Antiviral Compounds and Biopharmaceuticals. Molecules 2020, 25, 4049. [Google Scholar] [CrossRef]
- Fields, F.J.; Lejzerowicz, F.; Schroeder, D.; Ngoi, S.M.; Tran, M.; McDonald, D.; Jiang, L.; Chang, J.T.; Knight, R.; Mayfield, S. Effects of the Microalgae Chlamydomonas on Gastrointestinal Health. J. Funct. Foods 2020, 65, 103738. [Google Scholar] [CrossRef]
- Calatrava, V.; Tejada-Jimenez, M.; Sanz-Luque, E.; Fernandez, E.; Galvan, A.; Llamas, A. Chlamydomonas reinhardtii, a Reference Organism to Study Algal–Microbial Interactions: Why Can’t They Be Friends? Plants 2023, 12, 788. [Google Scholar] [CrossRef]
- Fakhimi, N.; Gonzalez-Ballester, D.; Fernández, E.; Galván, A.; Dubini, A. Algae-Bacteria Consortia as a Strategy to Enhance H2 Production. Cells 2020, 9, 1353. [Google Scholar] [CrossRef]
- Torres, M.; Gonzalez-Ballester, D.; Gomez-Osuna, A.; Galván, A.; Fernandez, E.; Dubini, A. Chlamydomonas-Methylobacterium oryzae Cooperation Leads to Increased Biomass, Nitrogen Removal, and Hydrogen Production. Bioresour. Technol. 2022, 352, 127088. [Google Scholar] [CrossRef]
- Kelterborn, S.; Boehning, F.; Sizova, I.; Baidukova, O.; Evers, H.; Hegemann, P. Gene Editing in Green Alga Chlamydomonas reinhardtii via CRISPR-Cas9 Ribonucleoproteins. Plant Synth. Biol. Methods Mol. Biol. 2022, 2379, 45–65. [Google Scholar] [CrossRef]
- Tejada-Jiménez, M.; Llamas, A.; Sanz-Luque, E.; Galván, A.; Fernández, E. A High-Affinity Molybdate Transporter in Eukaryotes. Proc. Natl. Acad. Sci. USA 2007, 104, 20126–20130. [Google Scholar] [CrossRef]
- Tejada-Jiménez, M.; Galván, A.; Fernández, E. Algae and Humans Share a Molybdate Transporter. Proc. Natl. Acad. Sci. USA 2011, 108, 6420–6425. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Llamas, A.; Tejada-Jiménez, M.; Schrader, N.; Kuper, J.; Ataya, F.S.; Galván, A.; Mendel, R.R.; Fernández, E.; Schwarz, G. Function and Structure of the Molybdenum Cofactor Carrier Protein from Chlamydomonas reinhardtii. J. Biol. Chem. 2006, 281, 30186–30194. [Google Scholar] [CrossRef] [PubMed]
- Ataya, F.S.; Witte, C.P.; Galván, A.; Igeño, M.I.; Fernández, E. Mcp1 Encodes the Molybdenum Cofactor Carrier Protein in Chlamydomonas reinhardtii and Participates in Protection, Binding, and Storage Functions of the Cofactor. J. Biol. Chem. 2003, 278, 10885–10890. [Google Scholar] [CrossRef] [PubMed]
- Llamas, A.; Tejada-Jimenez, M.; González-Ballester, D.; Higuera, J.J.; Schwarz, G.; Galván, A.; Fernández, E. Chlamydomonas Reinhardtii CNX1E Reconstitutes Molybdenum Cofactor Biosynthesis in Escherichia coli Mutants. Eukaryot. Cell 2007, 6, 1063–1067. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galván, A.; Fernandez, E. Nitrate Reductase Regulates Plant Nitric Oxide Homeostasis. Trends Plant Sci. 2017, 22, 163–174. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Ocaña-Calahorro, F.; Mariscal, V.; Carreras, A.; Barroso, J.B.; Galván, A.; Fernández, E. A Dual System Formed by the ARC and NR Molybdoenzymes Mediates Nitrite-Dependent NO Production in Chlamydomonas. Plant. Cell Environ. 2016, 39, 2097–2107. [Google Scholar] [CrossRef]
- Emsley, J. Nature’s Building Blocks; Oxford University Press: Oxford, UK, 2001. [Google Scholar]
- Lešková, A.; Javot, H.; Giehl, R.F.H. Metal Crossroads in Plants: Modulation of Nutrient Acquisition and Root Development by Essential Trace Metals. J. Exp. Bot. 2022, 73, 1751–1765. [Google Scholar] [CrossRef]
- Llamas, A.; Kalakoutskii, K.L.; Fernández, E. Molybdenum Cofactor Amounts in Chlamydomonas reinhardtii Depend on the Nit5 Gene Function Related to Molybdate Transport. Plant Cell Environ. 2000, 23, 1247–1255. [Google Scholar] [CrossRef]
- Mendel, R.R.; Haensh, R. Molybdoenzymes and Molybdenum Cofactor in Plants. J. Exp. Bot. 2002, 53, 1689–1698. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. Characterization of Chlamydomonas 102 and 104 Mutants Reveals Intermolecular Complementation in the Molybdenum Cofactor Protein CNX1E. Protist 2013, 164, 116–128. [Google Scholar] [CrossRef]
- Schwarz, G.; Schulze, J.; Bittner, F.; Eilers, T.; Kuper, J.; Bollmann, G.; Nerlich, A.; Brinkmann, H.; Mendel, R.R. The Molybdenum Cofactor Biosynthetic Protein Cnx1 Complements Molybdate-Repairable Mutants, Transfers Molybdenum to the Metal Binding Pterin, and Is Associated with the Cytoskeleton. Plant Cell 2000, 12, 2455–2471. [Google Scholar] [CrossRef]
- Tomatsu, H.; Takano, J.; Takahashi, H.; Watanabe-Takahashi, A.; Shibagaki, N.; Fujiwara, T. An Arabidopsis thaliana High-Affinity Molybdate Transporter Required for Efficient Uptake of Molybdate from Soil. Proc. Natl. Acad. Sci. USA 2007, 104, 18807–18812. [Google Scholar] [CrossRef]
- Ide, Y.; Kusano, M.; Oikawa, A.; Fukushima, A.; Tomatsu, H.; Saito, K.; Hirai, M.Y.; Fujiwara, T. Effects of Molybdenum Deficiency and Defects in Molybdate Transporter MOT1 on Transcript Accumulation and Nitrogen/Sulphur Metabolism in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 1483–1497. [Google Scholar] [CrossRef]
- Gao, J.-S.; Wu, F.-F.; Shen, Z.-L.; Meng, Y.; Cai, Y.-P.; Lin, Y. A Putative Molybdate Transporter LjMOT1 Is Required for Molybdenum Transport in Lotus japonicus. Physiol. Plant. 2016, 158, 331–340. [Google Scholar] [CrossRef]
- Tejada-Jiménez, M.; Gil-Díez, P.; León-Mediavilla, J.; Wen, J.; Mysore, K.S.; Imperial, J.; González-Guerrero, M. Medicago truncatula Molybdate Transporter Type 1 (MtMOT1.3) Is a Plasma Membrane Molybdenum Transporter Required for Nitrogenase Activity in Root Nodules under Molybdenum Deficiency. New Phytol. 2017, 216, 1223–1235. [Google Scholar] [CrossRef]
- Gil-Díez, P.; Tejada-Jiménez, M.; León-Mediavilla, J.; Wen, J.; Mysore, K.S.; Imperial, J.; González-Guerrero, M. MtMOT1.2 Is Responsible for Molybdate Supply to Medicago truncatula Nodules. Plant. Cell Environ. 2019, 42, 310–320. [Google Scholar] [CrossRef]
- Forsberg, S.K.G.; Andreatta, M.E.; Huang, X.Y.; Danku, J.; Salt, D.E.; Carlborg, Ö. The Multi-Allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance. PLoS Genet. 2015, 11, 1005648. [Google Scholar] [CrossRef]
- Li, W.; Fingrut, D.R.; Maxwell, D.P. Characterization of a Mutant of Chlamydomonas reinhardtii Deficient in the Molybdenum Cofactor. Physiol. Plant. 2009, 136, 336–350. [Google Scholar] [CrossRef]
- Tejada-Jiménez, M.; Chamizo-Ampudia, A.; Calatrava, V.; Galván, A.; Fernández, E.; Llamas, A. From the Eukaryotic Molybdenum Cofactor Biosynthesis to the Moonlighting Enzyme MARC. Molecules 2018, 23, 3287. [Google Scholar] [CrossRef]
- Schwarz, G.; Mendel, R.R. Molybdenum Cofactor Biosynthesis and Molybdenum Enzymes. Annu. Rev. Plant Biol. 2006, 57, 623–647. [Google Scholar] [CrossRef]
- Kruse, I.; Maclean, A.E.; Hill, L.; Balk, J. Genetic Dissection of Cyclic Pyranopterin Monophosphate Biosynthesis in Plant Mitochondria. Biochem. J. 2018, 475, 495–509. [Google Scholar] [CrossRef] [PubMed]
- González-Ballester, D.; de Montaigu, A.; Higuera, J.J.; Galván, A.; Fernández, E. Functional Genomics of the Regulation of the Nitrate Assimilation Pathway in Chlamydomonas. Plant Physiol. 2005, 137, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. The Chlamydomonas reinhardtii Molybdenum Cofactor Enzyme CrARC Has a Zn-Dependent Activity and Protein Partners Similar to Those of Its Human Homologue. Eukaryot. Cell 2011, 10, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Teschner, J.; Lachmann, N.; Schulze, J.; Geisler, M.; Selbach, K.; Santamaria-Araujo, J.; Balk, J.; Mendel, R.R.; Bittner, F. A Novel Role for Arabidopsis Mitochondrial ABC Transporter ATM3 in Molybdenum Cofactor Biosynthesis. Plant Cell 2010, 22, 468–480. [Google Scholar] [CrossRef]
- Kaufholdt, D.; Baillie, C.K.; Meinen, R.; Mendel, R.R.; Hänsch, R. The Molybdenum Cofactor Biosynthesis Network: In Vivo Protein-Protein Interactions of an Actin Associated Multi-Protein Complex. Front. Plant Sci. 2017, 8, 1946. [Google Scholar] [CrossRef]
- Nakai, Y.; Harada, A.; Hashiguchi, Y.; Nakai, M.; Hayashi, H. Arabidopsis Molybdopterin Biosynthesis Protein Cnx5 Collaborates with the Ubiquitin-like Protein Urm11 in the Thio-Modification of TRNA. J. Biol. Chem. 2012, 287, 30874–30884. [Google Scholar] [CrossRef]
- Veldman, A.; Santamaria-Araujo, J.A.; Sollazzo, S.; Pitt, J.; Gianello, R.; Yaplito-Lee, J.; Wong, F.; Ramsden, C.A.; Reiss, J.; Cook, I.; et al. Successful Treatment of Molybdenum Cofactor Deficiency Type A with CPMP. Pediatrics 2010, 125, e1249-54. [Google Scholar] [CrossRef]
- Farrell, S.; Karp, J.; Hager, R.; Wang, Y.; Adeniyi, O.; Wang, J.; Li, L.; Ma, L.; Peretz, J.; Summan, M.; et al. Regulatory News: Nulibry (Fosdenopterin) Approved to Reduce the Risk of Mortality in Patients with Molybdenum Cofactor Deficiency Type A: FDA Approval Summary. J. Inherit. Metab. Dis. 2021, 44, 1085–1087. [Google Scholar] [CrossRef]
- Kuper, J.; Llamas, A.; Hecht, H.-J.; Mendel, R.R.; Schwarz, G. Structure of the Molybdopterin-Bound Cnx1G Domain Links Molybdenum and Copper Metabolism. Nature 2004, 430, 803–806. [Google Scholar] [CrossRef]
- Kruse, T. Function of Molybdenum Insertases. Molecules 2022, 27, 5372. [Google Scholar] [CrossRef]
- Llamas, A.; Mendel, R.R.; Schwarz, G. Synthesis of Adenylated Molybdopterin: An Essential Step for Molybdenum Insertion. J. Biol. Chem. 2004, 279, 55241–55246. [Google Scholar] [CrossRef]
- Llamas, A.; Otte, T.; Multhaup, G.; Mendel, R.R.; Schwarz, G. The Mechanism of Nucleotide-Assisted Molybdenum Insertion into Molybdopterin. J. Biol. Chem. 2006, 281, 18343–18350. [Google Scholar] [CrossRef]
- Nichols, J.D.; Rajagopalan, K.V. In Vitro Molybdenum Ligation to Molybdopterin Using Purified Components. J. Biol. Chem. 2005, 280, 7817–7822. [Google Scholar] [CrossRef]
- Selles, B.; Moseler, A.; Caubrière, D.; Sun, S.K.; Ziesel, M.; Dhalleine, T.; Hériché, M.; Wirtz, M.; Rouhier, N.; Couturier, J. The Cytosolic Arabidopsis thaliana Cysteine Desulfurase ABA3 Delivers Sulfur to the Sulfurtransferase STR18. J. Biol. Chem. 2022, 298, 101749. [Google Scholar] [CrossRef]
- Mendel, R.R. The History of the Molybdenum Cofactor-A Personal View. Molecules 2022, 27, 4934. [Google Scholar] [CrossRef]
- Aguilar, M.; Cárdenas, J.; Fernández, E. Quantitation of Molybdopterin Oxidation Product in Wild-Type and Molybdenum Cofactor Deficient Mutants of Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1992, 1160, 269–274. [Google Scholar] [CrossRef]
- Witte, C.-P.; Igeño, M.I.; Mendel, R.; Schwarz, G.; Fernández, E. The Chlamydomonas reinhardtii MoCo Carrier Protein Is Multimeric and Stabilizes Molybdopterin Cofactor in a Molybdate Charged Form. FEBS Lett. 1998, 431, 205–209. [Google Scholar] [CrossRef]
- Aguilar, M.; Kalakoutskii, K.; Cárdenas, J.; Fernández, E. Direct Transfer of Molybdopterin Cofactor to Aponitrate Reductase from a Carrier Protein in Chlamydomonas reinhardtii. FEBS Lett 1992, 307, 162–163. [Google Scholar] [CrossRef]
- Krausze, J.; Hercher, T.W.; Archna, A.; Kruse, T. The Structure of the Moco Carrier Protein from Rippkaea orientalis. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2020, 76, 453–463. [Google Scholar] [CrossRef]
- Hercher, T.W.; Krausze, J.; Yang, J.; Kirk, M.L.; Kruse, T. Identification and Characterization of the Volvox carteri Moco Carrier Protein. Biosci. Rep. 2020, 40, BSR20202351. [Google Scholar] [CrossRef]
- Kruse, T.; Gehl, C.; Geisler, M.; Lehrke, M.; Ringel, P.; Hallier, S.; Hänsch, R.; Mendel, R.R. Identification and Biochemical Characterization of Molybdenum Cofactor-Binding Proteins from Arabidopsis thaliana. J. Biol. Chem. 2010, 285, 6623–6635. [Google Scholar] [CrossRef] [PubMed]
- Kruse, T. Moco Carrier and Binding Proteins. Molecules 2022, 27, 6571. [Google Scholar] [CrossRef] [PubMed]
- Lawson, D.M.; Williams, C.E.; White, D.J.; Choay, A.P.; Mitchenall, L.A.; Pau, R.N. Protein Ligands for Molybdate. Specificity and Charge Stabilisation at the Anion-Binding Sites of Periplasmic and Intracellular Molybdate-Binding Proteins of Azotobacter vinelandii. J. Chem. Soc. Dalt. Trans. 1997, 21, 3981–3984. [Google Scholar] [CrossRef]
- Fenske, D.; Gnida, M.; Schneider, K.; Meyer-Klaucke, W.; Schemberg, J.; Henschel, V.; Meyer, A.-K.; Knöchel, A.; Müller, A. A New Type of Metalloprotein: The Mo Storage Protein from Azotobacter vinelandii Contains a Polynuclear Molybdenum-Oxide Cluster. ChemBioChem 2005, 6, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Steinke, D.R.; Majak, W.; Sorensen, T.S.; Parvez, M. Chelation of Molybdenum in Medicago Sativa (Alfalfa) Grown on Reclaimed Mine Tailings. J. Agric. Food Chem. 2008, 56, 5437–5442. [Google Scholar] [CrossRef]
- Hille, R. Xanthine Oxidase-A Personal History. Molecules 2023, 28, 1921. [Google Scholar] [CrossRef]
- Pineda, M.; Cardenas, J. Transport and Assimilation of Purines in Chlamydomonas reinhardtii. Sci. Mar. 1996, 60, 195–201. [Google Scholar]
- Schmollinger, S.; Mühlhaus, T.; Boyle, N.R.; Blaby, I.K.; Casero, D.; Mettler, T.; Moseley, J.L.; Kropat, J.; Sommer, F.; Strenkert, D.; et al. Nitrogen-Sparing Mechanisms in Chlamydomonas Affect the Transcriptome, the Proteome, and Photosynthetic Metabolism. Plant Cell 2014, 26, 1410–1435. [Google Scholar] [CrossRef]
- Perez-Vicente, R.; Pineda, M.; Cardenas, J. Isolation and Characterization of Xanthine Dehydrogenase from Chlamydomonas reinhardtii. Physiol. Plant. 1988, 72, 101–107. [Google Scholar] [CrossRef]
- Seo, M.; Koiwai, H.; Akaba, S.; Komano, T.; Oritani, T.; Kamiya, Y.; Koshiba, T. Abscisic Aldehyde Oxidase in Leaves of Arabidopsis thaliana. Plant J. 2000, 23, 481–488. [Google Scholar] [CrossRef]
- Rodríguez-Trelles, F.; Tarrío, R.; Ayala, F.J. Convergent Neofunctionalization by Positive Darwinian Selection after Ancient Recurrent Duplications of the Xanthine Dehydrogenase Gene. Proc. Natl. Acad. Sci. USA 2003, 100, 13413–13417. [Google Scholar] [CrossRef]
- Abu-Ghosh, S.; Iluz, D.; Dubinsky, Z.; Miller, G. Exogenous Abscisic Acid Confers Salinity Tolerance in Chlamydomonas reinhardtii During Its Life Cycle. J. Phycol. 2021, 57, 1323–1334. [Google Scholar] [CrossRef]
- Al-Hijab, L.; Gregg, A.; Davies, R.; Macdonald, H.; Ladomery, M.; Wilson, I. Abscisic Acid Induced a Negative Geotropic Response in Dark-Incubated Chlamydomonas reinhardtii. Sci. Rep. 2019, 9, 12063. [Google Scholar] [CrossRef]
- Feng, C.; Tollin, G.; Enemark, J.H. Sulfite Oxidizing Enzymes. Biochim. Biophys. Acta-Proteins Proteom. 2007, 1774, 527–539. [Google Scholar] [CrossRef]
- Kirk, M.L.; Hille, R. Spectroscopic Studies of Mononuclear Molybdenum Enzyme Centers. Molecules 2022, 27, 4802. [Google Scholar] [CrossRef]
- Randewig, D.; Hamisch, D.; Herschbach, C.; Eiblmeier, M.; Gehl, C.; Jurgeleit, J.; Skerra, J.; Mendel, R.R.; Rennenberg, H.; Hänsch, R. Sulfite Oxidase Controls Sulfur Metabolism under SO2 Exposure in Arabidopsis thaliana. Plant. Cell Environ. 2012, 35, 100–115. [Google Scholar] [CrossRef]
- Mellis, A.T.; Roeper, J.; Misko, A.L.; Kohl, J.; Schwarz, G. Sulfite Alters the Mitochondrial Network in Molybdenum Cofactor Deficiency. Front. Genet. 2021, 11, 594828. [Google Scholar] [CrossRef]
- Nowak, K.; Luniak, N.; Witt, C.; Wüstefeld, Y.; Wachter, A.; Mendel, R.R.; Hänsch, R. Peroxisomal Localization of Sulfite Oxidase Separates It from Chloroplast-Based Sulfur Assimilation. Plant Cell Physiol. 2004, 45, 1889–1894. [Google Scholar] [CrossRef]
- Eilers, T.; Schwarz, G.; Brinkmann, H.; Witt, C.; Richter, T.; Nieder, J.; Koch, B.; Hille, R.; Hänsch, R.; Mendel, R.R. Identification and Biochemical Characterization of Arabidopsis thaliana Sulfite Oxidase: A New Player in Plant Sulfur Metabolism. J. Biol. Chem. 2001, 276, 46989–46994. [Google Scholar] [CrossRef]
- Gerin, S.; Mathy, G.; Blomme, A.; Franck, F.; Sluse, F.E. Plasticity of the Mitoproteome to Nitrogen Sources (Nitrate and Ammonium) in Chlamydomonas reinhardtii: The Logic of Aox1 Gene Localization. Biochim. Biophys. Acta-Bioenerg. 2010, 1797, 994–1003. [Google Scholar] [CrossRef]
- Coelho, C.; Romão, M.J. Structural and Mechanistic Insights on Nitrate Reductases. Protein Sci. 2015, 24, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, E.; Schnell, R.; Ranum, L.P.; Hussey, S.C.; Silflow, C.D.; Lefebvre, P.A. Isolation and Characterization of the Nitrate Reductase Structural Gene of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1989, 86, 6449–6453. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Alvarez, J.M.; Araus, V.; Riveras, E.; Brooks, M.D.; Krouk, G.; Ruffel, S.; Lejay, L.; Crawford, N.M.; Coruzzi, G.M.; et al. Nitrate in 2020: Thirty Years from Transport to Signaling Networks. Plant Cell 2020, 32, 2094–2119. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.V.; Harper, J.E. The Conversion of Nitrite to Nitrogen Oxide(s) by the Constitutive NAD(P)H-Nitrate Reductase Enzyme from Soybean. Plant Physiol. 1988, 88, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Ali, S.; Al Azzawi, T.N.I.; Yun, B.W. Nitric Oxide Acts as a Key Signaling Molecule in Plant Development under Stressful Conditions. Int. J. Mol. Sci. 2023, 24, 4782. [Google Scholar] [CrossRef]
- Gupta, K.J.; Kaladhar, V.C.; Fitzpatrick, T.B.; Fernie, A.R.; Møller, I.M.; Loake, G.J. Nitric Oxide Regulation of Plant Metabolism. Mol. Plant 2022, 15, 228–242. [Google Scholar] [CrossRef]
- Foresi, N.; Correa-Aragunde, N.; Parisi, G.; Caló, G.; Salerno, G.; Lamattina, L. Characterization of a Nitric Oxide Synthase from the Plant Kingdom: NO Generation from the Green Alga Ostreococcus tauri Is Light Irradiance and Growth Phase Dependent. Plant Cell 2010, 22, 3816–3830. [Google Scholar] [CrossRef]
- Santolini, J.; André, F.; Jeandroz, S.; Wendehenne, D. Nitric Oxide Synthase in Plants: Where Do We Stand? Nitric Oxide 2017, 63, 30–38. [Google Scholar] [CrossRef]
- Havemeyer, A.; Bittner, F.; Wollers, S.; Mendel, R.; Kunze, T.; Clement, B. Identification of the Missing Component in the Mitochondrial Benzamidoxime Prodrug-Converting System as a Novel Molybdenum Enzyme. J. Biol. Chem. 2006, 281, 34796–34802. [Google Scholar] [CrossRef]
- Hsieh, S.I.; Castruita, M.; Malasarn, D.; Urzica, E.; Erde, J.; Page, M.D.; Yamasaki, H.; Casero, D.; Pellegrini, M.; Merchant, S.S.; et al. The Proteome of Copper, Iron, Zinc, and Manganese Micronutrient Deficiency in Chlamydomonas reinhardtii. Mol. Cell. Proteom. 2013, 12, 65–86. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. Study of Different Variants of Mo Enzyme CrARC and the Interaction with Its Partners CrCytb5-R and CrCytb5-1. Int. J. Mol. Sci. 2017, 18, 670. [Google Scholar] [CrossRef]
- Ott, G.; Plitzko, B.; Krischkowski, C.; Reichmann, D.; Bittner, F.; Mendel, R.R.; Kunze, T.; Clement, B.; Havemeyer, A. Reduction of Sulfamethoxazole Hydroxylamine (SMX-HA) by the Mitochondrial Amidoxime Reducing Component (MARC). Chem. Res. Toxicol. 2014, 27, 1687–1695. [Google Scholar] [CrossRef]
- Kotthaus, J.; Wahl, B.; Havemeyer, A.; Schade, D.; Garbe-Schönberg, D.; Mendel, R.; Bittner, F.; Clement, B. Reduction of N(ω)-Hydroxy-L-Arginine by the Mitochondrial Amidoxime Reducing Component (MARC). Biochem. J. 2011, 433, 383–391. [Google Scholar] [CrossRef]
- Sparacino-Watkins, C.E.; Tejero, J.; Sun, B.; Gauthier, M.C.; Thomas, J.; Ragireddy, V.; Merchant, B.A.; Wang, J.; Azarov, I.; Basu, P.; et al. Nitrite Reductase and Nitric-Oxide Synthase Activity of the Mitochondrial Molybdopterin Enzymes MARC1 and MARC2. J. Biol. Chem. 2014, 289, 10345–10358. [Google Scholar] [CrossRef]
- Bender, D.; Schwarz, G. Nitrite-Dependent Nitric Oxide Synthesis by Molybdenum Enzymes. FEBS Lett. 2018, 592, 2126–2139. [Google Scholar] [CrossRef]
- Rockel, P.; Strube, F.; Rockel, A.; Wildt, J.; Kaiser, W.M. Regulation of Nitric Oxide (NO) Production by Plant Nitrate Reductase In Vivo and In Vitro. J. Exp. Bot. 2002, 53, 103–110. [Google Scholar] [CrossRef]
- Maiber, L.; Koprivova, A.; Bender, D.; Kopriva, S.; Fischer-Schrader, K. Characterization of the Amidoxime Reducing Components ARC1 and ARC2 from Arabidopsis thaliana. FEBS J. 2022, 289, 5656–5669. [Google Scholar] [CrossRef]
- Bellido-Pedraza, C.M.; Calatrava, V.; Llamas, A.; Fernandez, E.; Sanz-Luque, E.; Galvan, A. Nitrous Oxide Emissions from Nitrite Are Highly Dependent on Nitrate Reductase in the Microalga Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2022, 23, 9412. [Google Scholar] [CrossRef]
- Llamas, A.; Chamizo-Ampudia, A.; Tejada-Jimenez, M.; Galvan, A.; Fernandez, E. The Molybdenum Cofactor Enzyme MARC: Moonlighting or Promiscuous Enzyme? BioFactors 2017, 43, 486–494. [Google Scholar] [CrossRef]
- Gupta, M.N.; Uversky, V.N. Moonlighting Enzymes: When Cellular Context Defines Specificity. Cell. Mol. Life Sci. 2023, 80, 130. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).