Detection of Antagonistic Compounds Synthesized by Bacillus velezensis against Xanthomonas citri subsp. citri by Metabolome and RNA Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Antibacterial Activity of Ethyl Acetate Extracts of Bv-25 and Ba-2 in Landy Broth and YNB
2.3. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of Bv-25 and Ba-2 Extracts Cultured in Landy Broth and YNB
2.4. HPLC-MS Analysis of Ethyl Acetate Extracts from Bv-25 and Ba-2
2.5. Differential Gene Expression Analysis of Bv-25 Grown in Landy Broth and YNB
2.6. Antibacterial Activity Assay of Pure Surfactin and Fengycin against Xcc
3. Results
3.1. Antibacterial Activity of Ethyl Acetate Extracts of Bv-25 in Landy and YNB
3.2. Determination of MIC and MBC of the Ethyl Acetate Extracts of Bv-25 and Ba-2 Incubated in Landy Broth and YNB
3.3. Detection of Antimicrobial Compounds by HPLC-MS Analysis
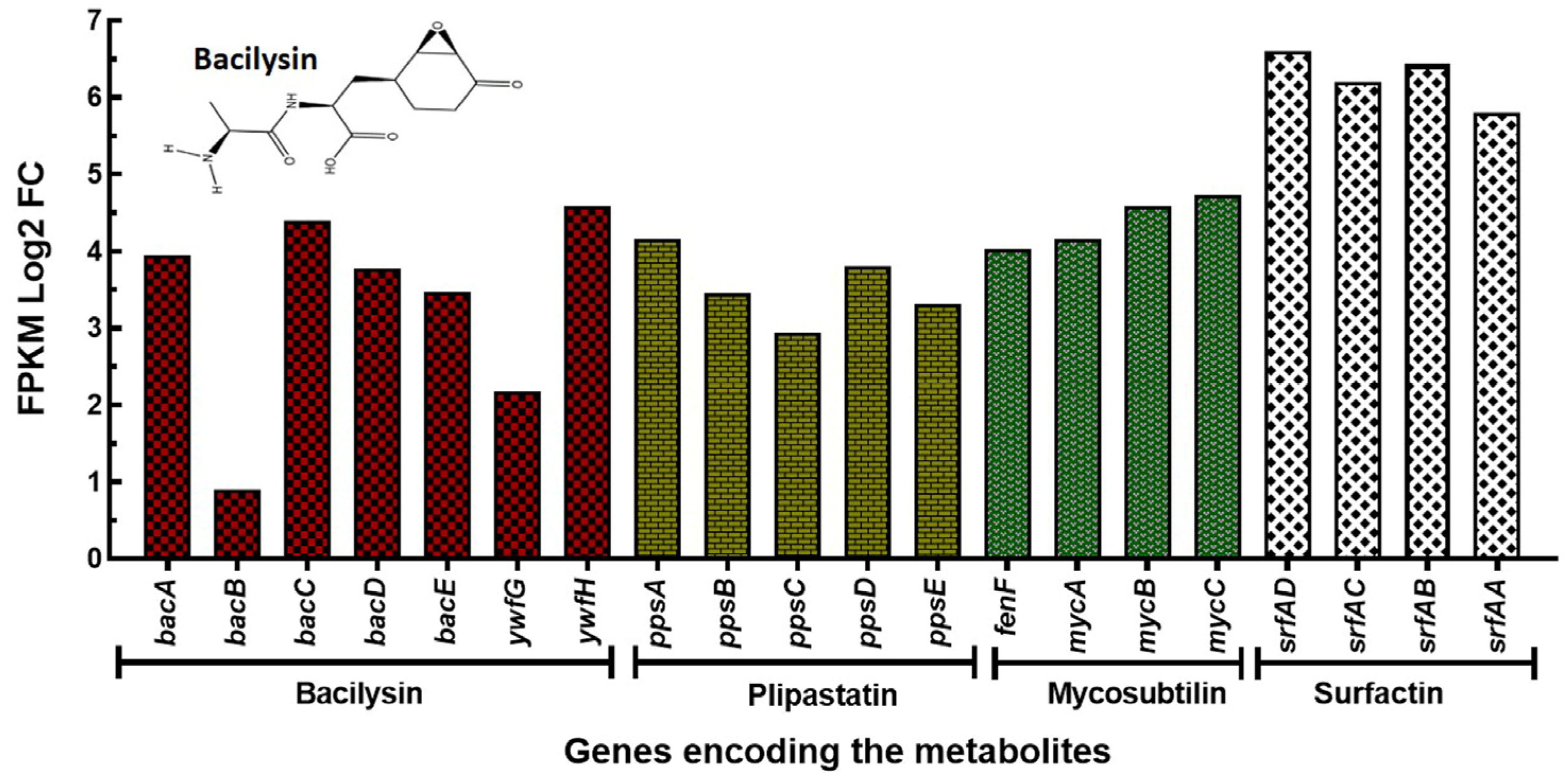
3.4. Differential Gene Expression Analysis of Bv-25 Grown in Landy and YNB Media
3.5. Antibacterial Activity Assay of Pure Surfactin and Fengycin against Xcc
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, N.J.; Bass, C.; Dixon, A.; Neve, P. The evolutionary origins of pesticide resistance. Biol. Rev. 2019, 94, 135–155. [Google Scholar] [CrossRef] [PubMed]
- Symonds, N. Antibiotic resistance in bacteria. Postgrad. Med. J. 1972, 48, 216–221. [Google Scholar] [CrossRef]
- Collinge, D.B.; Funck, D.; Mojgan, J.; Sabrina, R.; Shaw, M.W.; Shaw, R.H. Biological control of plant diseases-What has been achieved and what is the direction? Plant Pathol. 2022, 71, 1024–1047. [Google Scholar] [CrossRef]
- Graham, J.H.; Leite, R.P.; Yonce, H.D.; Myers, A.M. Streptomycin controls citrus canker on sweet orange in Brazil and reduces risk of copper burn on grapefruit in Florida. Proc. Fla. State Hort. Soc. 2008, 121, 118–123. [Google Scholar]
- Gottwald, T.R. Analysis of foci of Asiatic citrus canker in a Florida citrus orchard. Plant Dis. 1992, 76, 389–396. [Google Scholar] [CrossRef]
- Marin, T.G.S.; Galvanin, A.L.; Lanza, F.E.; Behlau, F. Description of copper tolerant Xanthomonas citri subsp. citri and genotypic comparison with sensitive and resistant strains. Plant Pathol. 2019, 68, 1088–1098. [Google Scholar] [CrossRef]
- Behlau, F.; Canteros, B.I.; Minsavage, G.V.; Jones, J.B.; Graham, J.H. Molecular characterization of copper resistance genes from Xanthomonas citri subsp. citri and Xanthomonas alfalfae subsp. citrumelonis. Appl. Environ. Microbiol. 2011, 77, 4089–4096. [Google Scholar] [CrossRef]
- Hyun, J.W.; Kim, H.J.; Yi, P.H.; Hwang, R.Y.; Park, E.W. Mode of action of streptomycin resistance in the citrus canker pathogen (Xanthomonas smithii subsp. citri) in Jeju island. Plant Pathol. J. 2012, 28, 207–211. [Google Scholar] [CrossRef]
- Klein-Gordon, J.M.; Xing, Y.; Garrett, K.A.; Abrahamian, P.; Paret, M.L.; Minsavage, G.V.; Strayer-Scherer, A.L.; Fulton, J.C.; Timilsina, S.; Jones, J.B.; et al. Assessing changes and associations in the Xanthomonas perforans population across Florida commercial tomato fields via a statewide survey. Phytopathology 2021, 111, 1029–1041. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [PubMed]
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A review of the microbial production of bioactive natural products and biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar] [CrossRef] [PubMed]
- Rabbee, M.F.; Baek, K.H. Antimicrobial activities of lipopeptides and polyketides of Bacillus velezensis for agricultural applications. Molecules 2020, 25, 4973. [Google Scholar] [CrossRef] [PubMed]
- Kunst, F.; Ogasawara, N.; Moszer, I.; Albertini, A.M.; Alloni, G.O.; Azevedo, V.; Bertero, M.G.; Bessières, P.; Bolotin, A.; Borchert, S.; et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 1997, 390, 249–256. [Google Scholar] [CrossRef]
- Zaid, D.S.; Cai, S.; Hu, C.; Li, Z.; Li, Y.; Gralnick, J.A. Comparative genome analysis reveals phylogenetic identity of Bacillus velezensis HNA3 and genomic insights into its plant growth promotion and biocontrol effects. Microbiol. Spectr. 2022, 10, e02169-21. [Google Scholar] [CrossRef]
- Chu, D.; Ilyas, N.; Peng, L.; Wang, X.; Wang, D.; Xu, Z.; Gao, Q.; Tan, X.; Zhang, C.; Li, Y.; et al. Genomic insights on fighting bacterial wilt by a novel Bacillus amyloliquefaciens strain Cas02. Microb. Biotechnol. 2022, 15, 1152–1167. [Google Scholar] [CrossRef]
- Xu, B.H.; Lu, Y.Q.; Ye, Z.W.; Zheng, Q.W.; Wei, T.; Lin, J.F.; Guo, L.Q. Genomics-guided discovery and structure identification of cyclic lipopeptides from the Bacillus siamensis JFL15. PLoS ONE 2018, 13, e0202893. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Ali, M.S.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K. Bacillus velezensis: A Valuable member of bioactive molecules within plant microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef]
- Pfeifer, B.A.; Khosla, C. Biosynthesis of polyketides in heterologous hosts. Microbiol. Mol. Biol. Rev. 2001, 65, 106–118. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2007, 16, 115–125. [Google Scholar] [CrossRef]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, C. Fengycins, cyclic lipopeptides from marine Bacillus subtilis strains, kill the plant-pathogenic fungus Magnaporthe grisea by Inducing reactive oxygen species production and chromatin condensation. Appl. Environ. Microbiol. 2018, 84, e00445-18. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, M.; Tang, W.; Guo, Z.; Zhang, Y.; Wang, M.; Horsman, G.P.; Zhong, J.; Lu, Z.; Chen, Y. Initiating polyketide biosynthesis by on-line methyl esterification. Nat. Commun. 2021, 12, 4499. [Google Scholar] [CrossRef]
- Gu, Q.; Yang, Y.; Yuan, Q.; Shi, G.; Wu, L.; Lou, Z.; Huo, R.; Wu, H.; Rainer Borriss, X.G. Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant-pathogenic fungus Fusarium graminearum. Appl. Environ. Microbiol 2017, 83, e01075-17. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Velandia, C.A.; Ongena, M.C.A. Effects of Fengycins and iturins on Fusarium oxysporum f. sp. physali and root colonization by Bacillus velezensis Bs006 protect golden berry against vascular wilt. Phytopathology 2021, 111, 2227–2237. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Ho, M.T.; Weselowski, B.; McDowell, T.; Solomon, O.; Renaud, J.; Yuan, Z.C. Characterization and complete genome analysis of the surfactin-producing, plant-protecting bacterium Bacillus velezensis 9D-6. BMC Microbiol. 2019, 19, 5. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Heng, J.; Qin, S.; Bian, K. A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS ONE 2018, 13, e0198560. [Google Scholar] [CrossRef]
- Chen, X.-H.; Vater, J.; Piel, J.; Franke, P.; Scholz, R.; Schneider, K.; Koumoutsi, A.; Hitzeroth, G.; Grammel, N.; Strittmatter, A.W.; et al. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 2006, 188, 4024–4036. [Google Scholar] [CrossRef]
- Wu, L.; Wu, H.; Chen, L.; Yu, X.; Borriss, R.; Gao, X. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 2015, 5, 12975. [Google Scholar] [CrossRef]
- Han, X.; Shen, D.; Xiong, Q.; Bao, B.; Zhang, W.; Dai, T.; Zhao, Y.; Borriss, R.; Fan, B. The plant-beneficial rhizobacterium Bacillus velezensis FZB42 controls the soybean pathogen Phytophthora sojae due to bacilysin production. Appl. Environ. Microbiol. 2021, 87, e01601-21. [Google Scholar] [CrossRef]
- Kalyon, B.; Helaly, S.E.; Scholz, R.; Nachtigall, J.; Vater, J.; Borriss, R.; Süssmuth, R.D. Plantazolicin A and B: Structure elucidation of ribosomally synthesized thiazole/oxazole peptides from Bacillus amyloliquefaciens FZB42. Org. Lett. 2011, 13, 2996–2999. [Google Scholar] [CrossRef] [PubMed]
- Scholz, R.; Vater, J.; Budiharjo, A.; Wang, Z.; He, Y.; Dietel, K.; Schwecke, T.; Herfort, S.; Lasch, P.; Borriss, R. Amylocyclicin, a novel circular bacteriocin produced by Bacillus amyloliquefaciens FZB42. J. Bacteriol. 2014, 196, 1842–1852. [Google Scholar] [CrossRef] [PubMed]
- Skaar, E.P. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010, 6, e1000949. [Google Scholar] [CrossRef] [PubMed]
- Rabbee, M.F.; Islam, N.; Baek, K.H. Biocontrol of citrus bacterial canker caused by Xanthomonas citri subsp. citri by Bacillus velezensis. Saudi J. Biol. Sci. 2022, 29, 2363–2371. [Google Scholar] [CrossRef] [PubMed]
- Rabbee, M.F.; Ali, S.; Baek, K. Endophyte Bacillus velezensis isolated from citrus spp. controls streptomycin-resistant Xanthomonas citri subsp. citri that causes citrus bacterial canker. Agronomy 2019, 9, 470. [Google Scholar] [CrossRef]
- Qureshi, K.A.; Bholay, A.D.; Rai, P.K.; Mohammed, H.A. Isolation, characterization, anti-MRSA evaluation, and in-silico multi-target anti-microbial validations of actinomycin X2 and actinomycin D produced by novel Streptomyces smyrnaeus UKAQ_23. Sci. Rep. 2021, 11, 14539. [Google Scholar] [CrossRef]
- Zhi, Y.; Wu, Q.; Xu, Y. Genome and transcriptome analysis of surfactin biosynthesis in Bacillus amyloliquefaciens MT45. Sci. Rep. 2017, 7, 40976. [Google Scholar] [CrossRef]
- Cho, M.S.; Jin, Y.J.; Kang, B.K.; Park, Y.K.; Kim, C.; Park, D.S. Understanding the ontogeny and succession of Bacillus velezensis and B. subtilis subsp. subtilis by focusing on kimchi fermentation. Sci. Rep. 2018, 8, 7045. [Google Scholar] [CrossRef]
- Cao, Y.; Pi, H.; Chandrangsu, P.; Li, Y.; Wang, Y.; Zhou, H.; Xiong, H.; Helmann, J.D.; Cai, Y. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 2018, 8, 4360. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Uhl, J.; Grosch, R.; Alquéres, S.; Pittroff, S.; Dietel, K.; Schmitt-Kopplin, P.; Borriss, R.; Hartmann, A. Cyclic lipopeptides of Bacillus amyloliquefaciens subsp. plantarum colonizing the lettuce rhizosphere enhance plant defense responses toward the bottom rot pathogen Rhizoctonia solani. Mol. Plant-Microbe Interact. 2015, 28, 984–995. [Google Scholar] [CrossRef]
- Zeriouh, H.; Romero, D.; García-Gutiérrez, L.; Cazorla, F.M.; De Vicente, A.; Pérez-García, A. The iturin-like lipopeptides are essential components in the biological control arsenal of Bacillus subtilis against bacterial diseases of cucurbits. Mol. Plant-Microbe Interact. 2011, 24, 1540–1552. [Google Scholar] [CrossRef]
- Hynek, M.; Marešov, H.; Ju, T. Killing Effect of Bacillus velezensis FZB42 on a Xanthomonas campestris pv. campestris (Xcc) strain newly isolated from cabbage Brassica oleracea Convar. Capitata (L.): A Metabolomic Study. Microorganisms 2021, 9, 1410. [Google Scholar]
- Wang, T.; Liu, X.H.; Wu, M.B.; Ge, S. Molecular insights into the antifungal mechanism of bacilysin. J. Mol. Model. 2018, 24, 118. [Google Scholar] [CrossRef] [PubMed]
- Gong, A.D.; Li, H.P.; Yuan, Q.S.; Song, X.S.; Yao, W.; He, W.J.; Zhang, J.B.; Liao, Y.C. Antagonistic mechanism of iturin a and plipastatin a from Bacillus amyloliquefaciens S76-3 from wheat spikes against Fusarium graminearum. PLoS ONE 2015, 10, e0116871. [Google Scholar] [CrossRef] [PubMed]
- Duitman, E.H.; Hamoen, L.W.; Rembold, M.; Venema, G.; Seitz, H.; Saenger, W.; Bernhard, F.; Reinhardt, R.; Schmidt, M.; Ullrich, C.; et al. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: A multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 1999, 96, 13294–13299. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Zhang, Z.; Li, Y.; Zhang, X.; Duan, Y.; Wang, Q. Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactin-mediated antibacterial activity and colonization. Front. Microbiol. 2017, 8, 1973. [Google Scholar] [CrossRef]
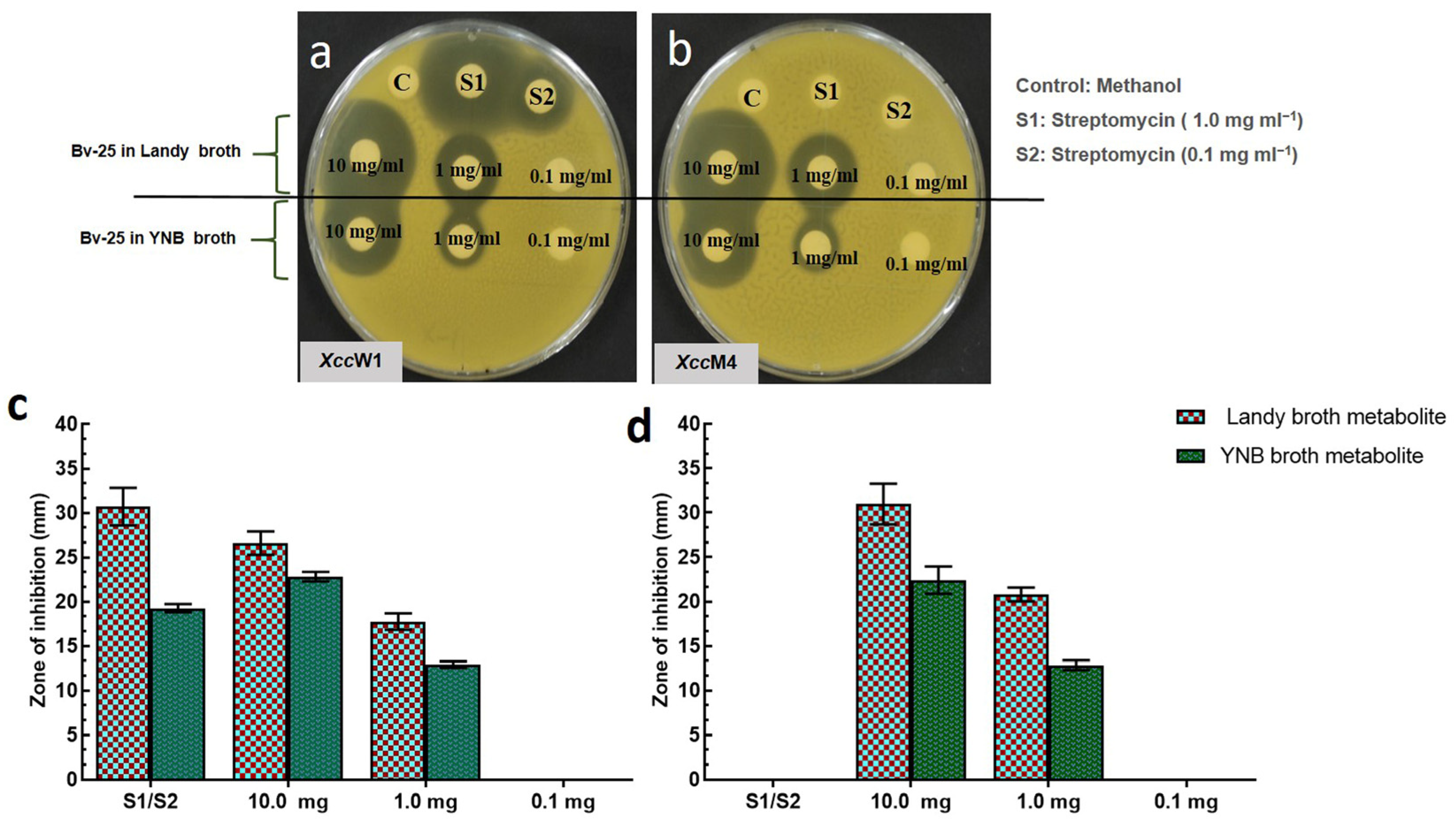
| Xcc Strains | Extract of Bv-25 Using Landy Broth | Extract of Bv-25 Using YNB | Streptomycin | |||
|---|---|---|---|---|---|---|
| MIC (µg mL−1) | MBC (µg mL−1) | MIC (µg mL−1) | MBC (µg mL−1) | MIC (µg mL−1) | MBC (µg mL−1) | |
| XccW1 | 31.25 | 62.5 | 62.5 | 125.0 | 1.25 | 2.5 |
| XccM4 | 31.25 | 62.5 | 62.5 | 125.0 | >500 | >1000 |
| Compound Name | M/Z | RT (min) | Ba-2 | Bv-25 | Fold Change (Bv-25/Ba-2) | p-Value |
|---|---|---|---|---|---|---|
| Oxydifficidin | 559.0 | 43.3 | 216,380.3 | 30,417,069.1 | 140.6 | 0.0027 |
| Bacillaene | 582.2 | 31.3 | 291,622.7 | 2,774,552.5 | 9.5 | 0.0001 |
| Macrolactin | 524.2 | 38.6 | 308,031.1 | 2,211,408.9 | 7.2 | 0.0046 |
| Surfactin | 1036.6 | 27.0 | 93,166.2 | 1,420,585.5 | 15.2 | 0.0000 |
| Fengycin A | 1475.9 | 30.5 | 128,474.2 | 1,092,443.5 | 8.5 | 0.0006 |
| Fengycin B | 1489.3 | 31.3 | 158,266.9 | 983,475.3 | 6.2 | 0.0005 |
| Bacillomycin D | 1118.6 | 40.5 | 92,804.4 | 19,132,037.6 | 206.2 | 0.0031 |
| Iturin | 1083.7 | 29.3 | 272,286.6 | 2,513,178.3 | 9.2 | 0.0036 |
| Active Compound | Gene | UniProt ID | Product |
|---|---|---|---|
| Bacilysin | bacA | Q8KWT6 | Prephenate decarboxylase |
| bacB | Q8KWT5 | 3-[(4R)-4-hydroxycyclohexa-1,5-dien-1-yl]-2-oxopropanoate isomerase | |
| bacC | Q8KWT4 | Dihydroanticapsin dehydrogenase | |
| bacD | Q8KWT3 | L-alanine-anticapsin ligase | |
| bacE | Q8KWT2 | Putative bacilysin exporter | |
| ywfG | P39643 | Probable aspartate aminotransferase | |
| ywfH | P39644 | Bacilysin biosynthesis oxidoreductase | |
| Plipastatin or Fengycin | ppsA | P39845 | Plipastatin synthase subunit A |
| ppsB | P39846 | Plipastatin synthase subunit B | |
| ppsC | P39847 | Plipastatin synthase subunit C | |
| ppsD | P94459 | Plipastatin synthase subunit D | |
| ppsE | O31827 | Plipastatin synthase subunit E | |
| Mycosubtilin | fenF | Q9R9J2 | Malonyl CoA-acyl carrier protein transacylase |
| mycA | Q9R9J1 | Mycosubtilin synthase subunit A | |
| mycB | Q9R9J0 | Mycosubtilin synthase subunit B | |
| mycC | Q9R9I9 | Mycosubtilin synthase subunit C | |
| Surfactin | srfAD | Q08788 | Surfactin synthase thioesterase subunit |
| srfAC | Q08787 | Surfactin synthase subunit 3 | |
| srfAB | Q04747 | Surfactin synthase subunit 2 | |
| srfAA | P27206 | Surfactin synthase subunit 1 |
| Bioactivity | Gene | Product | Log2 FC |
|---|---|---|---|
| Biofilm formation (Exopolysaccharide component) | epsC | Probable polysaccharide biosynthesis protein | −3.17 |
| epsD | Putative glycosyltransferase | −2.08 | |
| epsE | Putative glycosyltransferase | −4.00 | |
| epsF | Putative glycosyltransferase | −0.45 | |
| epsG | Transmembrane protein | 0.04 | |
| epsH | Putative glycosyltransferase | −1.25 | |
| epsI | Putative pyruvyl transferase | −0.85 | |
| epsJ | Uncharacterized glycosyltransferase | 0.51 | |
| epsK | Uncharacterized membrane protein | 0.20 | |
| epsL | Uncharacterized sugar transferase | −0.88 | |
| epsM | Putative acetyltransferase EpsM | −1.37 | |
| epsN | Putative pyridoxal phosphate-dependent aminotransferase | −0.55 | |
| Plant growth promotion and ISR induction (3- hydroxy-2-butanone) | alsR | HTH-type transcriptional regulator | 2.77 |
| alsS | Acetolactate synthase | −3.58 | |
| alsD | Alpha-acetolactate decarboxylase | −7.67 | |
| Plant growth promotion (Indole acetic acid) | ysnE | Uncharacterized N-acetyltransferase | inf |
| yhcX | Hydrolase | −0.48 | |
| Plant growth promotion (Trehalose) | treR | Trehalose operon transcriptional repressor | 1.91 |
| treA | Trehalose-6-phosphate hydrolase | 1.32 | |
| treP | PTS system trehalose-specific EIIBC component | 0.72 |
| Bioactivity | Gene | Product | Log2 FC |
|---|---|---|---|
| Bacillibactin for iron (Fe2+) assimilation | dhbA | 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase | 0.80 |
| dhbC | Isochorismate synthase | −1.52 | |
| dhbE | 2,3-dihydroxybenzoate-AMP ligase | −2.45 | |
| dhbB | Isochorismatase | −3.08 | |
| dhbF | Dimodular nonribosomal peptide synthase | −1.32 | |
| Manganese (Mn2+) assimilation | mntH | Divalent metal cation transporter | 3.71 |
| mntP | Putative manganese efflux pump | 0.69 | |
| mntR | Transcriptional regulator | −0.75 | |
| Magnesium (Mg2+) assimilation | mgtE | Magnesium transporter | 2.19 |
| corA | Magnesium transport protein | −2.66 | |
| Potassium (K) assimilation | ktrD | Ktr system potassium uptake protein D | 1.06 |
| ktrC | Ktr system potassium uptake protein C | −2.75 | |
| Nitrogen (N) assimilation | moaA | Cyclic pyranopterin monophosphate synthase | −1.03 |
| moaB | Molybdenum cofactor biosynthesis protein B | 1.59 | |
| moaC | Cyclic pyranopterin monophosphate synthase | 0.79 | |
| moaD | Molybdopterin synthase sulfur carrier subunit | 0.00 | |
| nasB | Assimilatory nitrate reductase electron transfer subunit | 1.34 | |
| nasD | Nitrite reductase [NAD(P)H] | 2.82 | |
| nasF | Uroporphyrinogen-III C-methyltransferase | 0.41 | |
| nrgB | Nitrogen regulatory PII-like protein | −3.03 | |
| narK | Nitrite extrusion protein | 3.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabbee, M.F.; Baek, K.-H. Detection of Antagonistic Compounds Synthesized by Bacillus velezensis against Xanthomonas citri subsp. citri by Metabolome and RNA Sequencing. Microorganisms 2023, 11, 1523. https://doi.org/10.3390/microorganisms11061523
Rabbee MF, Baek K-H. Detection of Antagonistic Compounds Synthesized by Bacillus velezensis against Xanthomonas citri subsp. citri by Metabolome and RNA Sequencing. Microorganisms. 2023; 11(6):1523. https://doi.org/10.3390/microorganisms11061523
Chicago/Turabian StyleRabbee, Muhammad Fazle, and Kwang-Hyun Baek. 2023. "Detection of Antagonistic Compounds Synthesized by Bacillus velezensis against Xanthomonas citri subsp. citri by Metabolome and RNA Sequencing" Microorganisms 11, no. 6: 1523. https://doi.org/10.3390/microorganisms11061523
APA StyleRabbee, M. F., & Baek, K.-H. (2023). Detection of Antagonistic Compounds Synthesized by Bacillus velezensis against Xanthomonas citri subsp. citri by Metabolome and RNA Sequencing. Microorganisms, 11(6), 1523. https://doi.org/10.3390/microorganisms11061523







