The Detrimental Effects of Peripartum Antibiotics on Gut Proliferation and Formula Feeding Injury in Neonatal Mice Are Alleviated with Lactobacillus rhamnosus GG
Abstract
1. Introduction
2. Materials and Methods
3. Results
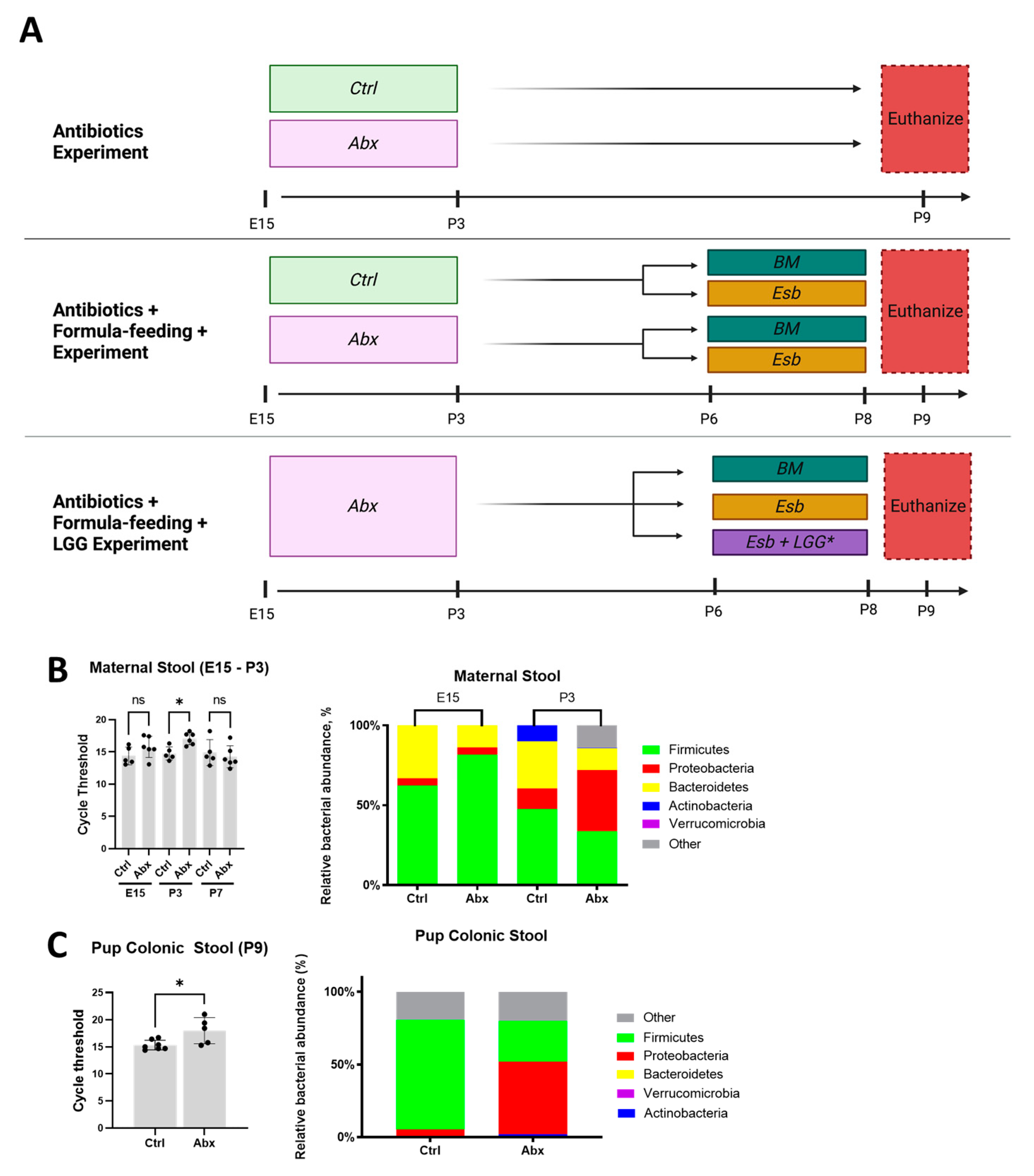
3.1. Administration of Peripartum Antibiotics Results in Maternal and Neonatal Gut Dysbiosis
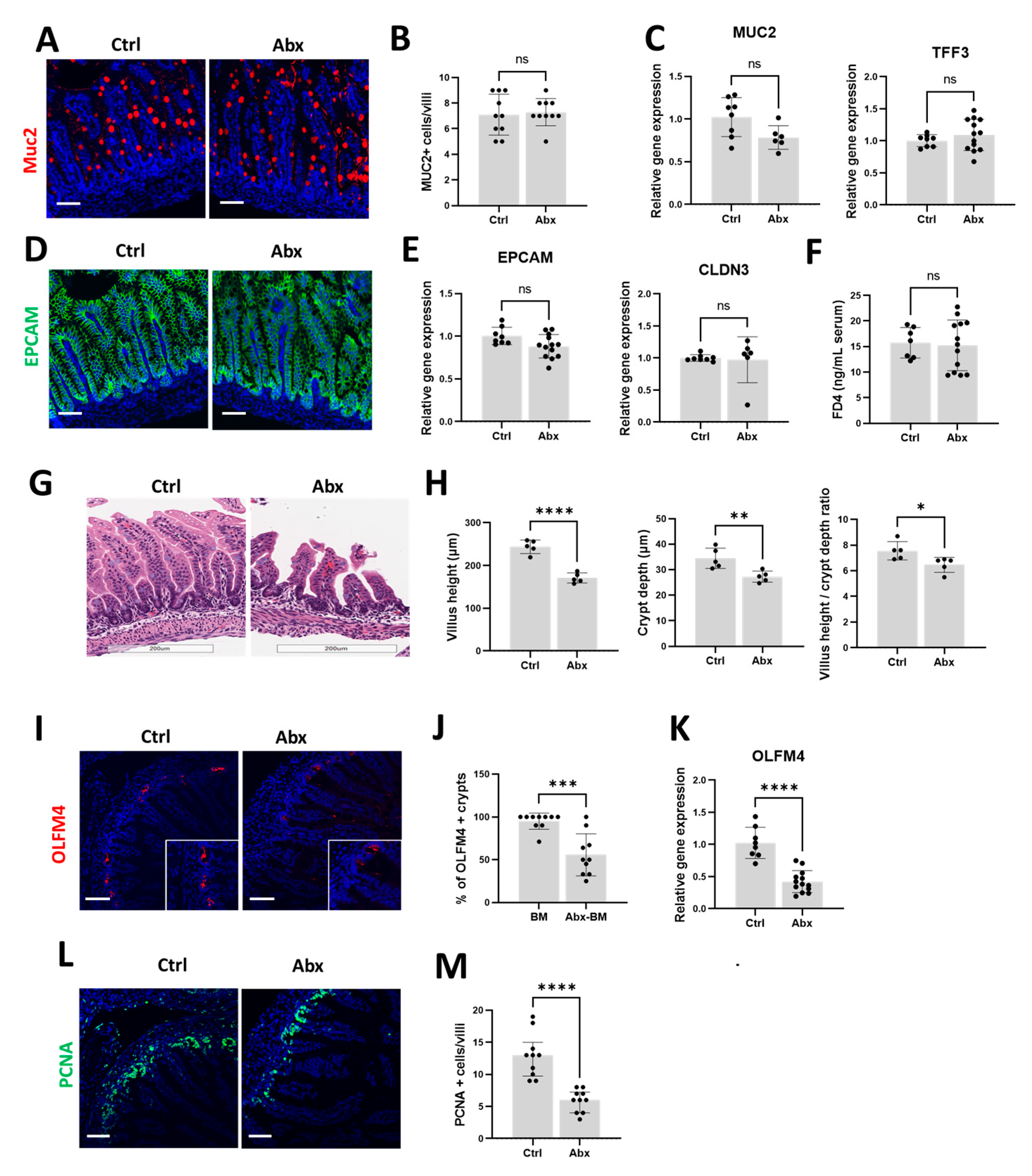
3.2. Peripartum Antibiotics Impair Intestinal Proliferation of the Developing Gut without Affecting the Gut Mucosal Barrier or Gut Permeability
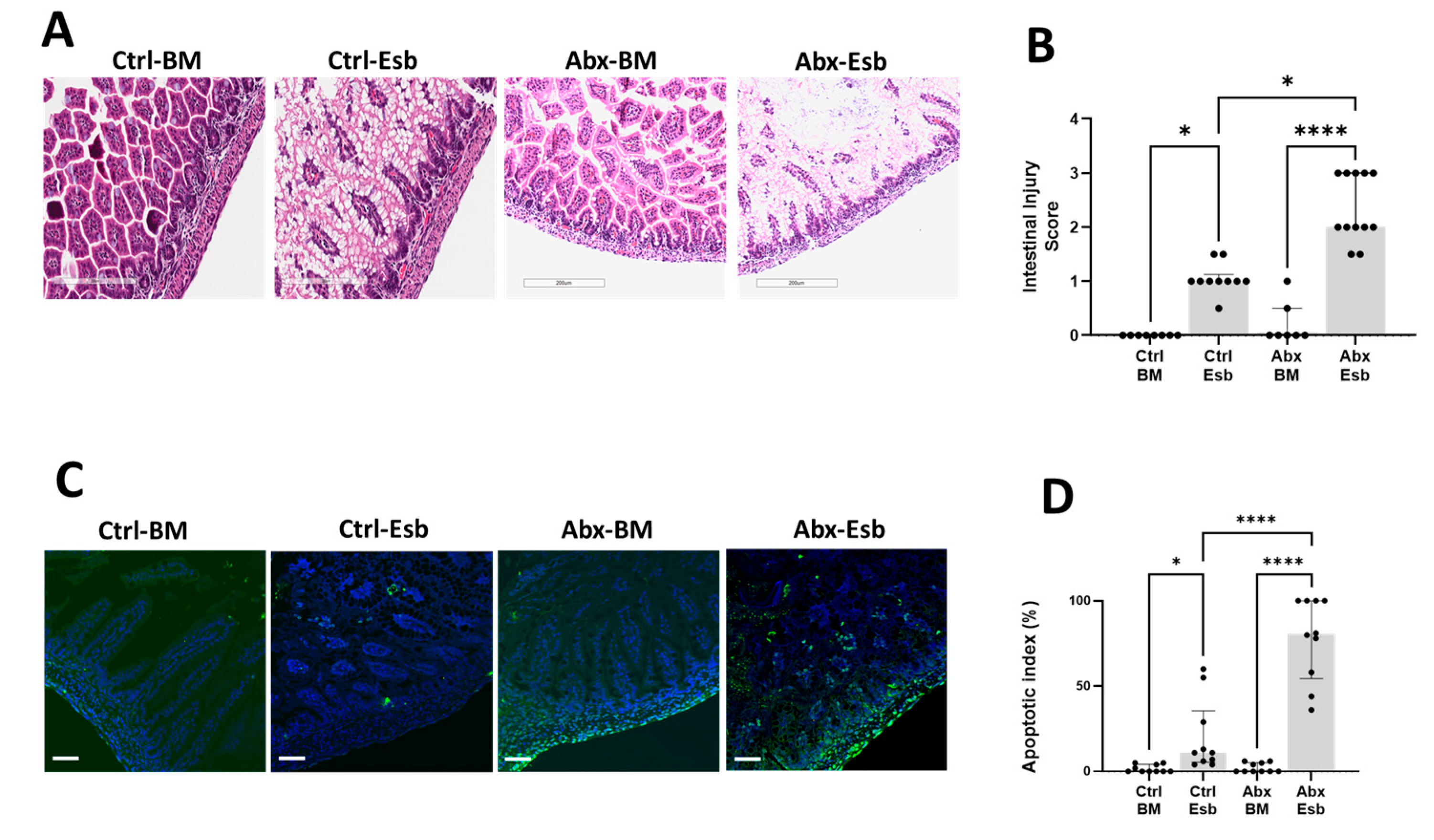
3.3. Peripartum Antibiotics Potentiate Neonatal Gut Injury from Formula Feeding
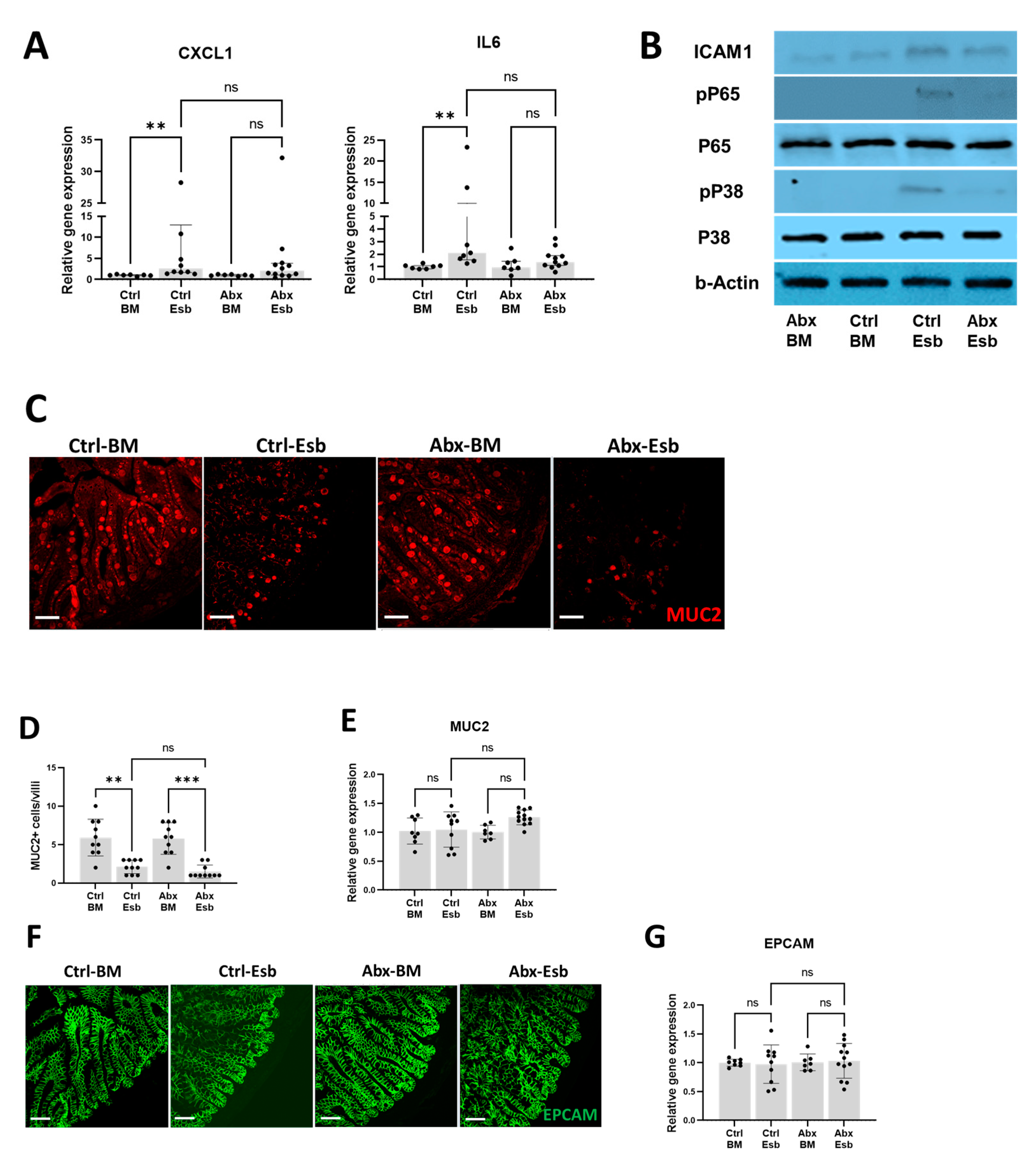
3.4. Worse Gut Injury with Peripartum Antibiotics Is Not Associated with Increased Inflammation Nor with Reduced Mucin or Tight Junction Protein Expression
3.5. Peripartum Antibiotics Potentiate NEC-like Injury by Causing Impairments in Intestinal Proliferation
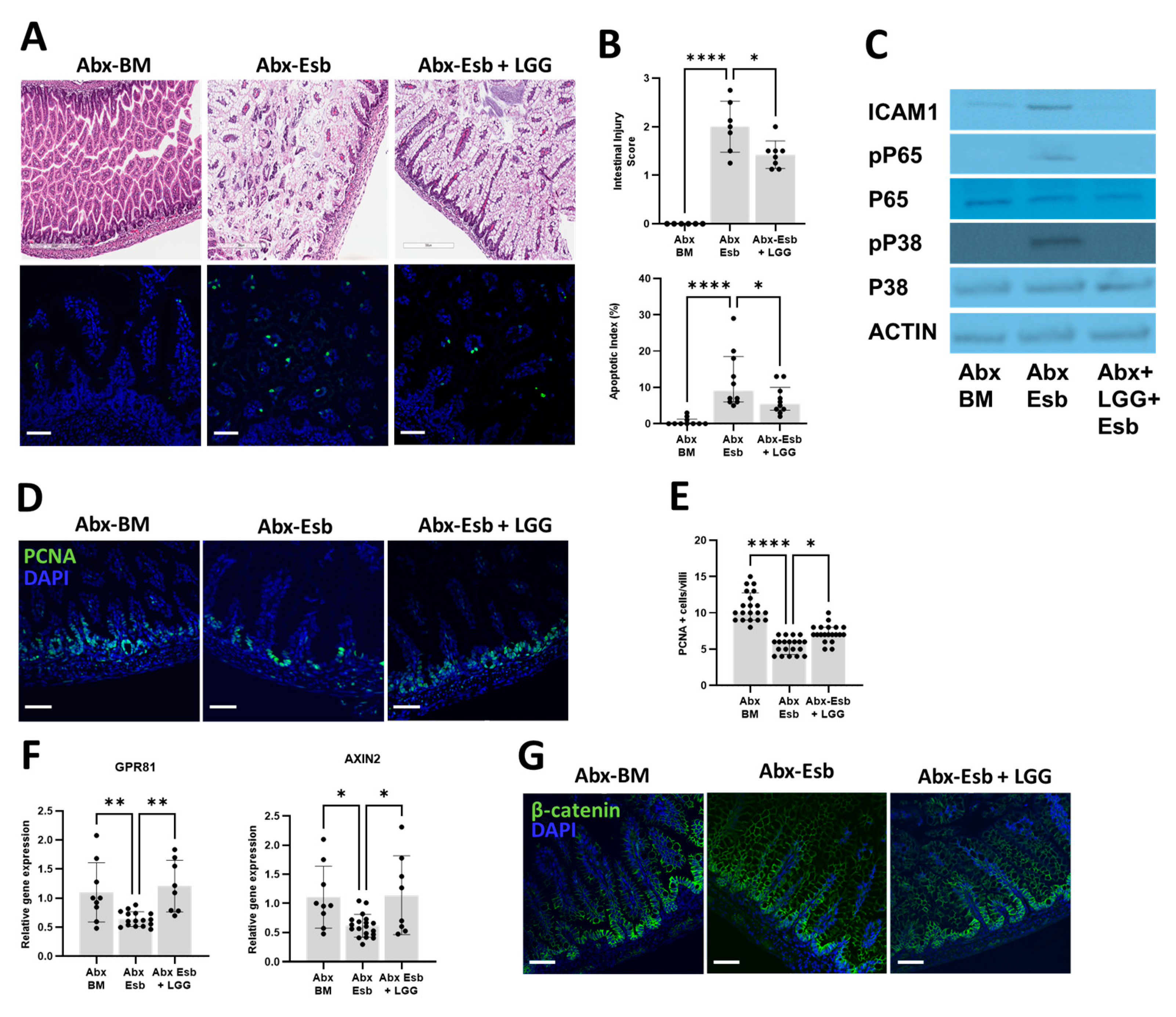
3.6. The Probiotic LGG Decreases NEC-like Injury Potentiated by Peripartum Antibiotics through Activation of the Gpr81-Wnt-β-Catenin Pathway
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stokholm, J.; Schjørring, S.; Pedersen, L.; Bischoff, A.L.; Følsgaard, N.; Carson, C.G.; Chawes, B.L.K.; Bønnelykke, K.; Mølgaard, A.; Krogfelt, K.A.; et al. Prevalence and predictors of antibiotic administration during pregnancy and birth. PLoS ONE 2013, 8, e82932. [Google Scholar] [CrossRef] [PubMed]
- Broe, A.; Pottegård, A.; Lamont, R.F.; Jørgensen, J.S.; Damkier, P. Increasing use of antibiotics in pregnancy during the period 2000–2010: Prevalence, timing, category, and demographics. BJOG Int. J. Obstet. Gynaecol. 2014, 121, 988–996. [Google Scholar] [CrossRef]
- Committee on Practice Bulletins-Obstetrics. ACOG Practice Bulletin No. 188: Prelabor Rupture of Membranes. Obstet. Gynecol. 2018, 131, e1–e14. [Google Scholar] [CrossRef]
- Conde-Agudelo, A.; Romero, R.; Jung, E.J.; Garcia Sánchez, Á.J. Management of clinical chorioamnionitis: An evidence-based approach. Am. J. Obstet. Gynecol. 2020, 223, 848–869. [Google Scholar] [CrossRef]
- Committee on Practice Bulletins-Obstetrics. ACOG Practice Bulletin No. 199: Use of Prophylactic Antibiotics in Labor and Delivery. Obstet. Gynecol. 2018, 132, e103–e119. [Google Scholar] [CrossRef] [PubMed]
- Prevention of Group B Streptococcal Early-Onset Disease in Newborns: ACOG Committee Opinion, Number 797. Obstet. Gynecol. 2020, 135, e51–e72. [CrossRef]
- Flannery, D.D.; Ross, R.K.; Mukhopadhyay, S.; Tribble, A.C.; Puopolo, K.M.; Gerber, J.S. Temporal Trends and Center Variation in Early Antibiotic Use among Premature Infants. JAMA Netw. Open 2018, 1, e180164. [Google Scholar] [CrossRef]
- Flannery, D.D.; Edwards, E.M.; Puopolo, K.M.; Horbar, J.D. Early-Onset Sepsis among Very Preterm Infants. Pediatrics 2021, 148, e2021052456. [Google Scholar] [CrossRef] [PubMed]
- Jokela, R.; Korpela, K.; Jian, C.; Dikareva, E.; Nikkonen, A.; Saisto, T.; Skogberg, K.; de Vos, W.M.; Kolho, K.-L.; Salonen, A. Quantitative insights into effects of intrapartum antibiotics and birth mode on infant gut microbiota in relation to well-being during the first year of life. Gut Microbes 2022, 14, 2095775. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Zhao, X.; Moeder, W.; Tun, H.M.; Simons, E.; Mandhane, P.J.; Moraes, T.J.; Turvey, S.E.; Subbarao, P.; Scott, J.A.; et al. Impact of Maternal Intrapartum Antibiotics, and Caesarean Section with and without Labour on Bifidobacterium and Other Infant Gut Microbiota. Microorganisms 2021, 9, 1847. [Google Scholar] [CrossRef]
- Nogacka, A.; Salazar, N.; Suárez, M.; Milani, C.; Arboleya, S.; Solís, G.; Fernández, N.; Alaez, L.; Hernández-Barranco, A.M.; de Los Reyes-Gavilán, C.G.; et al. Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. Microbiome 2017, 5, 93. [Google Scholar] [CrossRef]
- Gasparrini, A.J.; Wang, B.; Sun, X.; Kennedy, E.A.; Hernandez-Leyva, A.; Ndao, I.M.; Tarr, P.I.; Warner, B.B.; Dantas, G. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat. Microbiol. 2019, 4, 2285–2297. [Google Scholar] [CrossRef] [PubMed]
- Kummeling, I.; Stelma, F.F.; Dagnelie, P.C.; Snijders, B.E.P.; Penders, J.; Huber, M.; van Ree, R.; van den Brandt, P.A.; Thijs, C. Early life exposure to antibiotics and the subsequent development of eczema, wheeze, and allergic sensitization in the first 2 years of life: The KOALA Birth Cohort Study. Pediatrics 2007, 119, e225–e231. [Google Scholar] [CrossRef] [PubMed]
- Metzler, S.; Frei, R.; Schmaußer-Hechfellner, E.; von Mutius, E.; Pekkanen, J.; Karvonen, A.M.; Kirjavainen, P.V.; Dalphin, J.-C.; Divaret-Chauveau, A.; Riedler, J.; et al. Association between antibiotic treatment during pregnancy and infancy and the development of allergic diseases. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2019, 30, 423–433. [Google Scholar] [CrossRef]
- Chelimo, C.; Camargo, C.A.; Morton, S.M.B.; Grant, C.C. Association of Repeated Antibiotic Exposure Up to Age 4 Years with Body Mass at Age 4.5 Years. JAMA Netw. Open 2020, 3, e1917577. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.; Prince, J.M.; Wang, P. Gut microbiome and necrotizing enterocolitis: Understanding the connection to find a cure. Cell Host Microbe 2022, 30, 612–616. [Google Scholar] [CrossRef]
- Carlisle, E.M.; Morowitz, M.J. The intestinal microbiome and necrotizing enterocolitis. Curr. Opin. Pediatr. 2013, 25, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Cotten, C.M.; Taylor, S.; Stoll, B.; Goldberg, R.N.; Hansen, N.I.; Sánchez, P.J.; Ambalavanan, N.; Benjamin, D.K.; NICHD Neonatal Research Network. Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics 2009, 123, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Alexander, V.N.; Northrup, V.; Bizzarro, M.J. Antibiotic exposure in the newborn intensive care unit and the risk of necrotizing enterocolitis. J. Pediatr. 2011, 159, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Kuppala, V.S.; Meinzen-Derr, J.; Morrow, A.L.; Schibler, K.R. Prolonged initial empirical antibiotic treatment is associated with adverse outcomes in premature infants. J. Pediatr. 2011, 159, 720–725. [Google Scholar] [CrossRef]
- Abdel Ghany, E.A.; Ali, A.A. Empirical antibiotic treatment and the risk of necrotizing enterocolitis and death in very low birth weight neonates. Ann. Saudi Med. 2012, 32, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Esmaeilizand, R.; Shah, P.S.; Seshia, M.; Yee, W.; Yoon, E.W.; Dow, K.; Canadian Neonatal Network Investigators. Antibiotic exposure and development of necrotizing enterocolitis in very preterm neonates. Paediatr. Child Health 2018, 23, e56–e61. [Google Scholar] [CrossRef] [PubMed]
- Cantey, J.B.; Pyle, A.K.; Wozniak, P.S.; Hynan, L.S.; Sánchez, P.J. Early Antibiotic Exposure and Adverse Outcomes in Preterm, Very Low Birth Weight Infants. J. Pediatr. 2018, 203, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Gao, H.; Yuan, L.; Wang, L.; Deng, F. Prolonged antibiotic therapy increased necrotizing enterocolitis in very low birth weight infants without culture-proven sepsis. Front. Pediatr. 2022, 10, 949830. [Google Scholar] [CrossRef]
- Chen, W.-Y.; Lo, Y.-C.; Huang, P.-H.; Chen, Y.-X.; Tsao, P.-C.; Lee, Y.-S.; Jeng, M.-J.; Hung, M.-C. Increased antibiotic exposure in early life is associated with adverse outcomes in very low birth weight infants. J. Chin. Med. Assoc. JCMA 2022, 85, 939–943. [Google Scholar] [CrossRef]
- Vatne, A.; Hapnes, N.; Stensvold, H.J.; Dalen, I.; Guthe, H.J.; Støen, R.; Brigtsen, A.K.; Rønnestad, A.E.; Klingenberg, C. Early empirical antibiotics and adverse clinical outcomes in infants born very preterm: A population-based cohort. J. Pediatr. 2022, 253, 107–114.e5. [Google Scholar] [CrossRef]
- Morgan, R.L.; Preidis, G.A.; Kashyap, P.C.; Weizman, A.V.; Sadeghirad, B.; McMaster Probiotic, Prebiotic, and Synbiotic Work Group. Probiotics Reduce Mortality and Morbidity in Preterm, Low-Birth-Weight Infants: A Systematic Review and Network Meta-analysis of Randomized Trials. Gastroenterology 2020, 159, 467–480. [Google Scholar] [CrossRef]
- Sharif, S.; Meader, N.; Oddie, S.J.; Rojas-Reyes, M.X.; McGuire, W. Probiotics to prevent necrotising enterocolitis in very preterm or very low birth weight infants. Cochrane Database Syst. Rev. 2020, 10, CD005496. [Google Scholar] [CrossRef]
- Szajewska, H.; Berni Canani, R.; Domellöf, M.; Guarino, A.; Hojsak, I.; Indrio, F.; Lo Vecchio, A.; Mihatsch, W.A.; Mosca, A.; Orel, R.; et al. Probiotics for the Management of Pediatric Gastrointestinal Disorders: Position Paper of the ESPGHAN Special Interest Group on Gut Microbiota and Modifications. J. Pediatr. Gastroenterol. Nutr. 2023, 76, 232–247. [Google Scholar] [CrossRef]
- Marchand, V. Using probiotics in the paediatric population. Paediatr. Child Health 2012, 17, 575–576. [Google Scholar] [CrossRef]
- van den Akker, C.H.P.; van Goudoever, J.B.; Shamir, R.; Domellöf, M.; Embleton, N.D.; Hojsak, I.; Lapillonne, A.; Mihatsch, W.A.; Berni Canani, R.; Bronsky, J.; et al. Probiotics and Preterm Infants: A Position Paper by the European Society for Paediatric Gastroenterology Hepatology and Nutrition Committee on Nutrition and the European Society for Paediatric Gastroenterology Hepatology and Nutrition Working Group for Probiotics and Prebiotics. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 664–680. [Google Scholar] [CrossRef] [PubMed]
- Su, G.L.; Ko, C.W.; Bercik, P.; Falck-Ytter, Y.; Sultan, S.; Weizman, A.V.; Morgan, R.L. AGA Clinical Practice Guidelines on the Role of Probiotics in the Management of Gastrointestinal Disorders. Gastroenterology 2020, 159, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Ganguli, K.; Meng, D.; Rautava, S.; Lu, L.; Walker, W.A.; Nanthakumar, N. Probiotics prevent necrotizing enterocolitis by modulating enterocyte genes that regulate innate immune-mediated inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G132–G141. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.; Zhu, W.; Ganguli, K.; Shi, H.N.; Walker, W.A. Anti-inflammatory effects of Bifidobacterium longum subsp infantis secretions on fetal human enterocytes are mediated by TLR-4 receptors. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 311, G744–G753. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fatheree, N.Y.; Mangalat, N.; Rhoads, J.M. Lactobacillus reuteri strains reduce incidence and severity of experimental necrotizing enterocolitis via modulation of TLR4 and NF-κB signaling in the intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G608–G617. [Google Scholar] [CrossRef]
- Becker, H.M.; Apladas, A.; Scharl, M.; Fried, M.; Rogler, G. Probiotic Escherichia coli Nissle 1917 and commensal E. coli K12 differentially affect the inflammasome in intestinal epithelial cells. Digestion 2014, 89, 110–118. [Google Scholar] [CrossRef]
- Kern, M.; Aschenbach, J.R.; Tedin, K.; Pieper, R.; Loss, H.; Lodemann, U. Characterization of Inflammasome Components in Pig Intestine and Analysis of the Influence of Probiotic Enterococcus faecium during an Escherichia coli Challenge. Immunol. Investig. 2017, 46, 742–757. [Google Scholar] [CrossRef]
- Jeon, S.G.; Kayama, H.; Ueda, Y.; Takahashi, T.; Asahara, T.; Tsuji, H.; Tsuji, N.M.; Kiyono, H.; Ma, J.S.; Kusu, T.; et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012, 8, e1002714. [Google Scholar] [CrossRef]
- Reyes-Díaz, A.; Mata-Haro, V.; Hernández, J.; González-Córdova, A.F.; Hernández-Mendoza, A.; Reyes-Díaz, R.; Torres-Llanez, M.J.; Beltrán-Barrientos, L.M.; Vallejo-Cordoba, B. Milk Fermented by Specific Lactobacillus Strains Regulates the Serum Levels of IL-6, TNF-α and IL-10 Cytokines in a LPS-Stimulated Murine Model. Nutrients 2018, 10, 691. [Google Scholar] [CrossRef]
- Good, M.; Sodhi, C.P.; Ozolek, J.A.; Buck, R.H.; Goehring, K.C.; Thomas, D.L.; Vikram, A.; Bibby, K.; Morowitz, M.J.; Firek, B.; et al. Lactobacillus rhamnosus HN001 decreases the severity of necrotizing enterocolitis in neonatal mice and preterm piglets: Evidence in mice for a role of TLR9. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G1021–G1032. [Google Scholar] [CrossRef]
- Da Silva, S.; Robbe-Masselot, C.; Ait-Belgnaoui, A.; Mancuso, A.; Mercade-Loubière, M.; Salvador-Cartier, C.; Gillet, M.; Ferrier, L.; Loubière, P.; Dague, E.; et al. Stress disrupts intestinal mucus barrier in rats via mucin O-glycosylation shift: Prevention by a probiotic treatment. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G420–G429. [Google Scholar] [CrossRef] [PubMed]
- Engevik, M.A.; Luk, B.; Chang-Graham, A.L.; Hall, A.; Herrmann, B.; Ruan, W.; Endres, B.T.; Shi, Z.; Garey, K.W.; Hyser, J.M.; et al. Bifidobacterium dentium Fortifies the Intestinal Mucus Layer via Autophagy and Calcium Signaling Pathways. mBio 2019, 10, e01087-19. [Google Scholar] [CrossRef] [PubMed]
- Blackwood, B.P.; Yuan, C.Y.; Wood, D.R.; Nicolas, J.D.; Grothaus, J.S.; Hunter, C.J. Probiotic Lactobacillus Species Strengthen Intestinal Barrier Function and Tight Junction Integrity in Experimental Necrotizing Enterocolitis. J. Probiotics Health 2017, 5, 159. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Gillingham, T.; Guo, Y.; Meng, D.; Zhu, W.; Walker, W.A.; Ganguli, K. Secretions of Bifidobacterium infantis and Lactobacillus acidophilus Protect Intestinal Epithelial Barrier Function. J. Pediatr. Gastroenterol. Nutr. 2017, 64, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Fawley, J.; Cuna, A.; Menden, H.L.; McElroy, S.; Umar, S.; Welak, S.R.; Gourlay, D.M.; Li, X.; Sampath, V. Single-Immunoglobulin Interleukin-1-Related Receptor regulates vulnerability to TLR4-mediated necrotizing enterocolitis in a mouse model. Pediatr. Res. 2018, 83, 164–174. [Google Scholar] [CrossRef]
- Cuna, A.; Yu, W.; Menden, H.L.; Feng, L.; Srinivasan, P.; Chavez-Bueno, S.; Ahmed, I.; Umar, S.; Sampath, V. NEC-like intestinal injury is ameliorated by Lactobacillus rhamnosus GG in parallel with SIGIRR and A20 induction in neonatal mice. Pediatr. Res. 2020, 88, 546–555. [Google Scholar] [CrossRef]
- Yu, W.; Venkatraman, A.; Menden, H.L.; Martinez, M.; Umar, S.; Sampath, V. Short Chain Fatty Acids Ameliorate Necrotizing enterocolitis-like Intestinal Injury through Enhancing Notch1-mediated SIGIRR, TOLLIP and A20 induction. Am. J. Physiol. Gastrointest. Liver Physiol. 2022, 324, G24–G37. [Google Scholar] [CrossRef]
- Yan, F.; Polk, D.B. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J. Biol. Chem. 2002, 277, 50959–50965. [Google Scholar] [CrossRef]
- Sharma, K.; Pooranachithra, M.; Balamurugan, K.; Goel, G. Probiotic mediated colonization resistance against E. coli infection in experimentally challenged Caenorhabditis elegans. Microb. Pathog. 2019, 127, 39–47. [Google Scholar] [CrossRef]
- Liu, D.; Shao, L.; Zhang, Y.; Kang, W. Safety and efficacy of Lactobacillus for preventing necrotizing enterocolitis in preterm infants. Int. J. Surg. 2020, 76, 79–87. [Google Scholar] [CrossRef]
- Meyer, M.P.; Alexander, T. Reduction in necrotizing enterocolitis and improved outcomes in preterm infants following routine supplementation with Lactobacillus GG in combination with bovine lactoferrin. J. Neonatal-Perinat. Med. 2017, 10, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Jimeno, R.; Brailey, P.M.; Barral, P. Quantitative Polymerase Chain Reaction-based Analyses of Murine Intestinal Microbiota after Oral Antibiotic Treatment. J. Vis. Exp. JoVE 2018, 141, e58481. [Google Scholar] [CrossRef]
- Yang, Y.-W.; Chen, M.-K.; Yang, B.-Y.; Huang, X.-J.; Zhang, X.-R.; He, L.-Q.; Zhang, J.; Hua, Z.-C. Use of 16S rRNA Gene-Targeted Group-Specific Primers for Real-Time PCR Analysis of Predominant Bacteria in Mouse Feces. Appl. Environ. Microbiol. 2015, 81, 6749–6756. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Gunasekaran, A.; Eckert, J.; Burge, K.; Zheng, W.; Yu, Z.; Kessler, S.; de la Motte, C.; Chaaban, H. Hyaluronan 35 kDa enhances epithelial barrier function and protects against the development of murine necrotizing enterocolitis. Pediatr. Res. 2020, 87, 1177–1184. [Google Scholar] [CrossRef]
- van der Flier, L.G.; Haegebarth, A.; Stange, D.E.; van de Wetering, M.; Clevers, H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 2009, 137, 15–17. [Google Scholar] [CrossRef]
- Sampath, V.; Martinez, M.; Caplan, M.; Underwood, M.A.; Cuna, A. Necrotizing Enterocolitis in Premature Infants—A Defect in The Brakes? Evidence from Clinical and Animal Studies. Mucosal Immunol. 2023, 16, 208–220. [Google Scholar] [CrossRef]
- Jensen, E.C. Quantitative analysis of histological staining and fluorescence using ImageJ. Anat. Rec. 2013, 296, 378–381. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Kim, T.-Y.; Kim, Y.; Lee, S.-H.; Kim, S.; Kang, S.W.; Yang, J.-Y.; Baek, I.-J.; Sung, Y.H.; Park, Y.-Y.; et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe 2018, 24, 833–846.e6. [Google Scholar] [CrossRef]
- Chen, C.-M.; Chou, H.-C.; Yang, Y.-C.S.H. Maternal Antibiotic Treatment Disrupts the Intestinal Microbiota and Intestinal Development in Neonatal Mice. Front. Microbiol. 2021, 12, 684233. [Google Scholar] [CrossRef]
- Chaaban, H.; Patel, M.M.; Burge, K.; Eckert, J.V.; Lupu, C.; Keshari, R.S.; Silasi, R.; Regmi, G.; Trammell, M.; Dyer, D.; et al. Early Antibiotic Exposure Alters Intestinal Development and Increases Susceptibility to Necrotizing Enterocolitis: A Mechanistic Study. Microorganisms 2022, 10, 519. [Google Scholar] [CrossRef] [PubMed]
- Esaiassen, E.; Hjerde, E.; Cavanagh, J.P.; Pedersen, T.; Andresen, J.H.; Rettedal, S.I.; Støen, R.; Nakstad, B.; Willassen, N.P.; Klingenberg, C. Effects of Probiotic Supplementation on the Gut Microbiota and Antibiotic Resistome Development in Preterm Infants. Front. Pediatr. 2018, 6, 347. [Google Scholar] [CrossRef] [PubMed]
- Grazul, H.; Kanda, L.L.; Gondek, D. Impact of probiotic supplements on microbiome diversity following antibiotic treatment of mice. Gut Microbes 2016, 7, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Schaedler, R.W.; Dubos, R.; Costello, R. The Development of the Bacterial Flora in the Gastrointestinal Tract of Mice. J. Exp. Med. 1965, 122, 59–66. [Google Scholar] [CrossRef]
- Peña, J.A.; Li, S.Y.; Wilson, P.H.; Thibodeau, S.A.; Szary, A.J.; Versalovic, J. Genotypic and phenotypic studies of murine intestinal lactobacilli: Species differences in mice with and without colitis. Appl. Environ. Microbiol. 2004, 70, 558–568. [Google Scholar] [CrossRef]
- Abo, H.; Chassaing, B.; Harusato, A.; Quiros, M.; Brazil, J.C.; Ngo, V.L.; Viennois, E.; Merlin, D.; Gewirtz, A.T.; Nusrat, A.; et al. Erythroid differentiation regulator-1 induced by microbiota in early life drives intestinal stem cell proliferation and regeneration. Nat. Commun. 2020, 11, 513. [Google Scholar] [CrossRef]
- Kim, J.-E.; Li, B.; Fei, L.; Horne, R.; Lee, D.; Loe, A.K.; Miyake, H.; Ayar, E.; Kim, D.-K.; Surette, M.G.; et al. Gut microbiota promotes stem cell differentiation through macrophage and mesenchymal niches in early postnatal development. Immunity 2022, 55, 2300–2317.e6. [Google Scholar] [CrossRef]
- Flanagan, D.J.; Austin, C.R.; Vincan, E.; Phesse, T.J. Wnt Signalling in Gastrointestinal Epithelial Stem Cells. Genes 2018, 9, 178. [Google Scholar] [CrossRef]
- Venkatraman, A.; Yu, W.; Nitkin, C.; Sampath, V. Intestinal Stem Cell Development in the Neonatal Gut: Pathways Regulating Development and Relevance to Necrotizing Enterocolitis. Cells 2021, 10, 312. [Google Scholar] [CrossRef]
- Anand, R.J.; Leaphart, C.L.; Mollen, K.P.; Hackam, D.J. The role of the intestinal barrier in the pathogenesis of necrotizing enterocolitis. Shock 2007, 27, 124–133. [Google Scholar] [CrossRef]
- Patel, R.M.; Myers, L.S.; Kurundkar, A.R.; Maheshwari, A.; Nusrat, A.; Lin, P.W. Probiotic bacteria induce maturation of intestinal claudin 3 expression and barrier function. Am. J. Pathol. 2012, 180, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Cuna, A.; Morowitz, M.J.; Sampath, V. Early antibiotics and risk for necrotizing enterocolitis in premature infants: A narrative review. Front. Pediatr. 2023, 11, 1112812. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuna, A.; Nsumu, M.; Menden, H.L.; Chavez-Bueno, S.; Sampath, V. The Detrimental Effects of Peripartum Antibiotics on Gut Proliferation and Formula Feeding Injury in Neonatal Mice Are Alleviated with Lactobacillus rhamnosus GG. Microorganisms 2023, 11, 1482. https://doi.org/10.3390/microorganisms11061482
Cuna A, Nsumu M, Menden HL, Chavez-Bueno S, Sampath V. The Detrimental Effects of Peripartum Antibiotics on Gut Proliferation and Formula Feeding Injury in Neonatal Mice Are Alleviated with Lactobacillus rhamnosus GG. Microorganisms. 2023; 11(6):1482. https://doi.org/10.3390/microorganisms11061482
Chicago/Turabian StyleCuna, Alain, Marianne Nsumu, Heather L. Menden, Susana Chavez-Bueno, and Venkatesh Sampath. 2023. "The Detrimental Effects of Peripartum Antibiotics on Gut Proliferation and Formula Feeding Injury in Neonatal Mice Are Alleviated with Lactobacillus rhamnosus GG" Microorganisms 11, no. 6: 1482. https://doi.org/10.3390/microorganisms11061482
APA StyleCuna, A., Nsumu, M., Menden, H. L., Chavez-Bueno, S., & Sampath, V. (2023). The Detrimental Effects of Peripartum Antibiotics on Gut Proliferation and Formula Feeding Injury in Neonatal Mice Are Alleviated with Lactobacillus rhamnosus GG. Microorganisms, 11(6), 1482. https://doi.org/10.3390/microorganisms11061482




