The Impact of Anti-Inflammatory Drugs on the Prokaryotic Community Composition and Selected Bacterial Strains Based on Microcosm Experiments
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Determination of Physicochemical Parameters and Cell Count Values
2.3. Selective Cultivation of Bacteria
2.4. DNA Extraction from the Bacterial Strains, 16S rRNA Gene Amplification and Taxonomic Identification
2.5. DNA Extraction from Microcosms and Next-Generation DNA Sequencing
3. Results
3.1. Physicochemical Parameters of the Danube Water
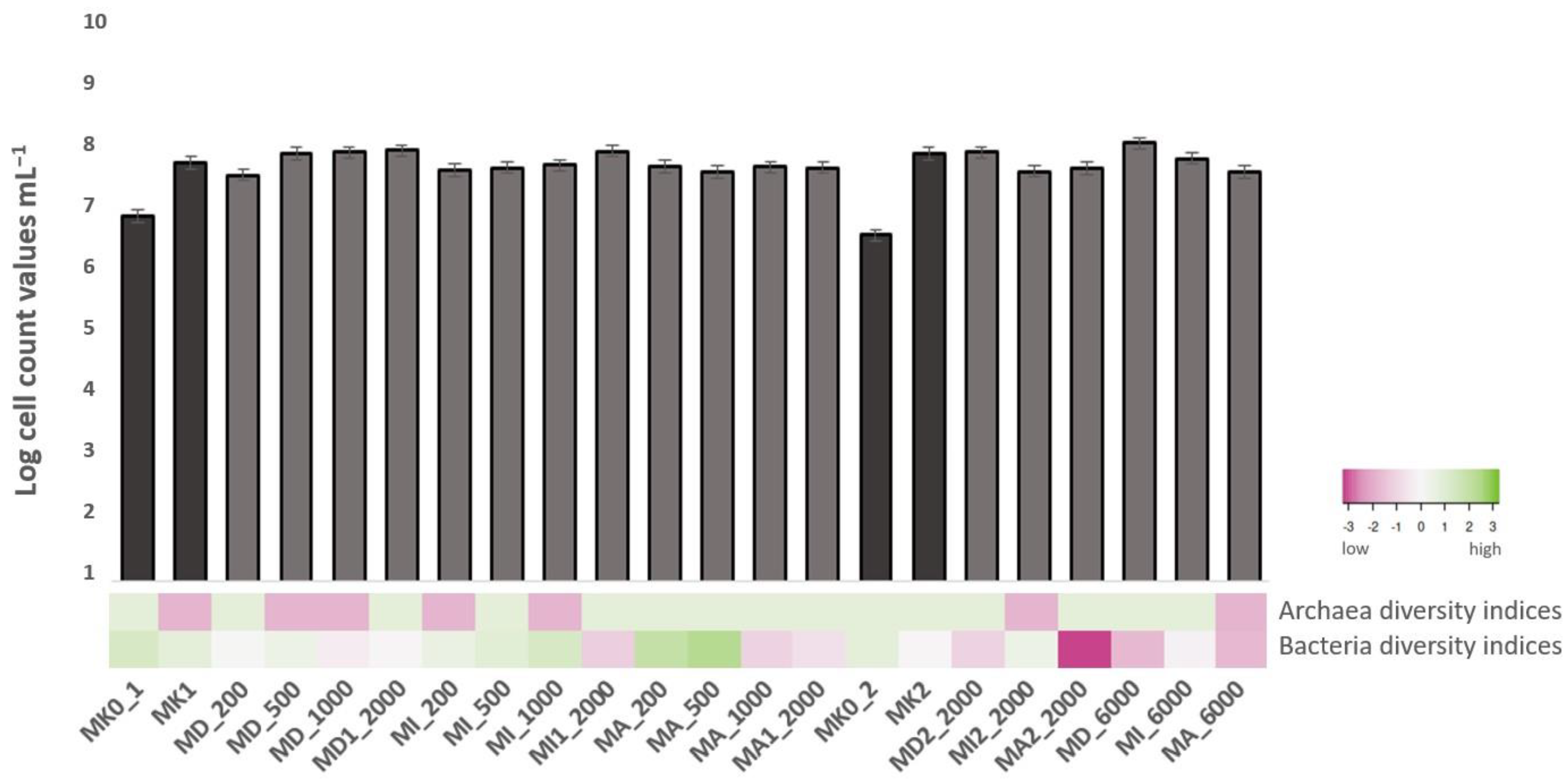
3.2. Microscopic Cell Counts and Diversity Indices of the Samples
3.3. Results of Selective Cultivation and Genomic Analysis of Bacterial Strains
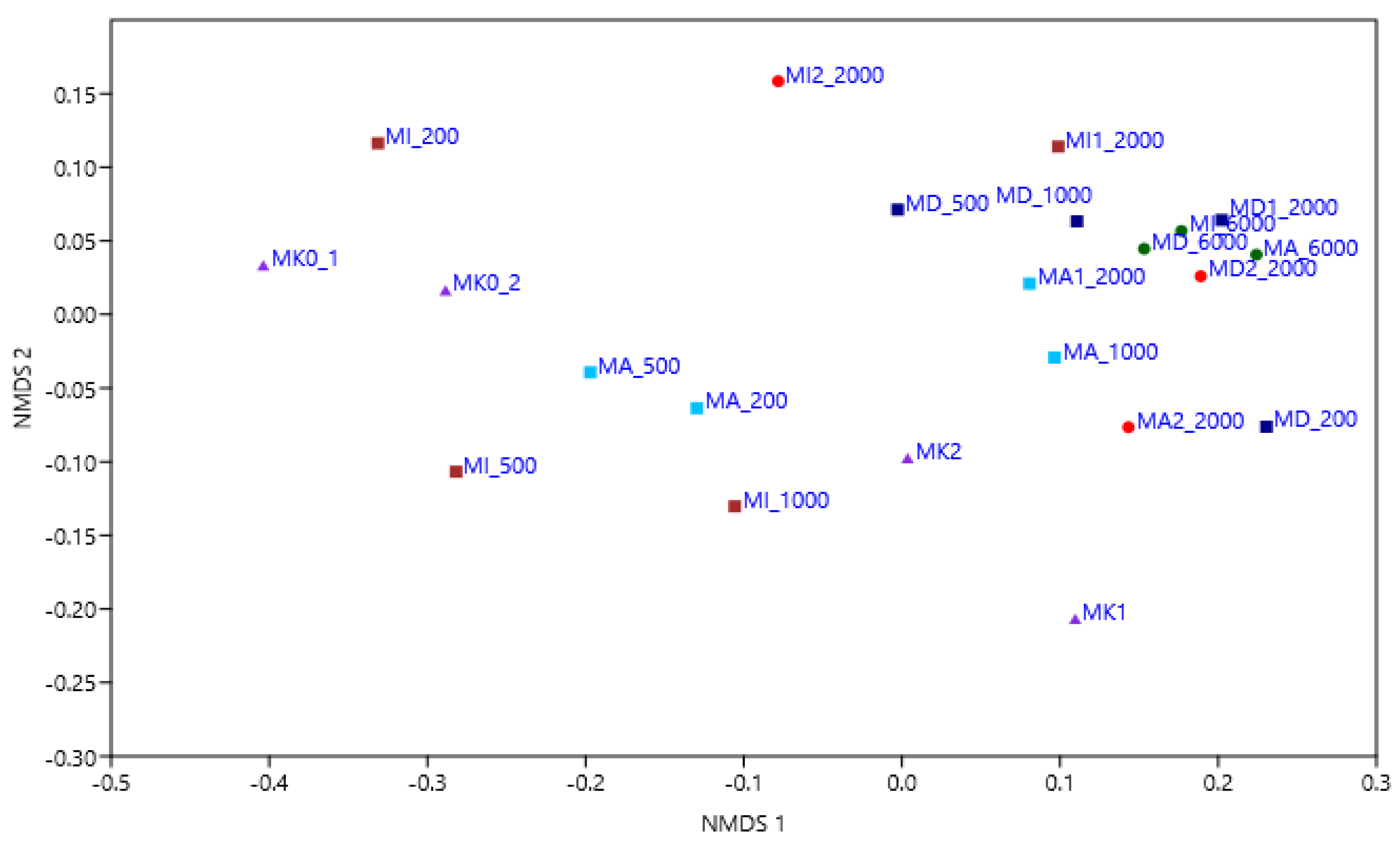
3.4. Results of Amplicon Sequencing
3.4.1. Bacterial Community Composition of the Microcosms Based on Amplicon Sequencing
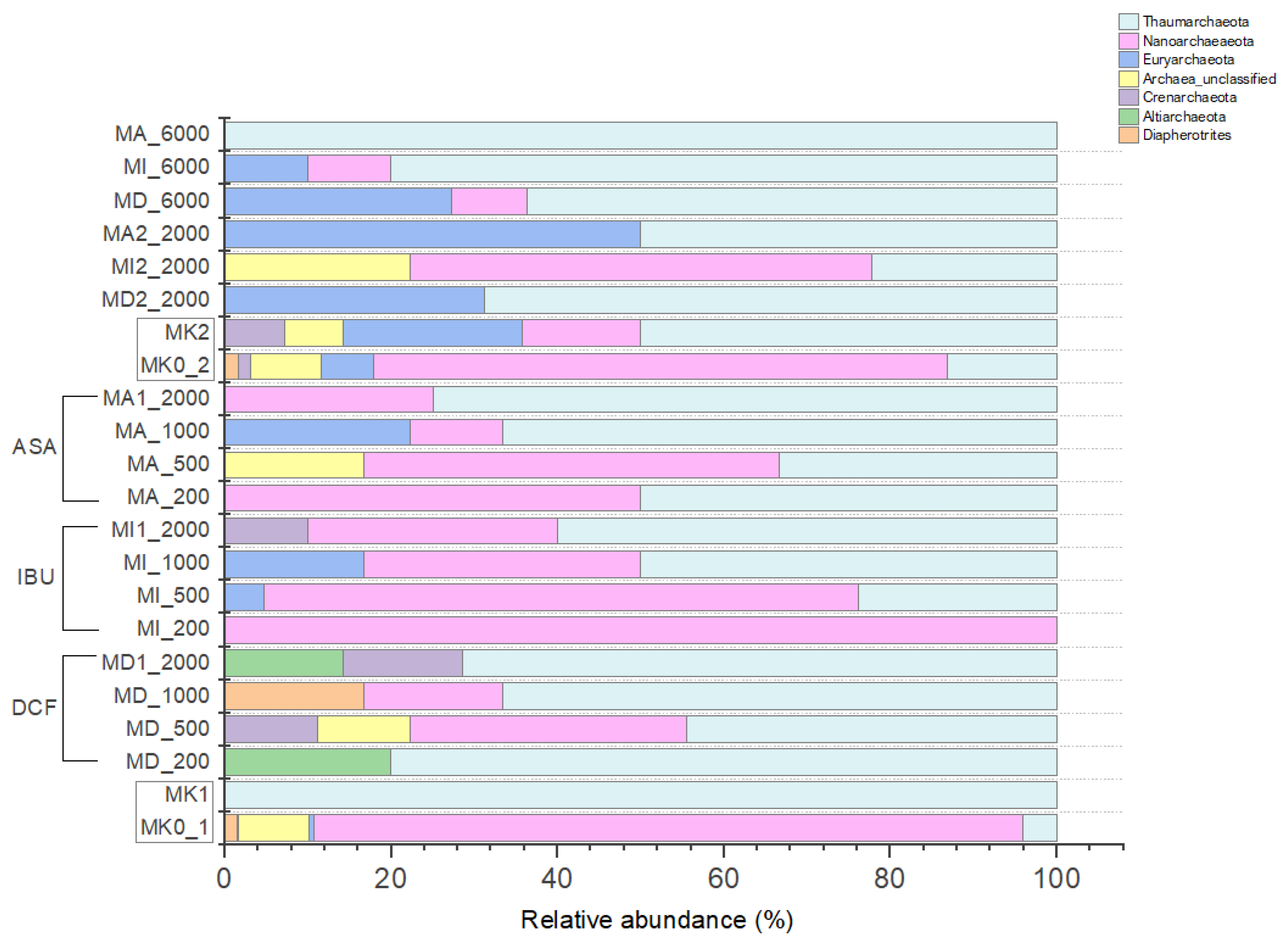
3.4.2. Archaeal Community Composition of the Microcosms Based on Amplicon Sequencing
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Aus der Beek, T.; Weber, F.-A.; Bergmann, A.; Hickmann, S.; Ebert, I.; Hein, A.; Küster, A. Pharmaceuticals in the environment—Global occurrences and perspectives. Environ. Toxicol. Chem. 2016, 35, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Loos, L.; Gawlik, B.M.; Locoro, G.; Rimaviciute, E.; Contini, S.; Bidoglio, G. EU-wide survey of polar organic persistent pollutants in European river waters. Environ. Pollut. 2009, 157, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Oliván, L.M.; Neri-Cruz, N.; Galar-Martínez, M.; Islas-Flores, H.; García-Medina, S. Binary mixtures of diclofenac with paracetamol, ibuprofen, naproxen, and acetylsalicylic acid and these pharmaceuticals in isolated form induce oxidative stress on Hyalella azteca. Environ. Monit. Assess. 2014, 186, 7259–7271. [Google Scholar] [CrossRef] [PubMed]
- Navrozidou, E.; Melidis, P.; Ntougias, S. Biodegradation aspects of ibuprofen and identification of ibuprofen-degrading microbiota in an immobilized cell bioreactor. Environ. Sci. Pollut. Res. 2019, 26, 14238–14249. [Google Scholar] [CrossRef]
- Fortunato, M.S.; Fuentes Abril, N.P.; Martinefski, M.; Trípodi, V.; Papalia, M.; Rádice, M.; Gutkind, G.; Gallego, A.; Korol, S.E. Aerobic degradation of ibuprofen in batch and continuous reactors by an indigenous bacterial community. Environ. Technol. 2016, 37, 2617–2626. [Google Scholar] [CrossRef]
- Żur, J.; Piński, A.; Wojcieszyńska, D.; Smułek, W.; Guzik, U. Diclofenac Degradation-Enzymes, Genetic Background and Cellular Alterations Triggered in Diclofenac-Metabolizing Strain Pseudomonas moorei KB4. Int. J. Mol. Sci. 2020, 21, 6786. [Google Scholar] [CrossRef]
- Candido, J.P.; Andrade, S.J.; Fonseca, A.L.; Silva, F.S.; Silva, M.R.A.; Kondo, M.M. Ibuprofen removal of heterogeneous photocatalysis and ecotoxicological evaluation of the treated solutions. Environ. Sci. Pollut. Res. 2017, 23, 19911–19920. [Google Scholar] [CrossRef]
- Bueno, M.J.M.; Gomez, M.J.; Herrera, S.; Hernando, M.D.; Agüera, A.; Fernández-Alba, A.R. Occurrence and persistence of organic emerging contaminants and priority pollutants in five sewage treatment plants of Spain: Two years pilot survey monitoring. Environ. Pollut. 2012, 164, 267–273. [Google Scholar] [CrossRef]
- Jonkers, N.; Sousa, A.; Galante-Oliveira, S.; Barroso, C.M.; Kohler, H.P.; Giger, W. Occurrence and sources of selected phenolic endocrine disruptors in Ria de Aveiro Portugal. Environ. Sci. Pollut. Res. Int. 2010, 17, 834–843. [Google Scholar] [CrossRef]
- Oketola, A.A.; Fagbemigun, T.K. Determination of nonylphenol, octylphenol, and bisphenol-A in water and sediments of two major rivers in Lagos, Nigeria. J. Environ. 2013, 4, 34127. [Google Scholar] [CrossRef]
- Bradley, P.M.; Barber, L.B.; Duris, J.W.; Foreman, W.T.; Furlong, E.T.; Hubbard, L.E.; Hutchinson, K.J.; Keefe, S.H.; Kolpin, D.W. Riverbank filtration potential of pharmaceuticals in a wastewater-impacted stream. Environ. Pollut. 2014, 193, 173–180. [Google Scholar] [CrossRef]
- Davoli, E.; Zuccato, E.; Castiglioni, S. Illicit drugs in drinking water. Curr. Opin. Environ. Sci. Health 2019, 7, 92–97. [Google Scholar] [CrossRef]
- JDS3. Available online: http://www.danubesurvey.org/jds3/jds3-files/nodes/documents/jds3_final_scientific_report_1.pdf (accessed on 25 April 2023).
- Kondor, A.C.; Jakab, G.; Vancsik, A.; Filep, T.; Szeberényi, J.; Szabó, L.; Maász, G.; Ferincz, Á.; Dobosy, P.; Szalai, Z. Occurrence of pharmaceuticals in the Danube and drinking water wells: Efficiency of riverbank filtration. Environ. Pollut. 2020, 265, 114893. [Google Scholar] [CrossRef]
- Daughton, C.G.; Ternes, T.A. Pharmaceuticals and personal care products in the environment: Agents of subtle change? Environ. Health Perspect. 1999, 107, 907–938. [Google Scholar] [CrossRef]
- Calisto, V.; Esteves, V.I. Psychiatric pharmaceuticals in the environment. Chemosphere 2009, 77, 1257–1274. [Google Scholar] [CrossRef]
- Santos, J.L.; Aparicio, I.; Callejón, M.; Alonso, E. Occurrence of pharmaceutically active compounds during a 1-year period in wastewaters from four wastewater treatment plants in Seville (Spain). J. Hazard. Mater. 2009, 164, 1509–1516. [Google Scholar] [CrossRef]
- Onesios, K.M.; Yu, J.T.; Bouwer, E.J. Biodegradation and removal of pharmaceuticals and personal care products in treatment systems: A review. Biodegradation 2009, 20, 441–466. [Google Scholar] [CrossRef]
- Caracciolo, A.; Topp, E.; Grenni, P. Pharmaceuticals in the environment: Biodegradation and effects on natural microbial communities. A review. J. Pharm. Biomed. Anal. 2015, 106, 25–36. [Google Scholar] [CrossRef]
- Gros, M.; Petrović, M.; Barceló, D. Wastewater treatment plants as a pathway for aquatic contamination by pharmaceuticals in the Ebro river basin (Northeast Spain). Environ. Toxicol. Chem. 2007, 26, 1553. [Google Scholar] [CrossRef]
- Dordio, A.V.; Carvalho, A.J.P. Organic xenobiotics removal in constructed wetlands, with emphasis on the importance of the support matrix. J. Hazard. Mater. 2013, 252–253, 272–292. [Google Scholar] [CrossRef]
- Vasquez, M.I.; Lambrianides, A.; Schneider, M.; Kümmerer, K.; Fatta-Kassinos, D. Environmental side effects of pharmaceutical cocktails: What we know and what we should know. J. Hazard. Mater. 2014, 279, 169–189. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, B.; Sellamuthu, B.; Ouarda, Y.; Drogui, P.; Tyagi, R.D.; Buelna, G. Review on fate and mechanism of removal of pharmaceutical pollutants from wastewater using biological approach. Bioresour. Technol. 2017, 224, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-S.; Keum, Y.-S.; Li, Q.X. Bacterial Degradation of Aromatic Compounds. Int. J. Environ. Res. Public Health 2009, 6, 278–309. [Google Scholar] [CrossRef] [PubMed]
- Leão, C.; Borges, A.; Simões, M. NSAIDs as a drug repurposing strategy for biofilm control. Antibiotics 2020, 9, 591. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Wu, T.; Xiong, Y.; Ni, H.; Ding, Y.; Zhang, W. Ibuprofen-mediated potential inhibition of biofilm development and quorum sensing in Pseudomonas aeruginosa. Life Sci. 2019, 237, 116947. [Google Scholar] [CrossRef]
- Gerbersdorf, S.U.; Hollert, H.; Brinkmann, M.; Wieprecht, S.; Schuttrumpf, H. Anthropogenic pollutants affect ecosystem services of freshwater sediments: The need for a “triad plus x” approach. J. Soils Sediments 2011, 11, 1099–1114. [Google Scholar] [CrossRef]
- Kéki, Z.; Makk, J.; Barkács, K.; Vajna, B.; Palatinszky, M.; Márialigeti, K.; Tóth, E. Critical point analysis and biocide treatment in a microbiologically contaminated water purification system of a power plant. SN Appl. Sci. 2019, 1, 820–831. [Google Scholar] [CrossRef]
- Szuróczki, S.; Kéki, Z.; Káli, S.; Lippai, A.; Márialigeti, K.; Tóth, E. Microbiological investigations on the water of a thermal bath at Budapest. Acta Microbiol. Immunol. Hung. 2016, 63, 229–241. [Google Scholar] [CrossRef]
- Kalwasinska, A.; Felföldi, T.; Walczak, M.; Kosobucki, P. Physiology and Molecular Phylogeny of Bacteria Isolated from Alkaline Distillery Lime. Pol. J. Microbiol. 2015, 64, 369–377. [Google Scholar] [CrossRef]
- Yoon, S.H.; Sung, M.H.; Soonjae, K.; Jeongmin, L.; Yeseul, K.; Hyungseok, S.; Jongsik, C. Introducing EzBioCloud: A Taxonomically United Database of 16S RRNA Gene Sequences and Whole-Genome Assemblies. Int. J. Sys. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, 353–361. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencingbased diversity studies. Nucleic Acid Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-source, platformindependent, community supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Zimmermann, P.; Curtis, N. Antimicrobial Effects of Antipyretics. Antimicrob. Agents Chemother. 2017, 61, e02268-16. [Google Scholar] [CrossRef]
- Laudy, A.E.; Mrowka, A.; Krajewska, J.; Tyski, S. The Influence of Efflux Pump Inhibitors on the Activity of Non-Antibiotic NSAIDS against Gram-Negative Rods. PLoS ONE 2016, 11, 0147131. [Google Scholar] [CrossRef]
- Murdoch, R.W.; Hay, A.G. Genetic and chemical characterization of ibuprofen degradation by Sphingomonas Ibu-2. Microbiology 2013, 159, 621–632. [Google Scholar] [CrossRef]
- Rocheleau, H.; Al-Harthi, R.; Ouellet, T. Degradation of salicylic acid by Fusarium graminearum. Fungal Biol. 2019, 123, 77–86. [Google Scholar] [CrossRef]
- Palyzová, A.; Marešová, H.; Novák, J.; Zahradník, J.; Řezanka, T. Effect of the anti-inflammatory drug diclofenac on lipid composition of bacterial strain Raoultella sp. KDF8. Folia Microbiol. 2020, 65, 763–773. [Google Scholar] [CrossRef] [PubMed]
- von Mering, C.; Hugenholtz, P.; Raes, J.; Tringe, S.G.; Doerks, T.; Jensen, L.J.; Ward, N.; Bork, P. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science 2007, 315, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Vasiliadou, I.A.; Molina, R.; Martinez, F.; Melero, J.A.; Stathopoulou, P.M.; Tsiamis, G. Toxicity assessment of pharmaceutical compounds on mixed culture from activated sludge using respirometric technique: The role of microbial community structure. Sci. Total Environ. 2018, 630, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Shahi, A.; Aydin, S.; Ince, B.; Ince, O. Reconstruction of bacterial community structure and variation for enhanced petroleum hydrocarbons degradation through biostimulation of oil contaminated soil. Chem. Eng. J. 2016, 306, 60–66. [Google Scholar] [CrossRef]
- Alexandrino, D.A.M.; Mucha, A.P.; Almeida, C.M.; Gao, W.; Jia, Z.; Carvalho, M.F. Biodegradation of the veterinary antibiotics enrofloxacin and ceftiofur and associated microbial community dynamics. Sci. Total Environ. 2017, 581–582, 359–368. [Google Scholar] [CrossRef]
- Carvalho, M.F.; De Marco, P.; Duque, A.F.; Pacheco, C.C.; Janssen, D.B.; Castro, P.M. Labrys portucalensis sp. nov., a fluorobenzene-degrading bacterium isolated from an industrially contaminated sediment in northern Portugal. Int. J. Syst. Evol. Microbiol. 2008, 58, 692–698. [Google Scholar] [CrossRef]
- Zeyer, J.; Wasserfallen, A.; Timmis, K.N. Microbial Mineralization of Ring-Substituted Anilines through an Ortho-Cleavage Pathway. Appl. Environ. Microbiol. 1985, 50, 447–453. [Google Scholar] [CrossRef]
- Moreira, I.S.; Bessa, V.S.; Murgolo, S.; Piccirillo, C.; Mascolo, G.; Castro, P.M.L. Biodegradation of Diclofenac by the bacterial strain Labrys portucalensis F11. Ecotoxicol. Environ. Saf. 2018, 15, 104–113. [Google Scholar] [CrossRef]
- Davids, M.; Gudra, D.; Radovica-Spalvina, I.; Fridmanis, D.; Bartkevics, V.; Muter, O. The effects of ibuprofen on activated sludge: Shift in bacterial community structure and resistance to ciprofloxacin. J. Hazard. Mater. 2017, 340, 291–299. [Google Scholar] [CrossRef]
- Jouanneau, Y.; Micoud, J.; Meyer, C. Purification and characterization of a from Sphingomonas sp. strain CHY-1 three-component salicylate 1-hydroxylase. Appl. Environ. Microbiol. 2007, 73, 7515–7521. [Google Scholar] [CrossRef]
- Murdoch, R.W.; Hay, A.G. The biotransformation of ibuprofen to trihydroxyibuprofen in activated sludge and by Variovorax Ibu-1. Biodegradation 2015, 26, 105–113. [Google Scholar] [CrossRef]
- Fernandes, G.L.; Shenoy, B.D.; Damare, S.R. Diversity of bacterial community in the oxygen minimum zones of Arabian Sea and Bay of Bengal as deduced by illumina sequencing. Front. Microbiol. 2020, 10, 3153. [Google Scholar] [CrossRef]
- Rutere, C.; Knoop, K.; Posselt, M.; Ho, A.; Horn, M.A. Ibuprofen Degradation and Associated Bacterial Communities in Hyporheic Zone Sediments. Microorganisms 2020, 8, 1245. [Google Scholar] [CrossRef]
- Chakrabarty, A.M. Genetic basis of the biodegradation of salicylate in Pseudomonas. J. Bacteriol. 1972, 112, 815–823. [Google Scholar] [CrossRef]
- Li, Y.; Wu, B.; Zhu, G.; Liu, Y.; Ng, W.J.; Appan, A.; Tan, S. High-throughput pyrosequencing analysis of bacteria relevant to cometabolic and metabolic degradation of ibuprofen in horizontal subsurface flow constructed wetlands. Sci. Total Environ. 2016, 562, 604–613. [Google Scholar] [CrossRef]
- Lanas, A.; Scarpignato, C. Microbial flora in NSAID-induced intestinal damage: A role for antibiotics? Digestion 2016, 73, 136–150. [Google Scholar] [CrossRef]
- Aissaoui, S.; Ouled-Haddar, H.; Sifour, M.; Harrouche, K.; Sghaier, H. Metabolic and co-metabolic transformation of diclofenac by Enterobacter hormaechei D15 isolated from activated sludge. Curr. Microbiol. 2017, 74, 381–388. [Google Scholar] [CrossRef]
- Gargouri, B.; Karray, F.; Mhiri, N.; Aloui, F.; Sayadi, S. Bioremediation of petroleum hydrocarbons-contaminated soil by bacterial consortium isolated from an industrial wastewater treatment plant. J. Chem. Technol. Biotech. 2014, 89, 978–987. [Google Scholar] [CrossRef]
- Drury, B.; Rosi-Marshall, E.; Kelly, J.J. Wastewater treatment effluent reduces the abundance and diversity of benthic bacterial communities in urban and suburban rivers. Appl. Environ. Microbiol. 2013, 79, 1897–1905. [Google Scholar] [CrossRef]
- Kang, D.; Jian, L.; Beihai, Z.; Rongfang, Y. Isolation of bacteria capable of removing 2-methylisoborneol and effect of cometabolism carbon on biodegradation. Environ. Eng. Res. 2016, 21, 256–264. [Google Scholar]
- de Bento Flores, C.E. Planctomycetes Cell Viability Studies: Perspectives of Toxicity Assessment Using Zeta Potential. Master’s Thesis, Dissertação de Mestrado apresentada à Faculdade de Ciências da Universidade do Porto, Centro Interdisciplinar de Investigação Marinha Ambiental, Biologia Celular e Molecular, Porto, Portugal, 2013. [Google Scholar]
- Yu, H.; Nie, E.; Xu, J.; Yan, S.; Cooper, W.J.; Song, W. Degradation of Diclofenac by Advanced Oxidation and Reduction Processes: Kinetic Studies, Degradation Pathways and Toxicity Assessments. Water Res. 2013, 47, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- Basik, A.A.; Sanglier, J.J.; Yeo, C.T.; Sudesh, K. Microbial Degradation of Rubber: Actinobacteria. Polymers 2021, 13, 1989. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Luo, J.; Lee, Z.M.P.; Maspolim, Y.; Gersberg, R.M.; Liu, Y.; Tan, S.K.; Ng, W.J. Characterization of bacterial communities in wetland mesocosms receiving pharmaceutical-enriched wastewater. Ecol. Eng. 2016, 90, 215–224. [Google Scholar] [CrossRef]
- Pala-Ozkok, I.; Rehman, A.; Ubay-Cokgor, E.; Jonas, D.; Orhon, D. Pyrosequencing reveals the inhibitory impact of chronic exposure to erythromycin on activated sludge bacterial community structure. Biochem. Eng. J. 2014, 90, 195–205. [Google Scholar] [CrossRef]
- Prior, J.E.; Shokati, T.; Christians, U.; Gill, R.T. Identification and characterization of a bacterial cytochrome P450 for the metabolism of diclofenac. Appl. Microbiol. Biotechnol. 2010, 85, 625–633. [Google Scholar] [CrossRef]
- Thelusmond, J.R.; Kawka, E.; Strathmann, T.J.; Cupples, A.M. Diclofenac, carbamazepine and triclocarban biodegradation in agricultural soils and the microorganisms and metabolic pathways affected. Sci. Total Environ. 2018, 640–641, 1393–1410. [Google Scholar] [CrossRef]
- Schlesner, H. Verrucomicrobium spinosum gen. nov., sp. nov.: A Fimbriated Prosthecate Bacterium. Syst. Appl. Microbiol. 1987, 10, 54–56. [Google Scholar] [CrossRef]
- Wagner, M.; Horn, M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 2006, 17, 241–249. [Google Scholar] [CrossRef]
- Lage, O.M.; Bondoso, J. Planctomycetes and macroalgae, a striking association. Front. Microbiol. 2014, 5, 267. [Google Scholar] [CrossRef]
- Bácsi, I.; B-Béres, V.; Kókai, Z.; Gonda, S.; Novák, Z.; Nagy, S.A.; Vasas, G. Effects of non-steroidal anti-inflammatory drugs on cyanobacteria and algae in laboratory strains and in natural algal assemblages. Environ. Pollut. 2016, 212, 508–518. [Google Scholar] [CrossRef]
- Lemmens, L.; Maklad, H.R.; Bervoets, I.; Peeters, E. Transcription regulators in Archaea: Homologies and differences with bacterial regulators. J. Mol. Biol. 2019, 431, 4132–4146. [Google Scholar] [CrossRef]
- Castelle, C.J.; Banfield, J.F. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell 2018, 172, 1181–1197. [Google Scholar] [CrossRef]
- Osorio, V.; Cruz-Alcalde, A.; Pérez, S. Nitrosation and nitration of diclofenac and structurally related nonsteroidal anti-inflammatory drugs (NSAIDs) in nitrifying activated sludge. Sci. Total Environ. 2022, 807, 150533. [Google Scholar] [CrossRef]
- Li, A.; Chu, Y.; Wang, X.; Ren, L.; Yu, J.; Liu, X.; Yan, J.; Zhang, L.; Wu, S.; Li, S. A pyrosequencing-based metagenomic study of methane-producing microbial community in solid-state biogas reactor. Biotechnol. Biofuels 2013, 6, 3. [Google Scholar] [CrossRef]
- Granatto, C.F.; Grosseli, G.M.; Sakamoto, I.K.; Fadini, P.S.; Varesche, M.B.A. Methanogenic potential of diclofenac and ibuprofen in sanitary sewage using metabolic cosubstrates. Sci. Total Environ. 2020, 742, 140530. [Google Scholar] [CrossRef]



| Names of the Samples | Controls and Experiments | Anti-Inflammatory Drug Content (ppm) | Anti-Inflammatory Drug Applied |
|---|---|---|---|
| MK0_1 | absolute control * (first) | 0 | none |
| MK1 | control ** (first) | 0 | none |
| MD_200 | first (July 2020) | 200 | DCF |
| MD_500 | first (July 2020) | 500 | DCF |
| MD_1000 | first (July 2020) | 1000 | DCF |
| MD1_2000 | first (July 2020) | 2000 | DCF |
| MI_200 | first (July 2020) | 200 | IBU |
| MI_500 | first (July 2020) | 500 | IBU |
| MI_1000 | first (July 2020) | 1000 | IBU |
| MI1_2000 | first (July 2020) | 2000 | IBU |
| MA_200 | first (July 2020) | 200 | ASA |
| MA_500 | first (July 2020) | 500 | ASA |
| MA_1000 | first (July 2020) | 1000 | ASA |
| MA1_2000 | first (July 2020) | 2000 | ASA |
| MK0_2 | absolute control * (second) | 0 | none |
| MK2 | control ** (second) | 0 | none |
| MD2_2000 | second (August 2020) | 2000 | DCF |
| MI2_2000 | second (August 2020) | 2000 | IBU |
| MA2_2000 | second (August 2020) | 2000 | ASA |
| MD_6000 | second (August 2020) | 6000 | DCF |
| MI_6000 | second (August 2020) | 6000 | IBU |
| MA_6000 | second (August 2020) | 6000 | ASA |
| Experiments | Location | T °C | Conductivity (μS m−1) | pH | O2 (%) | O2 (mg L−1) |
|---|---|---|---|---|---|---|
| first (July 2020) | 47.477357° N; 19.061891° E | 17 | 223 | 7.83 | 95.9 | 12.54 |
| second (August 2020) | 18.5 | 203 | 7.57 | 93.6 | 13.22 |
| Strain Designation | Taxonomic Identification | Strain | Similarity (%) |
|---|---|---|---|
| MA1 | Burkholderia ambifaria | AMMD | 99.79 |
| MA2 | Burkholderia ambifaria | AMMD | 99.81 |
| MA4 | Novosphingobium arvoryzae | JYI-02 | 100 |
| MA6 | Paenibacillus lautus | NBRC 15380 | 98.92 |
| MA7 | Burkholderia vietnamiensis | LMG 10929 | 99.89 |
| MA8 | Burkholderia vietnamiensis | LMG 10929 | 99.89 |
| MA9 | Pseudoduganella danionis | E3/2 | 98.54 |
| MA10 | Burkholderia vietnamiensis | LMG 10929 | 99.89 |
| MD7 | Sphingobium yanoikuyae | ATCC 51230 | 100 |
| MD8 | Ensifer adhaerens | Casida A | 100 |
| MD9 | Klebsiella grimontii | 06D021 | 100 |
| MI1 | Pseudomonas nitritireducens | WZBFD3-5A2 | 99.70 |
| MI3 | Novosphingobium barchaimii | LL02 | 99.43 |
| MI4 | Klebsiella pneumoniae subsp. ozaenae | ATCC 11296 | 99.90 |
| MI10 | Sphingobium yanoikuyae | ATCC 51230 | 100 |
| MI11 | Novosphingobium barchaimii | LL02 | 99.62 |
| MI12 | Pseudomonas nitroreducens | DSM 14399 | 99.77 |
| MI13 | Sphingobium yanoikuyae | ATCC 51230 | 100 |
| MI14 | Cupriavidus agavae | ASC-9842 | 98.70 |
| Strain Designation | Taxonomic Identification | Strain | Similarity (%) |
|---|---|---|---|
| MD1 | Aeromonas sanarellii | LMG 24682 | 99.90 |
| MD2 | Aeromonas sanarellii | LMG 24682 | 99.90 |
| MD6 | Rahnella aceris | SAP-19 | 98.49 |
| MD7_2 | Rahnella aceris | SAP-19 | 100.00 |
| MD8_2 | Aeromonas caviae | CECT 838 | 100.00 |
| MD10 | Klebsiella michiganensis | W14 | 98.92 |
| MD12 | Aeromonas sanarellii | LMG 24682 | 99.91 |
| MD13 | Rahnella aceris | SAP-19 | 100.00 |
| MD15 | Aeromonas caviae | CECT 838 | 98.95 |
| MD17 | Rahnella aceris | SAP-19 | 100.00 |
| MD18 | Rahnella aceris | SAP-19 | 100.00 |
| MD19 | Klebsiella michiganensis | W14 | 98.92 |
| MD21 | Citrobacter bitternis | SKKUI-TP7 | 99.81 |
| MD22 | Aeromonas caviae | CECT 838 | 98.95 |
| MD23 | Rahnella aceris | SAP-19 | 100.00 |
| MD24 | Phytobacter diazotrophicus | LS 8 | 99.39 |
| MD25 | Rahnella aceris | SAP-19 | 100.00 |
| MD26 | Klebsiella grimontii | 06D021 | 100.00 |
| MD27 | Klebsiella grimontii | 06D021 | 100.00 |
| MD28 | Rahnella aceris | SAP-19 | 100.00 |
| MD29 | Aeromonas caviae | CECT 838 | 98.95 |
| MD30 | Aeromonas taiwanensis | LMG 24683 | 99.50 |
| MD31 | Aeromonas sanarellii | LMG 24682 | 99.81 |
| MD32 | Aeromonas sanarellii | LMG 24682 | 99.90 |
| MD33 | Klebsiella michiganensis | W14 | 99.61 |
| MI1_2 | Klebsiella quasivariicola | KPN1705 | 99.80 |
| MI2 | Klebsiella pneumoniae subsp. pneumoniae | DSM 30104 | 99.61 |
| MI3_2 | Pseudomonas aeruginosa | JCM 5962 | 99.74 |
| MI6 | Klebsiella huaxiensis | WCHKl090001 ATCC 11296 | 99.84 |
| MI9 | Klebsiella pneumoniae subsp. ozaenae | ATCC 11296 | 99.90 |
| MI10_2 | Klebsiella quasivariicola | KPN1705 | 99.80 |
| MI11_2 | Klebsiella quasivariicola | KPN1705 | 99.80 |
| MI12_2 | Klebsiella huaxiensis | WCHKl090001 | 98.73 |
| MI14_2 | Klebsiella huaxiensis | WCHKl090001 W14 | 99.24 |
| MI15 | Klebsiella michiganensis | W14 | 99.65 |
| MI17 | Raoultella ornithinolytica | JCM 6096 | 100.00 |
| MI18 | Klebsiella huaxiensis | WCHKl090001 | 100.00 |
| MI21 | Klebsiella michiganensis | W14 | 99.44 |
| MI22 | Kosakonia radicincitans | DSM 16656 | 99.66 |
| MI23 | Klebsiella huaxiensis | WCHKl090001 | 100.00 |
| MI24 | Klebsiella huaxiensis | WCHKl090001 | 99.20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farkas, R.; Mireisz, T.; Toumi, M.; Abbaszade, G.; Sztráda, N.; Tóth, E. The Impact of Anti-Inflammatory Drugs on the Prokaryotic Community Composition and Selected Bacterial Strains Based on Microcosm Experiments. Microorganisms 2023, 11, 1447. https://doi.org/10.3390/microorganisms11061447
Farkas R, Mireisz T, Toumi M, Abbaszade G, Sztráda N, Tóth E. The Impact of Anti-Inflammatory Drugs on the Prokaryotic Community Composition and Selected Bacterial Strains Based on Microcosm Experiments. Microorganisms. 2023; 11(6):1447. https://doi.org/10.3390/microorganisms11061447
Chicago/Turabian StyleFarkas, Rózsa, Tamás Mireisz, Marwene Toumi, Gorkhmaz Abbaszade, Nóra Sztráda, and Erika Tóth. 2023. "The Impact of Anti-Inflammatory Drugs on the Prokaryotic Community Composition and Selected Bacterial Strains Based on Microcosm Experiments" Microorganisms 11, no. 6: 1447. https://doi.org/10.3390/microorganisms11061447
APA StyleFarkas, R., Mireisz, T., Toumi, M., Abbaszade, G., Sztráda, N., & Tóth, E. (2023). The Impact of Anti-Inflammatory Drugs on the Prokaryotic Community Composition and Selected Bacterial Strains Based on Microcosm Experiments. Microorganisms, 11(6), 1447. https://doi.org/10.3390/microorganisms11061447






