Copper-Contaminated Substrate Biosorption by Penicillium sp. Isolated from Kefir Grains
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Penicillium sp. from Kefir Grains
2.2. Culture Medium Preparation and Radial Growth Measurement
2.3. Identifying Undesirable Properties of Copper Compounds through Toxicological Forecast
2.4. Biomass Production of Penicillium sp. and Determination of Minimum Inhibitory Concentration (MIC)
2.5. Cultivation of Penicillium sp. in Solid Medium and Minimum Inhibitory Concentration (MIC)
2.6. Penicillium sp. Biomass and Quantification of Copper
2.7. Estimation of Residual Metals in the Culture Medium
2.8. Scanning Electron Microscopy (SEM) Analysis
2.9. Statistical Analysis
3. Results
3.1. Effects of Copper on the Growth of Penicillium sp.
3.2. Growth in pH 4.0, 7.0, and 9.0
3.3. Toxicological Prediction
3.4. Culture of Penicillium sp. in Liquid Medium
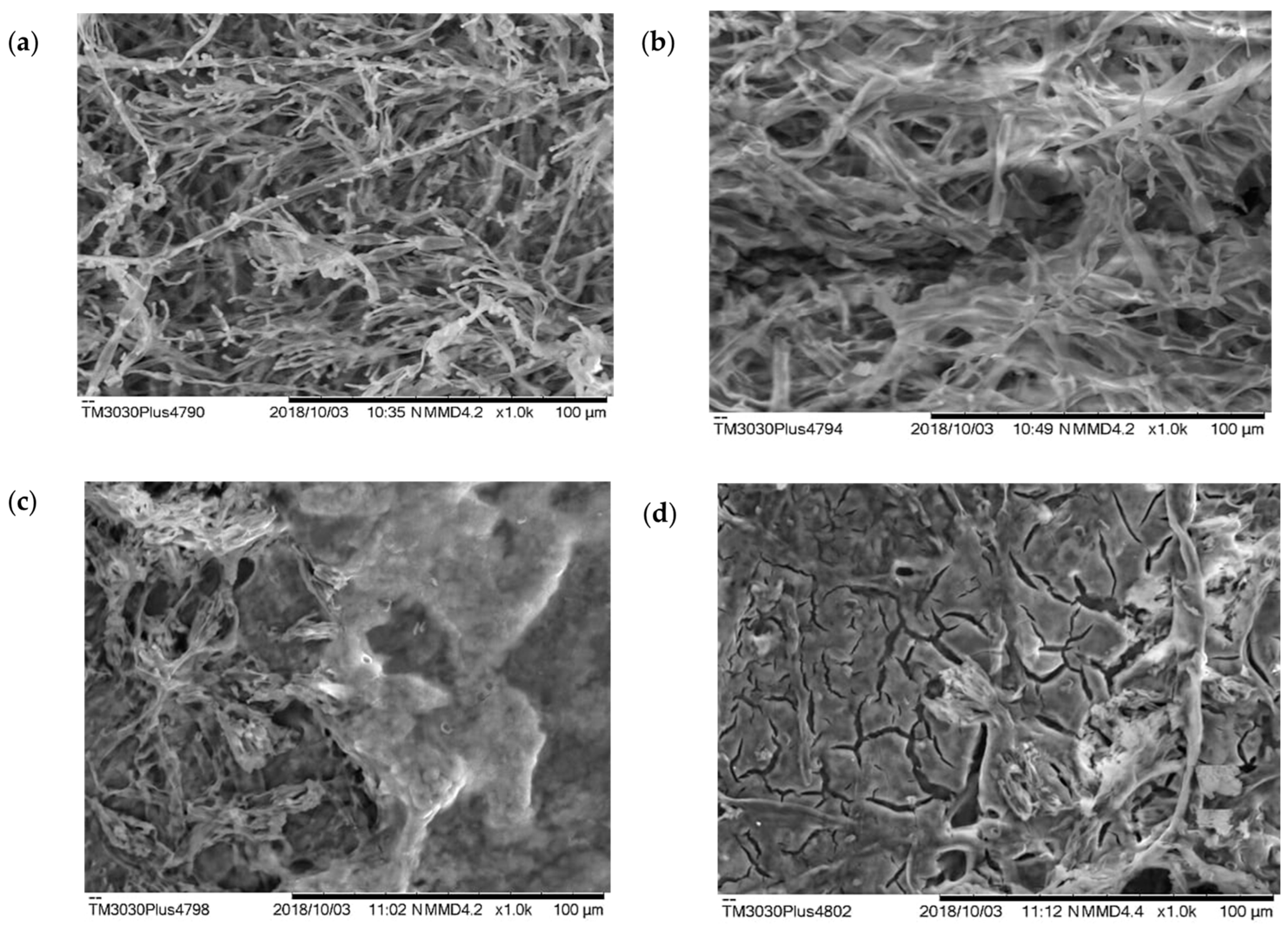
3.5. Scanning Electron Microscopy (SEM) Analysis of Penicillium sp. Biomass Isolated from Kefir Grains
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choińska-Pulit, A.; Sobolczyk-Bednarek, J.; Łaba, W. Optimization of Copper, Lead and Cadmium Biosorption onto Newly Isolated Bacterium Using a Box-Behnken Design. Ecotoxicol. Environ. Saf. 2018, 149, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Widmer, J.; Norgrove, L. Identifying Candidates for the Phytoremediation of Copper in Viticultural Soils: A Systematic Review. Environ. Res. 2023, 216, 114518. [Google Scholar] [CrossRef] [PubMed]
- Cornu, J.-Y.; Huguenot, D.; Jézéquel, K.; Lollier, M.; Lebeau, T. Bioremediation of Copper-Contaminated Soils by Bacteria. World J. Microbiol. Biotechnol. 2017, 33, 26. [Google Scholar] [CrossRef] [PubMed]
- Mohamadhasani, F.; Rahimi, M. Growth Response and Mycoremediation of Heavy Metals by Fungus Pleurotus Sp. Sci. Rep. 2022, 12, 19947. [Google Scholar] [CrossRef] [PubMed]
- Deniz, F.; Ersanli, E.T. A Natural Macroalgae Consortium for Biosorption of Copper from Aqueous Solution: Optimization, Modeling and Design Studies. Int. J. Phytoremed. 2018, 20, 362–368. [Google Scholar] [CrossRef]
- Naz, F.; Hamayun, M.; Rauf, M.; Arif, M.; Afzal Khan, S.; Ud-Din, J.; Gul, H.; Hussain, A.; Iqbal, A.; Kim, H.-Y.; et al. Molecular Mechanism of Cu Metal and Drought Stress Resistance Triggered by Porostereum Spadiceum AGH786 in Solanum Lycopersicum L. Front. Plant. Sci. 2022, 13, 1029836. [Google Scholar] [CrossRef]
- Palanivel, T.M.; Pracejus, B.; Novo, L.A.B. Bioremediation of Copper Using Indigenous Fungi Aspergillus Species Isolated from an Abandoned Copper Mine Soil. Chemosphere 2023, 314, 137688. [Google Scholar] [CrossRef]
- Fagnano, M.; Agrelli, D.; Pascale, A.; Adamo, P.; Fiorentino, N.; Rocco, C.; Pepe, O.; Ventorino, V. Copper Accumulation in Agricultural Soils: Risks for the Food Chain and Soil Microbial Populations. Sci. Total Environ. 2020, 734, 139434. [Google Scholar] [CrossRef]
- Kamal, N.; Parshad, J.; Saharan, B.S.; Kayasth, M.; Mudgal, V.; Duhan, J.S.; Mandal, B.S.; Sadh, P.K. Ecosystem Protection through Myco-Remediation of Chromium and Arsenic. J. Xenobiotics 2023, 13, 159–171. [Google Scholar] [CrossRef]
- Albert, Q.; Leleyter, L.; Lemoine, M.; Heutte, N.; Rioult, J.-P.; Sage, L.; Baraud, F.; Garon, D. Comparison of Tolerance and Biosorption of Three Trace Metals (Cd, Cu, Pb) by the Soil Fungus Absidia Cylindrospora. Chemosphere 2018, 196, 386–392. [Google Scholar] [CrossRef]
- Oliveira, A.; Maciel, A.; Florentino, A.; Fernandes, C.; Bezerra, R.; Góes, M.; Salcedo, M.; Zamora, R.; Carvalho, J.C. Study of Kefir Biofilm Associated with Hydroethanolic Extract of Euterpe Oleracea Mart. (Aai). Afr. J. Microbiol. Res. 2017, 11, 1474–1483. [Google Scholar] [CrossRef]
- de Oliveira, A.F.; dos Santos, C.B.R.; Ferreira, A.M.; Bezerra, R.M.; Zamora, R.R.M.; Cruz, R.A.S.; Amado, J.R.R.; Carvalho, J.C.T. A Viability Study for the Production of Biofilms and In Silico Predictions of Major Compounds in Kefir. J. Comput. Theor. Nanosci. 2017, 14, 2915–2926. [Google Scholar] [CrossRef]
- Gams, W.; Bisset, T. Morphology and Identification of Trichoderma. In Trichoderma and Gliocladium: Basic Biology Taxonomy and Genetics Harman GE and CP Kubicek; Harman, G., Kubicek, C., Eds.; Taylor and Francis: London, UK, 1998; Volume 1, pp. 3–34. [Google Scholar]
- Bandmann, O.; Weiss, K.H.; Kaler, S.G. Wilson’s disease and other neurological copper disorders. Lancet Neurol. 2015, 14, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Ortúzar, M.; Trujillo, M.E.; Román-Ponce, B.; Carro, L. Micromonospora Metallophores: A Plant Growth Promotion Trait Useful for Bacterial-Assisted Phytoremediation? Sci. Total Environ. 2020, 739, 139850. [Google Scholar] [CrossRef] [PubMed]
- Sevcikova, M.; Modra, H.; Slaninova, A.; Svobodova, Z. Metals as a Cause of Oxidative Stress in Fish: A Review. Vet. Med. 2011, 56, 537–554. [Google Scholar] [CrossRef]
- Subramaniyam, V.; Subashchandrabose, S.R.; Thavamani, P.; Chen, Z.; Krishnamurti, G.S.R.; Naidu, R.; Megharaj, M. Toxicity and Bioaccumulation of Iron in Soil Microalgae. J. Appl. Phycol. 2016, 28, 2767–2776. [Google Scholar] [CrossRef]
- Torres, E.M.; Hess, D.; McNeil, B.T.; Guy, T.; Quinn, J.C. Impact of Inorganic Contaminants on Microalgae 440 Productivity and Bioremediation Potential. Ecotoxicol. Environ. Saf. 2017, 139, 367–376, 441. [Google Scholar] [CrossRef]
- Schümann, K.; Classen, H.; Dieter, H.; König, J.; Multhaup, G.; Rükgauer, M.; Summer, K.; Bernhardt, J.; Biesalski, H. Hohenheim Consensus Workshop: Copper. Eur. J. Clin. Nutr. 2002, 56, 469–483. [Google Scholar] [CrossRef]
- Nishimuta, M.; Masui, K.; Yamamoto, T.; Ikarashi, Y.; Tokushige, K.; Hashimoto, E.; Nagashima, Y.; Shibata, N. 445 Copper Deposition in Oligodendroglial Cells in an Autopsied Case of Hepatolenticular Degeneration. Neuropathology 2018, 38, 321–328. [Google Scholar] [CrossRef]
- Monteiro, C.M.; Castro, P.M.L.; Malcata, F.X. Metal Uptake by Microalgae: Underlying Mechanisms and Practical Applications. Biotechnol. Prog. 2012, 28, 299–311. [Google Scholar] [CrossRef]
- Pereira, A.; Freitas, D.A. Uso de Micro-organismos Para a Biorremediação de Ambientes Impactados. Rev. Eletrônica Em Gestão Educ. E Tecnol. Ambient. 2012, 6, 995–1006. [Google Scholar] [CrossRef]
- Sahu, O. Reduction of Organic and Inorganic Pollutant from Waste Water by Algae. Int. Lett. Nat. Sci. 2014, 8, 1–8. [Google Scholar] [CrossRef]
- Nascimento, R.; Lima, A.; Vidal, B.; Melo, D.; Raulino, G. Adsorção: Aspectos Teóricos e Aplicações Ambientais, 1st ed.; Impressão Universitária: Fortaleza City, Brazil, 2020; Volume 1. [Google Scholar]
- Worms, I.; Simon, D.F.; Hassler, C.S.; Wilkinson, K.J. Bioavailability of Trace Metals to Aquatic Microorganisms: Importance of Chemical, Biological and Physical Processes on Biouptake. Biochimie 2006, 88, 1721–1731. [Google Scholar] [CrossRef]
- Diep, P.; Mahadevan, R.; Yakunin, A.F. Heavy Metal Removal by Bioaccumulation Using Genetically Engineered Microorganisms. Front. Bioeng. Biotechnol. 2018, 6, 157. [Google Scholar] [CrossRef] [PubMed]
- Zagatto, P.; Bertoletti, E. Ecotoxicologia Aquática: Princípios e Aplicações, 2nd ed.; Rima: São Carlos, Brazil, 2014; Volume 1. [Google Scholar]
- Dorr, F.; Guaratini, T.; Cardoso, K.; Pavanelli, D.; Colepicolo, P.; Pinto, E. Toxicologia Ambiental. In Fundamentos de Toxicologia; Oga, S., Camargo, M., Batistuzzo, J., Eds.; Atheneu: São Paulo, Brazil, 2014; Volume 1. [Google Scholar]
- Purchase, D.; Scholes, L.N.L.; Revitt, D.M.; Shutes, R.B.E. Effects of Temperature on Metal Tolerance and the Accumulation of Zn and Pb by Metal-Tolerant Fungi Isolated from Urban Runoff Treatment Wetlands. J. Appl. Microbiol. 2009, 106, 1163–1174. [Google Scholar] [CrossRef] [PubMed]
- Jakovljević, V.; Grujić, S.; Simić, Z.; Ostojić, A.; Radojević, I. Finding the Best Combination of Autochthonous Microorganisms with the Most Effective Biosorption Ability for Heavy Metals Removal from Wastewater. Front. Microbiol. 2022, 13, 1017372. [Google Scholar] [CrossRef]
- Inès, M.; Mekki, S.; Ghribi, D. Treatment of Heavy Metals Contaminated Water: Use of B. Mojavensis BI2 Derived Lipopeptide and Palm Waste Flour. Water Sci. Technol. 2022, 86, 1083–1094. [Google Scholar] [CrossRef]
- Lotlikar, N.; Damare, S.; Meena, R.M.; Jayachandran, S. Variable protein expression in marine-derived filamentous fungus Penicillium chrysogenum in response to varying copper concentrations and salinity. Metallomics 2020, 12, 1083–1093. [Google Scholar] [CrossRef]
- Kugler, A.; Brigmon, R.L.; Friedman, A.; Coutelot, F.M.; Polson, S.W.; Seaman, J.C.; Simpson, W. Bioremediation of Copper in Sediments from a Constructed Wetland Ex Situ with the Novel Bacterium Cupriavidus Basilensis SRS. Sci. Rep. 2022, 12, 17615. [Google Scholar] [CrossRef]
- Lacerda, E.C.M.; dos Passos, G.B.M.; Reis, T.A.; Nascimento, C.A.O.; Côrrea, B.; Gimenes, L.J. Copper biosorption from an aqueous solution by the dead biomass of Penicillium ochrochloron. Environ. Monit. Assess. 2019, 191, 247. [Google Scholar] [CrossRef]
- Viana, L.F.; Kummrow, F.; Cardoso, C.A.L.; Lima, N.A.; Crispim, B.A.; Bafuratti, A.; Florentino, A.C. Metal bioaccumulation in fish from the Araguari River (Amazon biome) and human health risks from fish consumption. Environ. Sci. Pollut. Res. 2023, 30, 4111–4122. [Google Scholar] [CrossRef]
- Lau, P.S.; Lee, H.Y.; Tsang, C.C.K.; Tam, N.F.Y.; Wong, Y.S. Effect of Metal Interference, PH and Temperature on Cu and Ni Biosorption by Chlorella Vulgaris and Chlorella Miniata. Environ. Technol. 1999, 20, 953–961. [Google Scholar] [CrossRef]
- Brinza, L.; Dring, M.J.; Gavrilescu, M. Marine Micro and Macro Algal Species as Biosorbents for Heavy Metals. Environ. Eng. Manag. J. 2007, 6, 237–251. [Google Scholar] [CrossRef]
- Kumar, S.; Vijay, M.A.; Kumar, K.P.S. Biosorption of Lead(II) and Chromium(VI) by Immobilized Cells of Microalga Isochrysis galbana. J. Algal Biomass Utln. 2013, 1, 42–50. [Google Scholar]
- Indhumathi, P.; Syed Shabudeen, P.; Shoba, U.; Saraswathy, C. The Removal of Chromium from Aqueous Solution by Using Green Micro Algae. J. Chem. Pharm. Res. 2014, 6, 799–808. [Google Scholar]
- Monteiro, C.M.; Marques, A.P.G.C.; Castro, P.M.L.; Xavier Malcata, F. Characterization of Desmodesmus pleiomorphus Isolated from a Heavy Metal-Contaminated Site: Biosorption of Zinc. Biodegradation 2009, 20, 629–641. [Google Scholar] [CrossRef]
- Bishnoi, N.R.; Kumar, R.; Kumar, S.; Rani, S. Biosorption of Cr(III) from Aqueous Solution Using Algal Biomass Spirogyra Spp. J. Hazard. Mater. 2007, 145, 142–147. [Google Scholar] [CrossRef]
- Martinelli, P.; Santos, J. Microscopia Eletrônica de Varredura de Fungos Nematófagos Associados a Tylenchulus Semipenetrans e Pratylenchus Jaehni Scanning Electron Microscopy of Nematophagous Fungi Associated Tylenchulus Semipenetrans and Pratylenchus Jaehni. Biosci. J. 2010, 26, 809–816. [Google Scholar]
- Juříková, T.; Luptáková, D.; Kofroňová, O.; Škríba, A.; Novák, J.; Marešová, H.; Palyzová, A.; Petřík, M.; Havlíček, V.; Benada, O. Bringing SEM and MSI Closer Than Ever Before: Visualizing Aspergillus and Pseudomonas Infection in the Rat Lungs. J. Fungi 2020, 6, 257. [Google Scholar] [CrossRef]
| CTR mg/L−1 | 400 mg/L ns | 500 g/L * | 600 mg/L *** | 800 mg/L *** | ANOVA | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Penicillium sp. | D (mm) | D (mm) | IC (%) | D (mm) | IC (%) | D (mm) | IC (%) | D (mm) | IC (%) | F |
| 90 a | 73 ± 1.67 a | 19 | 51.50 b ± 0.50 | 43 | 40.33 c ± 1.89 | 55 | 22.17 d ± 1.75 | 75 | 8.57 *** | |
| Treatment | 48 h | IC% | 72 h | IC% | 96 h | IC% | 120 h | IC% | F |
|---|---|---|---|---|---|---|---|---|---|
| CTR | 20.00 ± 1.26 | 45.00 ± 6.33 | 18.00 ± 1.28 | 90.00 | 932.85 ** | ||||
| Cu pH 4.0 | 07.25 ± 1.18 | 6 | 12.00 ± 0.58 | 6 | 14.00 ± 1.76 | 5 | 24.40 ± 0.5 | 73 | |
| Cu pH 7.0 | 6.00 ± 1.06 | 28 | 13.00 ± 1.0 | 3 | 19.00 ± 1.83 | 2 | 22.50 ± 1.9 | 75 | |
| Cu pH 9.0 | 5.00 ± 0.65 | 24 | 14.00 ± 20.10 | 19 | 15.00 ± 1.16 | 18 | 20.66 ± 0.76 | 77 |
| Toxicological Prediction | |
|---|---|
| Toxicological class | 2 |
| LC50 | 25 mg·kg−1 |
| Molecular weight | 187.56 |
| Number of hydrogen acceptors | 6 |
| Number of atoms | 9 |
| Number of connections | 6 |
| Molecular polar surface area (PSA) | 137.76 |
| Treatment | Cu(NO3)2 | Biomass (µg) | Inhibition (%) | pH |
|---|---|---|---|---|
| 1 | (control group) | 578 ± 2.75 | 7.00 | |
| 2 | Cu2+ 400 mg | 257 ± 3.00 | 56% | 4.33 ± 0.032 |
| 3 | Cu2+ 500 mg | 156.67 ± 2.52 | 73% | 4.19 ± 0.025 |
| 4 | Cu2+ 600 mg | 55.33 ± 0.58 | 90% | 4.08 ± 0.003 |
| 5 | Cu2+ 800 mg | 41.67 ± 2.08 | 93% | 3.37 ± 0.078 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, A.F.d.; Machado, R.B.; Ferreira, A.M.; Sena, I.d.S.; Silveira, M.E.; Almeida, A.M.S.d.; Braga, F.S.; Rodrigues, A.B.L.; Bezerra, R.M.; Ferreira, I.M.; et al. Copper-Contaminated Substrate Biosorption by Penicillium sp. Isolated from Kefir Grains. Microorganisms 2023, 11, 1439. https://doi.org/10.3390/microorganisms11061439
Oliveira AFd, Machado RB, Ferreira AM, Sena IdS, Silveira ME, Almeida AMSd, Braga FS, Rodrigues ABL, Bezerra RM, Ferreira IM, et al. Copper-Contaminated Substrate Biosorption by Penicillium sp. Isolated from Kefir Grains. Microorganisms. 2023; 11(6):1439. https://doi.org/10.3390/microorganisms11061439
Chicago/Turabian StyleOliveira, Antonio Ferreira de, Raquellyne Baia Machado, Adriana Maciel Ferreira, Iracirema da Silva Sena, Maria Eduarda Silveira, Ana Maria Santos de Almeida, Francinaldo S. Braga, Alex Bruno Lobato Rodrigues, Roberto Messias Bezerra, Irlon Maciel Ferreira, and et al. 2023. "Copper-Contaminated Substrate Biosorption by Penicillium sp. Isolated from Kefir Grains" Microorganisms 11, no. 6: 1439. https://doi.org/10.3390/microorganisms11061439
APA StyleOliveira, A. F. d., Machado, R. B., Ferreira, A. M., Sena, I. d. S., Silveira, M. E., Almeida, A. M. S. d., Braga, F. S., Rodrigues, A. B. L., Bezerra, R. M., Ferreira, I. M., & Florentino, A. C. (2023). Copper-Contaminated Substrate Biosorption by Penicillium sp. Isolated from Kefir Grains. Microorganisms, 11(6), 1439. https://doi.org/10.3390/microorganisms11061439








