Effects of Degradation on Microbial Communities of an Amazonian Mangrove
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Site Description
2.2. DNA Extraction and Sequencing of 16S rRNA Genes
2.3. Data Analysis
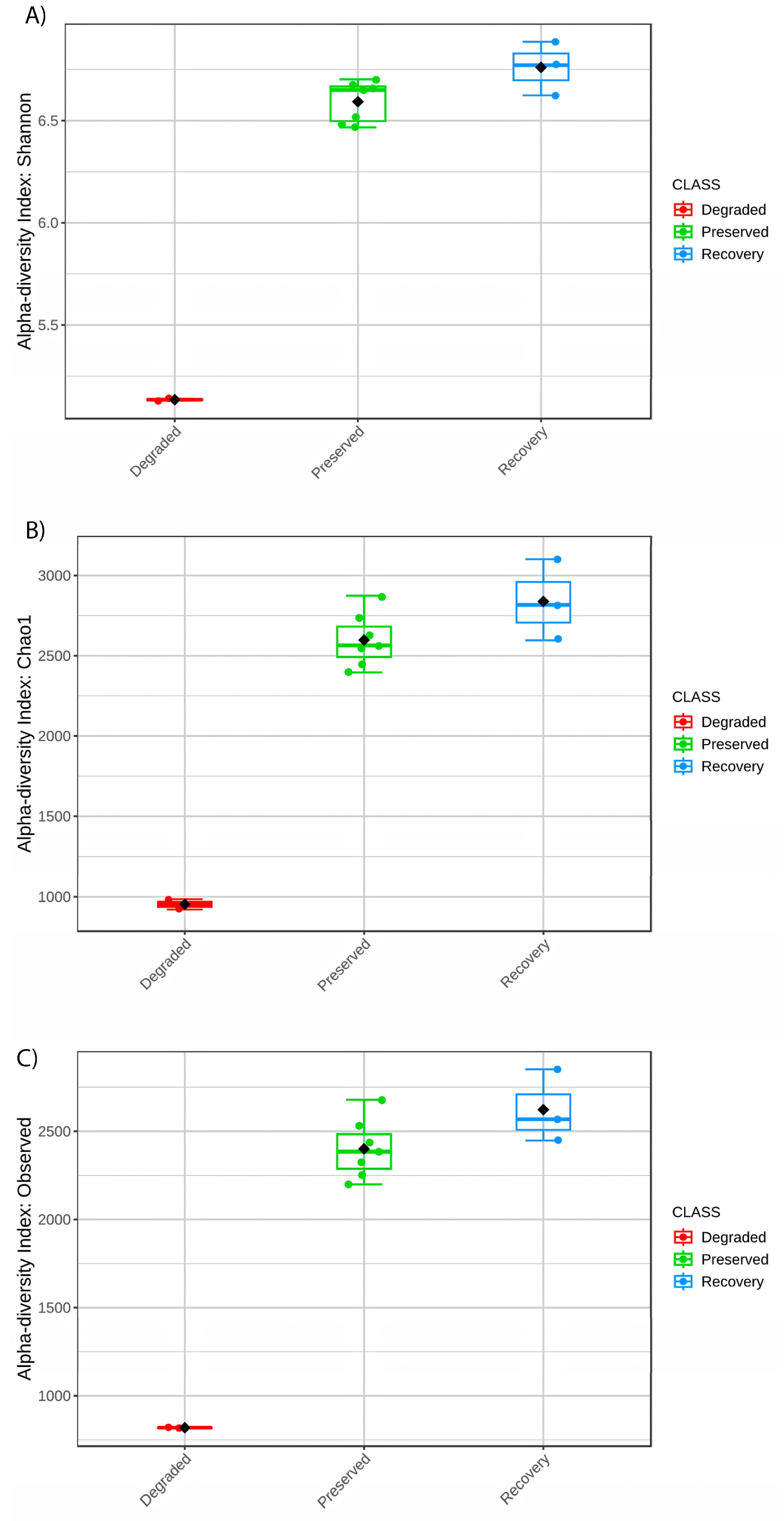
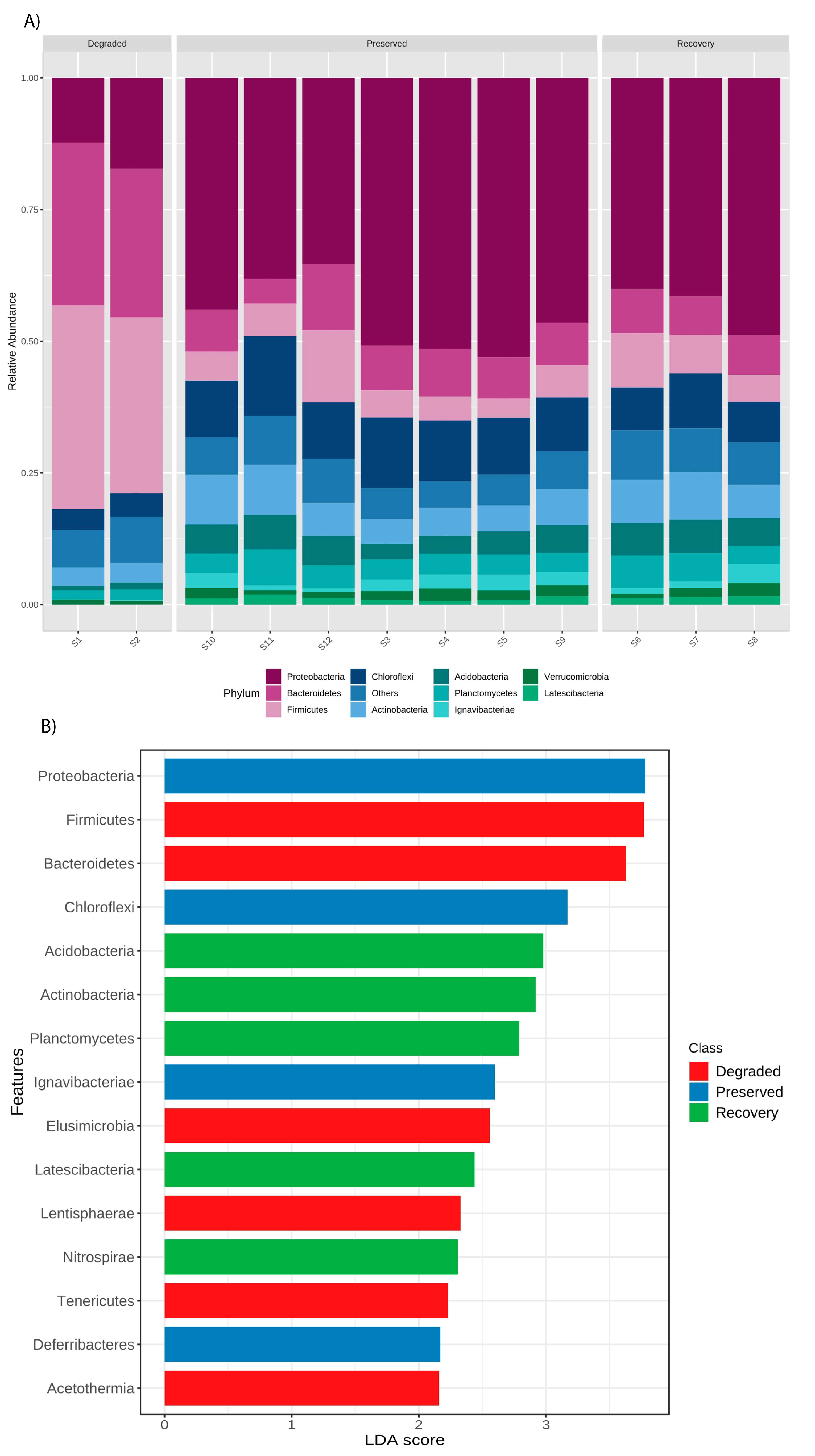
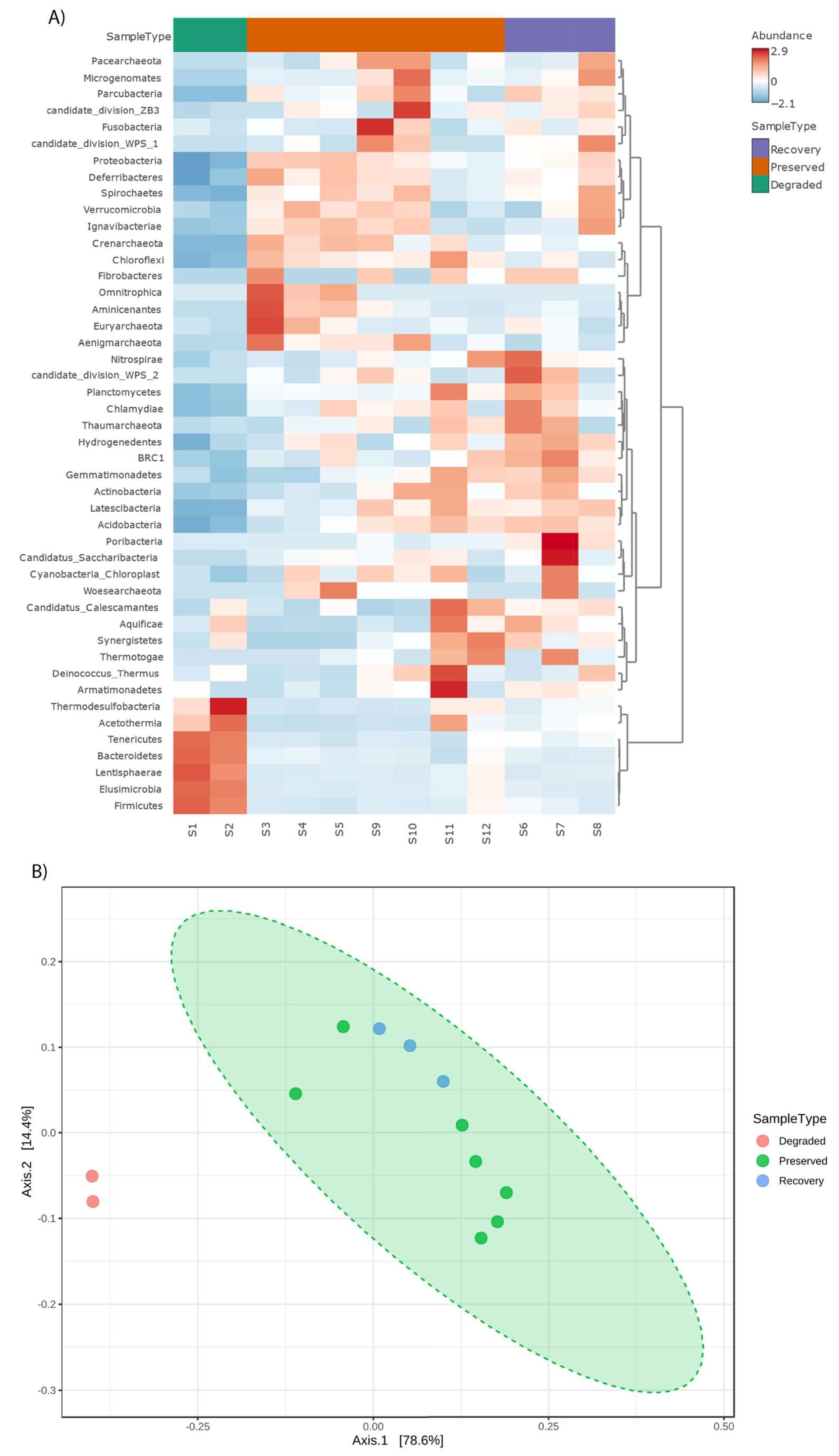
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sahoo, K.; Dhal, N.K. Potential Microbial Diversity in Mangrove Ecosystems:A Review. Indian J. Mar. Sci. 2009, 38, 249–256. [Google Scholar]
- Holguin, G.; Vazquez, P.; Bashan, Y. The Role of Sediment Microorganisms in the Productivity, Conservation, and Rehabilitation of Mangrove Ecosystems: An Overview. Biol. Fertil. Soils 2001, 33, 265–278. [Google Scholar] [CrossRef]
- Ferreira, T.O.; Otero, X.L.; de Souza Junior, V.S.; Vidal-Torrado, P.; Macías, F.; Firme, L.P. Spatial Patterns of Soil Attributes and Components in a Mangrove System in Southeast Brazil (São Paulo). J. Soils Sediments 2010, 10, 995–1006. [Google Scholar] [CrossRef]
- Lu, X.; Taylor, A.E.; Myrold, D.D.; Neufeld, J.D. Expanding Perspectives of Soil Nitrification to Include Ammonia-Oxidizing Archaea and Comammox Bacteria. Soil. Sci. Soc. Am. J. 2020, 84, 287–302. [Google Scholar] [CrossRef]
- Tian, J.; Ge, F.; Zhang, D.; Deng, S.; Liu, X. Roles of Phosphate Solubilizing Microorganisms from Managing Soil Phosphorus Deficiency to Mediating Biogeochemical P Cycle. Biology 2021, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Klotz, M.G.; Bryant, D.A.; Hanson, T.E. The Microbial Sulfur Cycle. Front. Microbiol. 2011, 2, 241. [Google Scholar] [CrossRef]
- Ottoni, F.P.; Hughes, R.M.; Katz, A.M.; Rangel-Pereira, F.d.S.; de Bragança, P.H.N.; Fernandes, R.; Palmeira-Nunes, A.R.O.; Nunes, J.L.S.; Dos Santos, R.R.; Piorski, N.M.; et al. Brazilian Mangroves at Risk. Biota Neotrop. 2021, 21, e20201172. [Google Scholar] [CrossRef]
- Rovai, A.S.; Twilley, R.R.; Worthington, T.A.; Riul, P. Brazilian Mangroves: Blue Carbon Hotspots of National and Global Relevance to Natural Climate Solutions. Front. For. Glob. Chang. 2022, 4, 217. [Google Scholar] [CrossRef]
- Nóbrega, M.S.; Silva, B.S.; Tschoeke, D.A.; Appolinario, L.R.; Calegario, G.; Venas, T.M.; Macedo, L.; Asp, N.; Cherene, B.; Marques, J.S.J.; et al. Mangrove Microbiome Reveals Importance of Sulfur Metabolism in Tropical Coastal Waters. Sci. Total Environ. 2022, 813, 151889. [Google Scholar] [CrossRef]
- Tavares, T.C.L.; Bezerra, W.M.; Normando, L.R.O.; Rosado, A.S.; Melo, V.M.M. Brazilian Semi-Arid Mangroves-Associated Microbiome as Pools of Richness and Complexity in a Changing World. Front. Microbiol. 2021, 12, 2485. [Google Scholar] [CrossRef]
- Palit, K.; Rath, S.; Chatterjee, S.; Das, S. Microbial Diversity and Ecological Interactions of Microorganisms in the Mangrove Ecosystem: Threats, Vulnerability, and Adaptations. Environ. Sci. Pollut. Res. 2022, 29, 32467–32512. [Google Scholar] [CrossRef]
- Mendes, L.W.; Tsai, S.M. Variations of Bacterial Community Structure and Composition in Mangrove Sediment at Different Depths in Southeastern Brazil. Diversity 2014, 6, 827–843. [Google Scholar] [CrossRef]
- Huergo, L.F.; Rissi, D.V.; Elias, A.S.; Gonçalves, M.V.; Gernet, M.V.; Barreto, F.; Dahmer, G.W.; Reis, R.A.; Pedrosa, F.O.; Souza, E.M.; et al. Influence of Ancient Anthropogenic Activities on the Mangrove Soil Microbiome. Sci. Total Environ. 2018, 645, 1–9. [Google Scholar] [CrossRef] [PubMed]
- do Carmo Linhares, D.; Saia, F.T.; Duarte, R.T.D.; Nakayama, C.R.; de Melo, I.S.; Pellizari, V.H. Methanotrophic Community Detected by DNA-SIP at Bertioga’s Mangrove Area, Southeast Brazil. Microb. Ecol. 2021, 81, 954–964. [Google Scholar] [CrossRef]
- Mendes, L.W.; Tsai, S.M. Distinct Taxonomic and Functional Composition of Soil Microbiomes along the Gradient Forest-Restinga-Mangrove in Southeastern Brazil. Antonie Van Leeuwenhoek 2018, 111, 101–114. [Google Scholar] [CrossRef]
- Andreote, F.D.; Jiménez, D.J.; Chaves, D.; Dias, A.C.F.; Luvizotto, D.M.; Dini-Andreote, F.; Fasanella, C.C.; Lopez, M.V.; Baena, S.; Taketani, R.G.; et al. The Microbiome of Brazilian Mangrove Sediments as Revealed by Metagenomics. PLoS ONE 2012, 7, e38600. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.C.L.; Lara, R.J. Temporal Changes of Mangrove Vegetation Boundaries in Amazonia: Application of GIS and Remote Sensing Techniques. Wetl. Ecol. Manag. 2003, 11, 223–231. [Google Scholar] [CrossRef]
- Fernandes, S.O.; Kirchman, D.L.; Michotey, V.D.; Bonin, P.C.; Lokabharathi, P.A. Bacterial Diversity in Relatively Pristine and Anthropogenically-Influenced Mangrove Ecosystems (Goa, India). Braz. J. Microbiol. 2014, 45, 1161–1171. [Google Scholar] [CrossRef]
- Cardenas, S.M.M.; Cohen, M.C.L.; Ruiz, D.P.C.; Souza, A.V.; Gomez-Neita, J.S.; Pessenda, L.C.R.; Culligan, N. Death and Regeneration of an Amazonian Mangrove Forest by Anthropic and Natural Forces. Remote Sens. 2022, 14, 6197. [Google Scholar] [CrossRef]
- Oliveira, M.V.C.; Henrique, M.C. In the Middle of the Road, There Was a Swamp: The Social and Environmental Impacts of the Bragança-Ajuruteua Highway, Pará, Brazil. Hist. Cienc. Saude Manguinhos 2018, 25, 497–514. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/2019 (accessed on 9 April 2023).
- Edgar, R.C. SEARCH_16S: A New Algorithm for Identifying 16S Ribosomal RNA Genes in Contigs and Chromosomes. bioRxiv 2017, 124131. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M. Vegan: Community Ecology Package. R Package Version 2.5-7. Community Ecol. Package 2020, 10, 31. [Google Scholar]
- Khleborodova, A. Lefser: R Implementation of the LEfSE Method for Microbiome Biomarker Discovery. R Package Version 1.8.0. Available online: https://github.com/waldronlab/lefser.2022 (accessed on 9 April 2023).
- Nogueira, V.L.R.; Rocha, L.L.; Colares, G.B.; Angelim, A.L.; Normando, L.R.O.; Cantão, M.E.; Agnez-Lima, L.F.; Andreote, F.D.; Melo, V.M.M. Microbiomes and Potential Metabolic Pathways of Pristine and Anthropized Brazilian Mangroves. Reg. Stud. Mar. Sci. 2015, 2, 56–64. [Google Scholar] [CrossRef]
- Haldar, S.; Nazareth, S.W. Taxonomic Diversity of Bacteria from Mangrove Sediments of Goa: Metagenomic and Functional Analysis. 3 Biotech 2018, 8, 436. [Google Scholar] [CrossRef]
- Wu, P.; Xiong, X.; Xu, Z.; Lu, C.; Cheng, H.; Lyu, X.; Zhang, J.; He, W.; Deng, W.; Lyu, Y.; et al. Bacterial Communities in the Rhizospheres of Three Mangrove Tree Species from Beilun Estuary, China. PLoS ONE 2016, 11, e0164082. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Cao, H.; Peng, C.; Perčulija, V.; Tong, G.; Fang, H.; Wei, X.; Ouyang, S. High-Throughput Sequencing and Analysis of Microbial Communities in the Mangrove Swamps along the Coast of Beibu Gulf in Guangxi, China. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Sadaiappan, B.; Prasannakumar, C.; Subramanian, K.; Subramanian, M. Metagenomic Data of Vertical Distribution and Abundance of Bacterial Diversity in the Hypersaline Sediments of Mad Boon-Mangrove Ecosystem, Bay of Bengal. Data Brief. 2019, 22, 716–721. [Google Scholar] [CrossRef]
- Luis, P.; Saint-Genis, G.; Vallon, L.; Bourgeois, C.; Bruto, M.; Marchand, C.; Record, E.; Hugoni, M. Contrasted Ecological Niches Shape Fungal and Prokaryotic Community Structure in Mangroves Sediments. Environ. Microbiol. 2019, 21, 1407–1424. [Google Scholar] [CrossRef]
- Cotta, S.R.; Cadete, L.L.; van Elsas, J.D.; Andreote, F.D.; Dias, A.C.F. Exploring Bacterial Functionality in Mangrove Sediments and Its Capability to Overcome Anthropogenic Activity. Mar. Pollut. Bull. 2019, 141, 586–594. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, L.; Zhang, Y.; Liu, H.; Jing, H. Comparative Metagenomics Study Reveals Pollution Induced Changes of Microbial Genes in Mangrove Sediments. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Ullah, R.; Yasir, M.; Khan, I.; Bibi, F.; Sohrab, S.S.; Al-Ansari, A.; Al-Abbasi, F.; Al-Sofyani, A.A.; Daur, I.; Lee, S.W.; et al. Comparative Bacterial Community Analysis in Relatively Pristine and Anthropogenically Influenced Mangrove Ecosystems on the Red Sea. Can. J. Microbiol. 2017, 63, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Mushinski, R.M.; Zhou, Y.; Gentry, T.J.; Boutton, T.W. Bacterial Metataxonomic Profile and Putative Functional Behavior Associated with C and N Cycle Processes Remain Altered for Decades after Forest Harvest. Soil. Biol. Biochem. 2018, 119, 184–193. [Google Scholar] [CrossRef]
- Naether, A.; Foesel, B.U.; Naegele, V.; Wüst, P.K.; Weinert, J.; Bonkowski, M.; Alt, F.; Oelmann, Y.; Polle, A.; Lohaus, G.; et al. Environmental Factors Affect Acidobacterial Communities below the Subgroup Level in Grassland and Forest Soils. Appl. Environ. Microbiol. 2012, 78, 7398–7406. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Diamond, S.; Butterfield, C.N.; Thomas, B.C.; Banfield, J.F. Novel Soil Bacteria Possess Diverse Genes for Secondary Metabolite Biosynthesis. Nature 2018, 558, 440–444. [Google Scholar] [CrossRef]
- Kielak, A.M.; Castellane, T.C.L.; Campanharo, J.C.; Colnago, L.A.; Costa, O.Y.A.; Da Silva, M.L.C.; Van Veen, J.A.; Lemos, E.G.M.; Kuramae, E.E. Characterization of Novel Acidobacteria Exopolysaccharides with Potential Industrial and Ecological Applications. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Taketani, R.G.; Moitinho, M.A.; Mauchline, T.H.; Melo, I.S. Co-Occurrence Patterns of Litter Decomposing Communities in Mangroves Indicate a Robust Community Resistant to Disturbances. PeerJ 2018, 2018. [Google Scholar] [CrossRef]
- Dunfield, P.F.; Yuryev, A.; Senin, P.; Smirnova, A.V.; Stott, M.B.; Hou, S.; Ly, B.; Saw, J.H.; Zhou, Z.; Ren, Y.; et al. Methane Oxidation by an Extremely Acidophilic Bacterium of the Phylum Verrucomicrobia. Nature 2007, 450, 879–882. [Google Scholar] [CrossRef]
- Mohammadi, S.S.; Pol, A.; van Alen, T.; Jetten, M.S.M.; den Camp, H.J.M.O. Ammonia Oxidation and Nitrite Reduction in the Verrucomicrobial Methanotroph Methylacidiphilum Fumariolicum SolV. Front. Microbiol. 2017, 8, 1901. [Google Scholar] [CrossRef]
- Dias, A.C.F.; Dini-Andreote, F.; Taketani, R.G.; Tsai, S.M.; Azevedo, J.L.; de Melo, I.S.; Andreote, F.D. Archaeal Communities in the Sediments of Three Contrasting Mangroves. J. Soils Sediments 2011, 11, 1466–1476. [Google Scholar] [CrossRef]
- Prosser, J.I.; Nicol, G.W. Archaeal and Bacterial Ammonia-Oxidisers in Soil: The Quest for Niche Specialisation and Differentiation. Trends Microbiol. 2012, 20, 523–531. [Google Scholar] [CrossRef]
- Iino, T. Ignavibacteriae. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: New York, NY, USA, 2018; p. 1. [Google Scholar] [CrossRef]
- Quintero, I.J.; Castillo, A.M.; Mejía, L.C. Diversity and Taxonomy of Soil Bacterial Communities in Urban and Rural Mangrove Forests of the Panama Bay. Microorganisms 2022, 10, 2191. [Google Scholar] [CrossRef] [PubMed]
- Farag, I.F.; Youssef, N.H.; Elshahed, M.S. Global Distribution Patterns and Pangenomic Diversity of the Candidate Phylum “Latescibacteria” (WS3). Appl. Environ. Microbiol. 2017, 83, e00521-17. [Google Scholar] [CrossRef] [PubMed]
- Youssef, N.H.; Farag, I.F.; Rinke, C.; Hallam, S.J.; Woyke, T.; Elshahed, M.S. In Silico Analysis of the Metabolic Potential and Niche Specialization of Candidate Phylum “Latescibacteria” (WS3). PLoS ONE 2015, 10, e0127499. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.L.; Biggerstaff, J.; Eichhorn, A.; Ewing, E.; Shahan, R.; Soriano, D.; Stewart, S.; VanMol, K.; Walker, R.; Walters, P.; et al. Genomic Characterization of Three Novel Desulfobacterota Classes Expand the Metabolic and Phylogenetic Diversity of the Phylum. Environ. Microbiol. 2021, 23, 4326–4343. [Google Scholar] [CrossRef] [PubMed]
- Waite, D.W.; Chuvochina, M.; Pelikan, C.; Parks, D.H.; Yilmaz, P.; Wagner, M.; Loy, A.; Naganuma, T.; Nakai, R.; Whitman, W.B.; et al. Proposal to Reclassify the Proteobacterial Classes Deltaproteobacteria and Oligoflexia, and the Phylum Thermodesulfobacteria into Four Phyla Reflecting Major Functional Capabilities. Int. J. Syst. Evol. Microbiol. 2020, 70, 5972–6016. [Google Scholar] [CrossRef]
- Zhang, Y.; Gui, H.; Zhang, S.; Li, C. Diversity and Potential Function of Prokaryotic and Eukaryotic Communities from Different Mangrove Sediments. Sustainability 2022, 14, 3333. [Google Scholar] [CrossRef]
- Mori, K.; Suzuki, K.I. The Family Thioalkalispiraceae. Prokaryotes Gammaproteobacteria 2014, 9783642389221, 653–658. [Google Scholar] [CrossRef]
- Wiegand, S.; Jogler, M.; Jogler, C. On the Maverick Planctomycetes. FEMS Microbiol. Rev. 2018, 42, 739–760. [Google Scholar] [CrossRef]
- Wang, H.; Qi, Z.; Zheng, P.; Jiang, C.; Diao, X. Abundant and Rare Microbiota Assembly and Driving Factors between Mangrove and Intertidal Mudflats. Appl. Soil. Ecol. 2022, 174, 104438. [Google Scholar] [CrossRef]
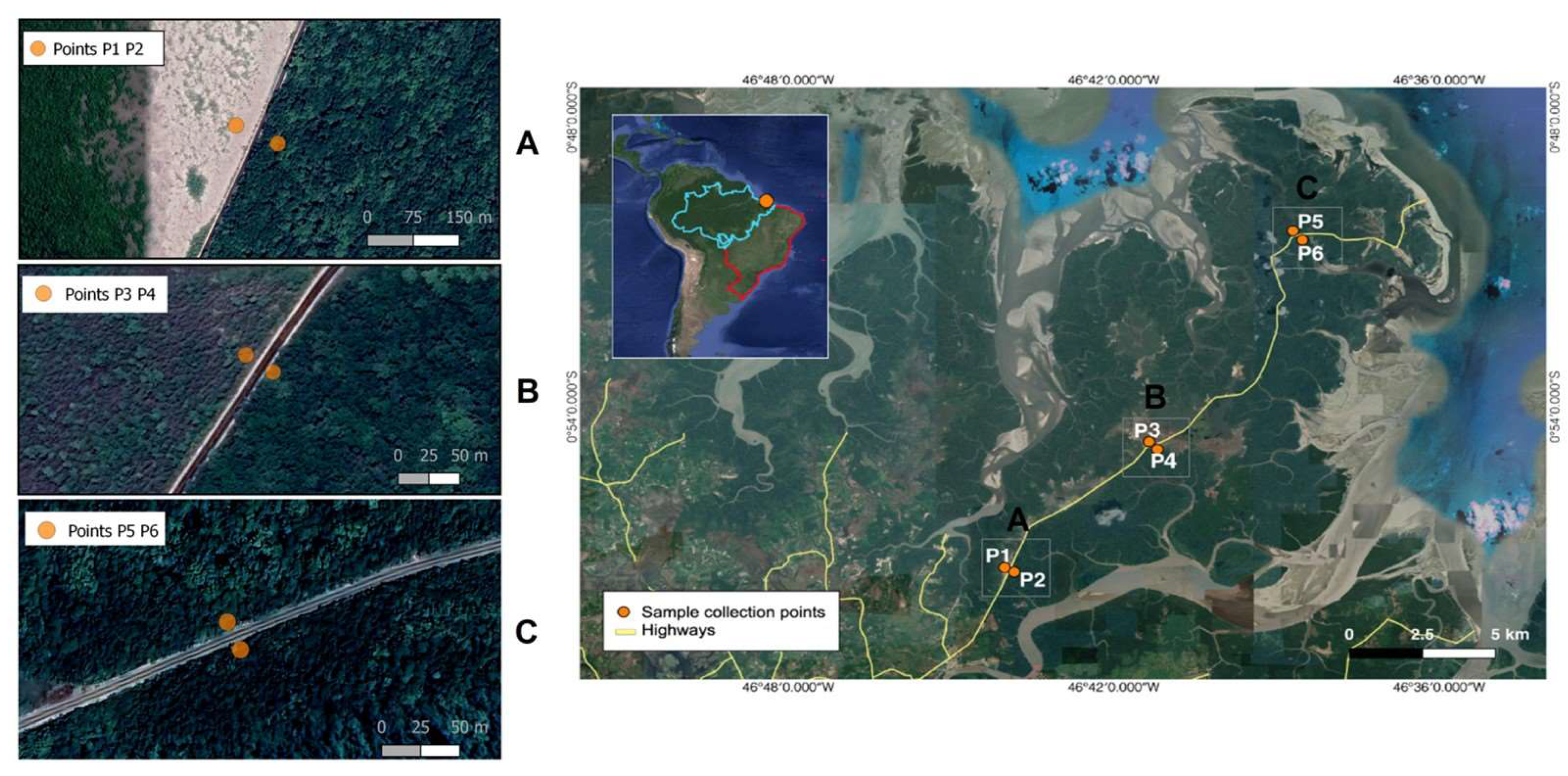
| Samples | C | OM | N | N | Ratio | P | K | Na | Al | Ca | Ca + Mg | pH | H + Al | CEC | Saturation | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | g/kg | g/kg | % | g/kg | C/N | mg/dm3 | cmol/dm3 | Water | cmolc/dm3 | Total (cmol/dm3) | Effective (cmol/dm3) | Base (V%) | Aluminum (m%) | ||||
| P1 (Degraded) | 10.8 | 18.7 | 0.08 | 0.81 | 13.3 | 91 | 1303 | 23,072 | 0 | 5 | 33.2 | 5.9 | 0 | 136.81 | 136.83 | 100 | 0.01 |
| P2 (Preserved) | 16.5 | 28.5 | 0.07 | 0.73 | 22.8 | 16 | 444 | 3957 | 0.1 | 2.5 | 8.5 | 6.4 | 3.82 | 30.66 | 26.91 | 87.55 | 0.26 |
| P3 (Recovery) | 10.4 | 18 | 0.1 | 1.03 | 10.1 | 85 | 1391 | 19,967 | 0 | 6.1 | 29.9 | 5.6 | 0.49 | 120.75 | 120.29 | 99.6 | 0.02 |
| P4 (Preserved) | 24.8 | 42.8 | 0.13 | 1.29 | 19.2 | 43 | 788 | 10,957 | 0 | 3.6 | 17.4 | 5.8 | 2.09 | 69.14 | 67.07 | 96.97 | 0.04 |
| P5 (Preserved) | 21.7 | 37.5 | 0.07 | 0.73 | 30 | 34 | 919 | 11,976 | 0 | 3.2 | 15.7 | 5.1 | 0.79 | 70.93 | 70.18 | 98.89 | 0.04 |
| P6 (Preserved) | 8.4 | 14.5 | 0.07 | 0.7 | 11.9 | 28 | 876 | 12,082 | 0 | 3.1 | 15.2 | 5.5 | 0.9 | 70.89 | 70.03 | 98.73 | 0.04 |
| Sample | Collection Point | Mangrove Zone | Library Size | OTUs | Chao1 | Shannon | Simpson |
|---|---|---|---|---|---|---|---|
| S1 | P1 | Degraded | 90,508 | 1846 | 2221.686 | 5.642208 | 0.9890705 |
| S2 | P1 | Degraded | 131,794 | 1672 | 2213.535 | 5.476673 | 0.9883609 |
| S3 | P2 | Preserved | 72,723 | 4307 | 5320.197 | 6.945422 | 0.9969483 |
| S4 | P2 | Preserved | 93,865 | 4876 | 6218.262 | 7.089495 | 0.9973286 |
| S5 | P2 | Preserved | 65,004 | 4663 | 5628.641 | 7.091619 | 0.9972898 |
| S6 | P3 | Recovery | 43,328 | 4491 | 5379.287 | 7.155558 | 0.9974265 |
| S7 | P3 | Recovery | 124,523 | 5185 | 6733.596 | 7.296043 | 0.9975854 |
| S8 | P3 | Recovery | 89,942 | 4284 | 5545.846 | 6.936312 | 0.9966584 |
| S9 | P4 | Preserved | 80,006 | 4738 | 5771.278 | 7.065384 | 0.9965857 |
| S10 | P4 | Preserved | 84,816 | 4634 | 6056.463 | 7.051824 | 0.9972325 |
| S11 | P5 | Preserved | 67,836 | 4125 | 5065.642 | 6.937517 | 0.9961980 |
| S12 | P5 | Preserved | 55,652 | 4140 | 5362.078 | 6.842138 | 0.9962226 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, G.M.d.; Costa, S.S.; Baraúna, R.A.; Castilho, B.P.; Pinheiro, I.C.; Silva, A.; Schaan, A.P.; Ribeiro-dos-Santos, Â.; Graças, D.A.d. Effects of Degradation on Microbial Communities of an Amazonian Mangrove. Microorganisms 2023, 11, 1389. https://doi.org/10.3390/microorganisms11061389
Costa GMd, Costa SS, Baraúna RA, Castilho BP, Pinheiro IC, Silva A, Schaan AP, Ribeiro-dos-Santos Â, Graças DAd. Effects of Degradation on Microbial Communities of an Amazonian Mangrove. Microorganisms. 2023; 11(6):1389. https://doi.org/10.3390/microorganisms11061389
Chicago/Turabian StyleCosta, Gleyciane Machado da, Sávio Souza Costa, Rafael Azevedo Baraúna, Bruno Pureza Castilho, Izabel Cruz Pinheiro, Artur Silva, Ana Paula Schaan, Ândrea Ribeiro-dos-Santos, and Diego Assis das Graças. 2023. "Effects of Degradation on Microbial Communities of an Amazonian Mangrove" Microorganisms 11, no. 6: 1389. https://doi.org/10.3390/microorganisms11061389
APA StyleCosta, G. M. d., Costa, S. S., Baraúna, R. A., Castilho, B. P., Pinheiro, I. C., Silva, A., Schaan, A. P., Ribeiro-dos-Santos, Â., & Graças, D. A. d. (2023). Effects of Degradation on Microbial Communities of an Amazonian Mangrove. Microorganisms, 11(6), 1389. https://doi.org/10.3390/microorganisms11061389






