Ecoinformatic Analysis of the Gut Ecological Diversity of Wild and Captive Long-Tailed Gorals Using Improved ITS2 Region Primers to Support Their Conservation
Abstract
1. Introduction
2. Materials and Methods
2.1. Fecal Sample Collection
2.2. Evaluating and Improving the Performance of the In Silico ITS2 Region Primers
2.3. Gut Eco-Information Analysis: Fungi and Undigested DNA of Viridiplantae
3. Results
3.1. In Silico Improving the Fungal ITS2 Region Primers
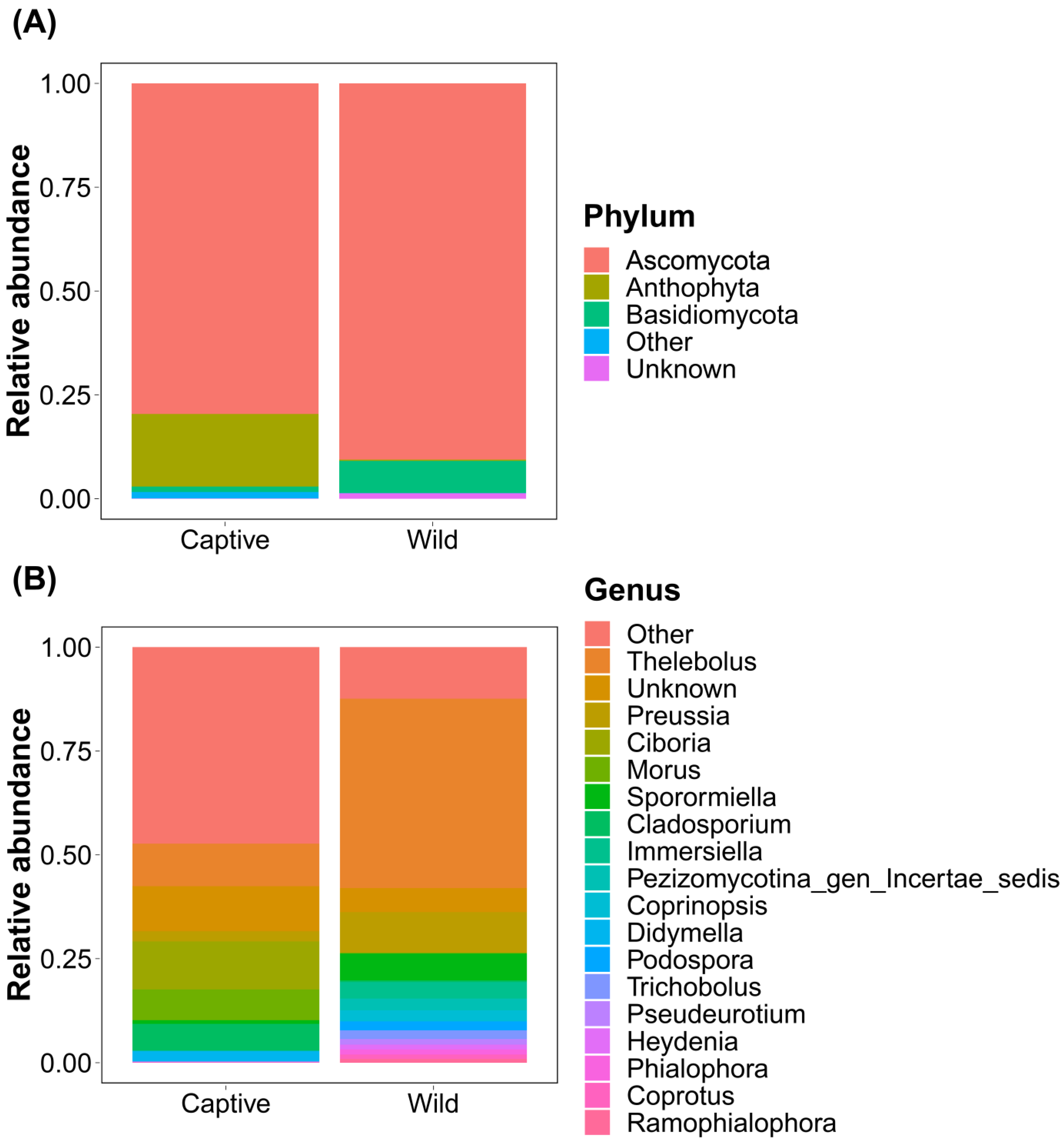
3.2. Gut Eco-Information: Fungi and Undigested DNA of Viridiplantae
3.3. Alpha Diversity
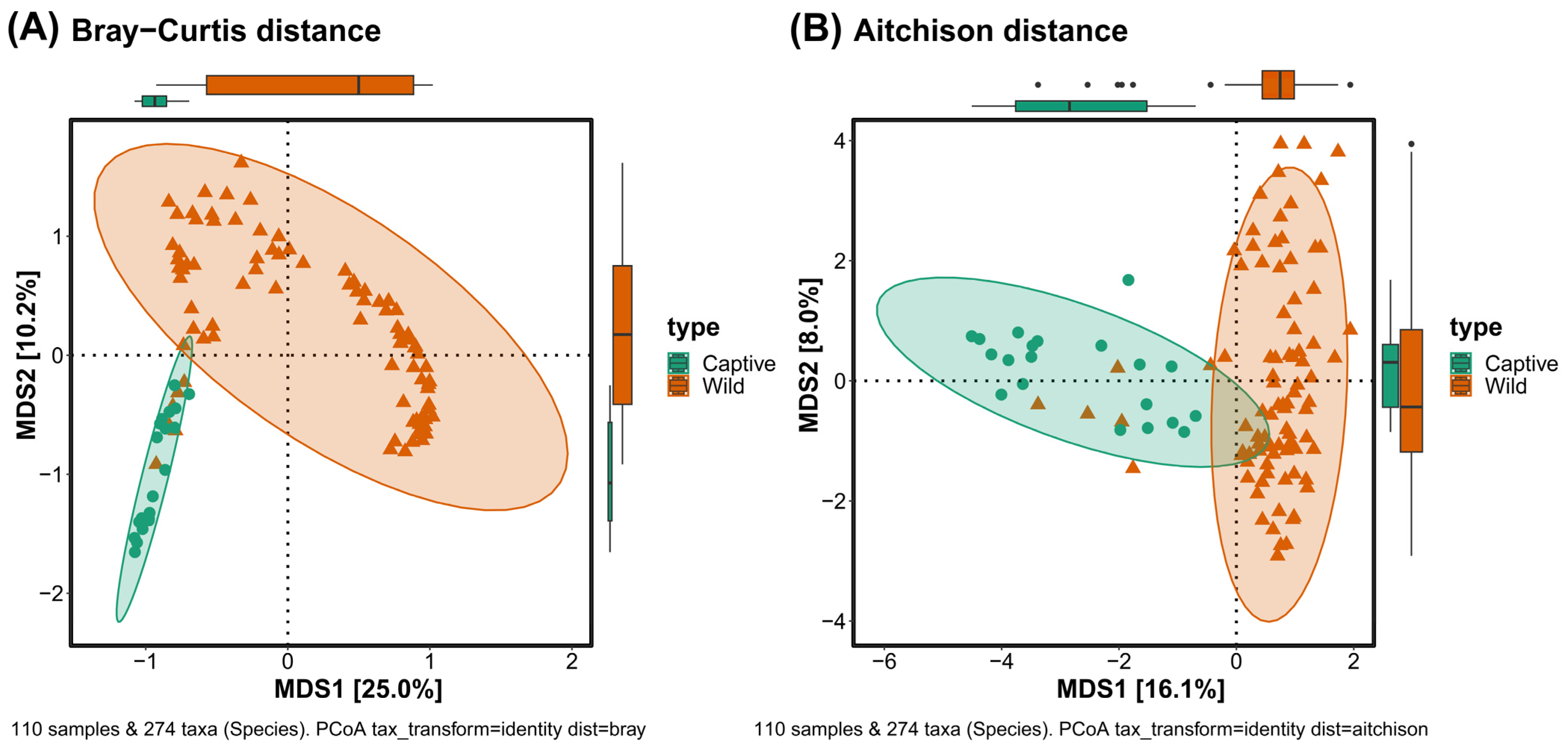
3.4. Beta Diversity
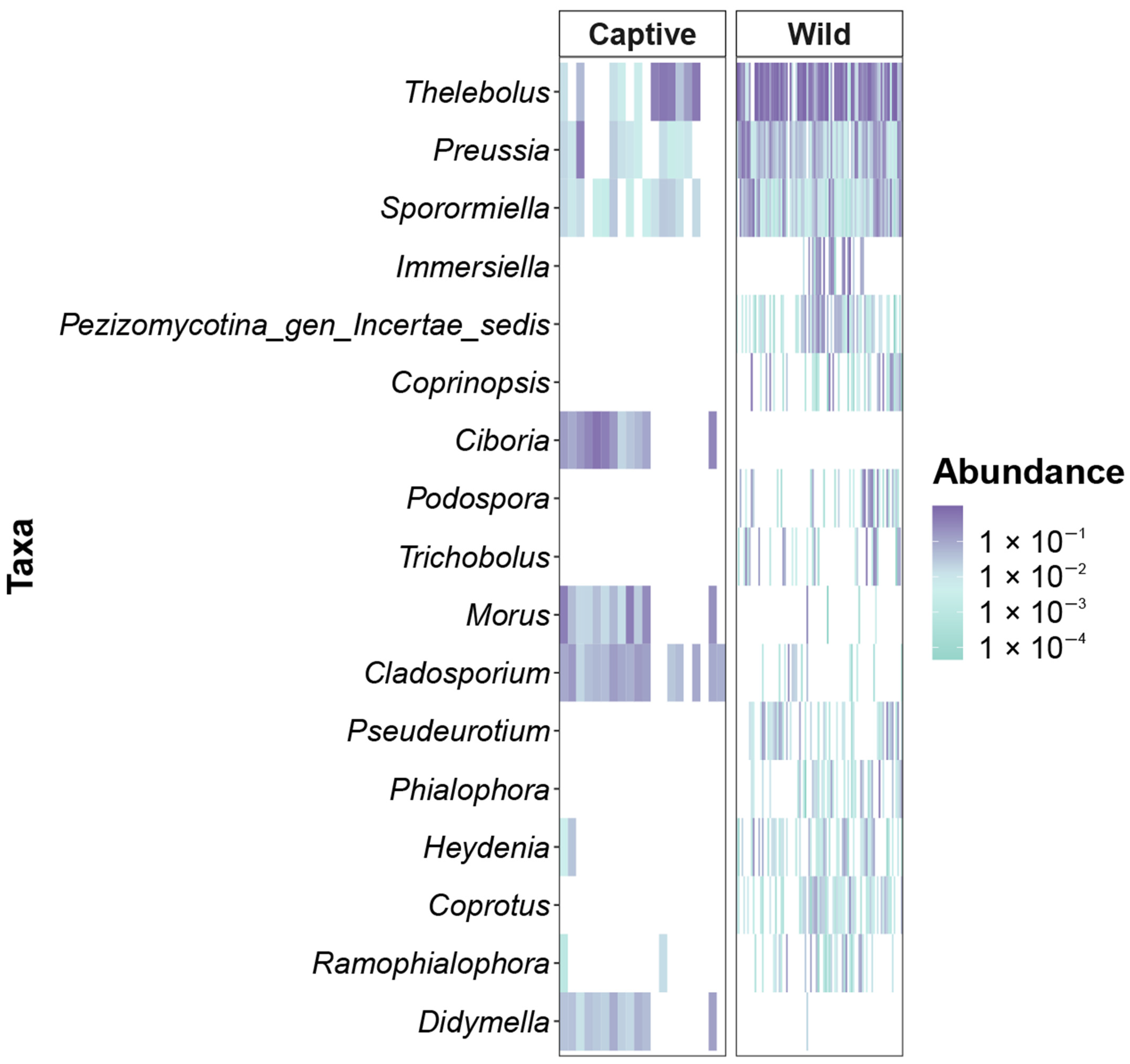
3.5. Heatmap Analysis
3.6. Undigested DNA of Viridiplantae
4. Discussion
4.1. Validating and Improving the Fungal ITS2 Region Primers
4.2. Gut Eco-Information of Long-Tailed GORALS
4.2.1. Gut Ecological Diversity
4.2.2. Food Sources of Long-Tailed Gorals from Gut Eco-Information
4.2.3. Efforts to Reintroduce Gorals in the Future
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karesh, W.B.; Cook, R.A. Applications of veterinary medicine to in situ conservation efforts. Oryx 1995, 29, 244–252. [Google Scholar] [CrossRef]
- Kaplan, G. Casting the net widely for change in animal welfare: The plight of birds in zoos, ex situ conservation, and conservation fieldwork. Animals 2021, 12, 31. [Google Scholar] [CrossRef] [PubMed]
- Hale, R.; Mac Nally, R.; Blumstein, D.T.; Swearer, S.E. Evaluating where and how habitat restoration is undertaken for animals. Restor. Ecol. 2019, 27, 775–781. [Google Scholar] [CrossRef]
- Genes, L.; Fernandez, F.A.S.; Vaz-de-Mello, F.Z.; da Rosa, P.; Fernandez, E.; Pires, A.S. Effects of howler monkey reintroduction on ecological interactions and processes. Conserv. Biol. 2019, 33, 88–98. [Google Scholar] [CrossRef]
- Wearn, O.R.; Glover-Kapfer, P. Snap happy: Camera traps are an effective sampling tool when compared with alternative methods. R. Soc. Open Sci. 2019, 6, 181748. [Google Scholar] [CrossRef]
- Mellish, J.-A.; Hennen, D.; Thomton, J.; Petrauskas, L.; Atkinson, S.; Calkins, D. Permanent marking in an endangered species: Physiological response to hot branding in Steller sea lions (Eumetopias jubatus). Wildl. Res. 2007, 34, 43–47. [Google Scholar] [CrossRef]
- Peixoto, R.S.; Harkins, D.M.; Nelson, K.E. Advances in microbiome research for animal health. Annu. Rev. Anim. Biosci. 2021, 9, 289–311. [Google Scholar] [CrossRef]
- Law, R.; Illian, J.; Burslem, D.F.; Gratzer, G.; Gunatilleke, C.V.S.; Gunatilleke, I.A.U.N. Ecological information from spatial patterns of plants: Insights from point process theory. J. Ecol. 2009, 97, 616–628. [Google Scholar] [CrossRef]
- Michener, W.K.; Jones, M.B. Ecoinformatics: Supporting ecology as a data-intensive science. Trends Ecol. Evol. 2012, 27, 85–93. [Google Scholar] [CrossRef]
- Krapivin, V.F.; Varotsos, C.A.; Soldatov, V.Y. New Ecoinformatics Tools in Environmental Science; Springer: Vienna, Austria, 2015; Volume 903. [Google Scholar] [CrossRef]
- Park, H.-B.; Park, H.C.; Cowan, P.E.; Hong, S. Comparison of hair-trapping methods for the long-tailed goral. Wildl. Soc. Bull. 2018, 42, 310–313. [Google Scholar] [CrossRef]
- Park, H.-B.; Hong, S. Habitat characteristics coincidence of dead and living long-tailed gorals (Naemorhedus caudatus) according to extreme snowfall. Animals 2021, 11, 997. [Google Scholar] [CrossRef]
- Lavrinienko, A.; Scholier, T.; Bates, S.T.; Miller, A.N.; Watts, P.C. Defining gut mycobiota for wild animals: A need for caution in assigning authentic resident fungal taxa. Anim. Microbiome 2021, 3, 75. [Google Scholar] [CrossRef]
- Moorhouse-Gann, R.J.; Dunn, J.C.; de Vere, N.; Goder, M.; Cole, N.; Hipperson, H.; Symondson, W.O.C. New universal ITS2 primers for high-resolution herbivory analyses using DNA metabarcoding in both tropical and temperate zones. Sci. Rep. 2018, 8, 8542. [Google Scholar] [CrossRef]
- Chiang, T.Y.; Schaal, B.A. Phylogeography of North American populations of the moss species Hylocomium splendens based on the nucleotide sequence of internal transcribed spacer 2 of nuclear ribosomal DNA. Mol. Ecol. 1999, 8, 1037–1042. [Google Scholar] [CrossRef]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Neves, D.M. Ecoinformatics for conservation biology. Nat. Ecol. Evol. 2022, 6, 1595–1596. [Google Scholar] [CrossRef]
- Vancov, T.; Keen, B. Amplification of soil fungal community DNA using the ITS86F and ITS4 primers. FEMS Microbiol. Lett. 2009, 296, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.; Brown, G.; Tatusova, T.; Maglott, D. The reference sequence (RefSeq) database. In The NCBI Handbook; National Center for Biotechnology Information: Bethesda, MD, USA, 2012; Volume 2. [Google Scholar]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021, 49, D10–D17. [Google Scholar] [CrossRef] [PubMed]
- Python, W. Python. Python Releases Wind 2021 24. Available online: https://citeseerx.ist.psu.edu/document?repid=rep1&type=pdf&doi=1f2ee3831eebfc97bfafd514ca2abb7e2c5c86bb (accessed on 28 March 2023).
- Lee, Y.H.; Kang, G.-U.; Jeon, S.Y.; Tagele, S.B.; Pham, H.Q.; Kim, M.-S.; Ahmad, S.; Jung, D.-R.; Park, Y.-J.; Han, H.S.; et al. Vaginal microbiome-based bacterial signatures for predicting the severity of cervical intraepithelial neoplasia. Diagnostics 2020, 10, 1013. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.-U.; Jung, D.-R.; Lee, Y.H.; Jeon, S.Y.; Han, H.S.; Chong, G.O.; Shin, J.-H. Dynamics of fecal microbiota with and without invasive cervical cancer and its application in early diagnosis. Cancers 2020, 12, 3800. [Google Scholar] [CrossRef]
- Kim, M.-S.; Kim, S.-H.; Jeong, M.; Park, M.-K.; Jo, Y.; Kang, G.-U.; Jung, D.-R.; Lee, C.-I.; Shin, J.-H. Fermented coffee grounds diminish livestock odors: A microbiome study. Agronomy 2021, 11, 1914. [Google Scholar] [CrossRef]
- Fentie, E.G.; Jeong, M.; Emire, S.A.; Demsash, H.D.; Kim, M.A.; Jeon, H.-J.; Lee, S.-E.; Tagele, S.B.; Park, Y.-J.; Shin, J.-H. Microbiome dataset of spontaneously fermented Ethiopian honey wine, Tej. Data Brief 2022, 42, 108022. [Google Scholar] [CrossRef]
- Azimirad, M.; Jo, Y.; Kim, M.-S.; Jeong, M.; Shahrokh, S.; Aghdaei, H.A.; Zali, M.R.; Lee, S.; Yadegar, A.; Shin, J.-H. Alterations and prediction of functional profiles of gut microbiota after fecal microbiota transplantation for Iranian recurrent Clostridioides difficile infection with underlying inflammatory bowel disease: A pilot study. J. Inflamm. Res. 2022, 15, 105–116. [Google Scholar] [CrossRef]
- Parson, W.; Strobl, C.; Huber, G.; Zimmermann, B.; Gomes, S.M.; Souto, L.; Fendt, L.; Delport, R.; Langit, R.; Wootton, S.; et al. Evaluation of next generation mtGenome sequencing using the ion torrent personal genome machine (PGM). Forensic Sci. Int. Genet. 2013, 7, 543–549. [Google Scholar] [CrossRef]
- Hall, M.; Beiko, R.G. 16S rRNA gene analysis with QIIME2. In Microbiome Analysis: Methods and Protocols; Humana Press: New York, NY, USA, 2018; Volume 1849, pp. 113–129. [Google Scholar]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. Union of Needletrades, Industrial & Textile Employees. QIIME Release for Eukaryotes 2; UNITE Community: Slough, UK, 2022. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Gómez-Rubio, V. ggplot2-elegant graphics for data analysis. J. Stat. Softw. 2017, 77, 1–3. [Google Scholar] [CrossRef]
- Wickham, H.; Chang, W.; Wickham, M.H. Package ‘ggplot2’. Create Elegant Data Visualisations Using the Grammar of Graphics, Version 2. 2016, pp. 1–189. Available online: https://rdrr.io/cran/ggplot2/ (accessed on 28 March 2023).
- Beck, M.W. ggord: Ordination Plots with ggplot2. R Package Version 1. 2017. Available online: https://zenodo.org/badge/latestdoi/35334615 (accessed on 28 March 2023).
- Ahlmann-Eltze, C.; Ahlmann-Eltze, M.C. Package ‘Ggsignif’. 2017. Available online: https://github.com/const-ae/ggsignif (accessed on 28 March 2023).
- Ahlmanneltze, C. Ggsignif: Significance Bars for ‘ggplot2’. 2017. Available online: https://github.com/Artjom-Metro/ggsignif (accessed on 28 March 2023).
- Lahti, L.; Shetty, S. Introduction to the microbiome R package. Bioconductor 2018. Available online: https://www.bioconductor.org/packages/release/bioc/html/microbiome.html (accessed on 15 October 2022).
- Shetty, S.A.; Lahti, L.; de Vos, W.M.; Smidt, H. Microbiomeutilities: An R package for utilities to guide in-depth marker gene amplicon data analysis. Ecophysiol. Insights Hum. Intest. Microbiota Single Strains Defin. Consortia 2018, 95. Available online: https://edepot.wur.nl/468013#page=97 (accessed on 28 March 2023).
- Barnett, D.J.; Arts, I.C.; Penders, J. microViz: An R package for microbiome data visualization and statistics. J. Open Source Softw. 2021, 6, 3201. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Bisanz, J.E. qiime2R. Importing QIIME2 Artifacts and Associated Data into R Sessions. Version 0.99. 2018, Volume 13. Available online: https://github.com/jbisanz/qiime2R (accessed on 28 March 2023).
- Gorzelak, M.; McAmmond, B.M.; Van Hamme, J.D.; Birnbaum, C.; Thomsen, C.; Hart, M. Soil microbial communities in long-term soil storage for sand mine reclamation. Ecol. Restor. 2020, 38, 13–23. [Google Scholar] [CrossRef]
- Hart, M.; Gorzelak, M.A.; McAmmond, B.M.; Van Hamme, J.D.; Stevens, J.; Abbott, L.; Whiteley, A.S.; Nevill, P. Fungal communities resist recovery in sand mine restoration. Front. For. Glob. Change 2019, 2, 78. [Google Scholar] [CrossRef]
- Stevens, V.; Thijs, S.; Bongaerts, E.; Nawrot, T.; Marchal, W.; Van Hamme, J.; Vangronsveld, J. Ambient air pollution shapes bacterial and fungal ivy leaf communities. Microorganisms 2021, 9, 2088. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Park, C.E.; Cho, B.-J.; Kim, M.-J.; Park, H.C.; Shin, J.-H. Geographical relationships between long-tailed goral (Naemorhedus caudatus) populations based on gut microbiome analysis. Microorganisms 2021, 9, 2002. [Google Scholar] [CrossRef]
- Quijada, L.; Matočec, N.; Kušan, I.; Tanney, J.B.; Johnston, P.R.; Mešić, A.; Pfister, D.H. Apothecial Ancestry, Evolution, and Re-Evolution in Thelebolales (Leotiomycetes, Fungi). Biology 2022, 11, 583. [Google Scholar] [CrossRef]
- Tong, Y.; Wu, J.; Guo, W.; Yang, Z.; Wang, H.; Liu, H.; Gao, Y.; Sun, M.; Yue, C. The Effect of Combining Millet and Corn Straw as Source Forage for Beef Cattle Diets on Ruminal Degradability and Fungal Community. Animals 2023, 13, 548. [Google Scholar] [CrossRef]
- Bohmann, K.; Evans, A.; Gilbert, M.T.P.; Carvalho, G.R.; Creer, S.; Knapp, M.; Yu, D.W.; de Bruyn, M. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 2014, 29, 358–367. [Google Scholar] [CrossRef]
- Yoccoz, N.G. The future of environmental DNA in ecology. Mol. Ecol. 2012, 21, 2031–2038. [Google Scholar] [CrossRef]
- Shokralla, S.; Spall, J.L.; Gibson, J.F.; Hajibabaei, M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 2012, 21, 1794–1805. [Google Scholar] [CrossRef]
- Zaumyslova, O.Y.; Bondarchuk, S.N. The use of camera traps for monitoring the population of long-tailed gorals. Achiev. Life Sci. 2015, 9, 15–21. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Mason, G.J. Species differences in responses to captivity: Stress, welfare and the comparative method. Trends Ecol. Evol. 2010, 25, 713–721. [Google Scholar] [CrossRef]
- Bahrndorff, S.; Alemu, T.; Alemneh, T.; Lund Nielsen, J. The microbiome of animals: Implications for conservation biology. Int. J. Genom. 2016, 2016, 5304028. [Google Scholar] [CrossRef]

| Regions | No. of Samples | Type of Samples |
|---|---|---|
| Seoraksan National Park | 26 | Wild |
| Odaesan National Park | 8 | Wild |
| Taebaeksan National Park | 3 | Wild |
| Woraksan National Park | 9 | Wild |
| Juwangsan National Park | 2 | Wild |
| Wangpicheon Conservation Area | 14 | Wild |
| Unnamed mountain in Samcheok | 28 | Wild |
| Northern Conservation Center | 11 | Captive |
| Association of Korean Goral Conservation | 9 | Captive |
| Total | 110 |
| Regions | Sequences | Counts | Proportions (%) |
|---|---|---|---|
| ITS86F | GTGAATCATCGAATCTTTGAA | 7700 | 79.4224 |
| GTGAATCATCGAGTCTTTGAA | 888 | 9.1594 | |
| GTGAGTCATCGAATCTTTGAA | 253 | 2.6096 | |
| GTGAATCATTGAATCTTTGAA | 108 | 1.1140 | |
| GTGAACCATCGAATCTTTGAA | 96 | 0.9902 | |
| ITS4 | TCCTCCGCTTATTGATATGC | 2058 | 93.6732 |
| CCTCCGGCTTATTGATATGC | 16 | 0.7283 | |
| CCTCCCGCTTATTGATATGC | 12 | 0.5462 | |
| TCCTCTGCTTATTGATATGC | 10 | 0.4552 | |
| TCCTCCGCTGACTGATATGC | 8 | 0.3641 |
| Primers | Sequences | Melting Temperature (°C) | Matching Rate (%) |
|---|---|---|---|
| CEP-ITS86F | GTGARTCATYGARTCTTTGAA | 53–59 | 92.3053 |
| CEP-ITS86F-GCG | GCGARTCATCGARTCTTTGAA | 58–62 | 0.9593 |
| CEP-ITS86F-CTG | CTGAATCATCRAATYTTTGAA | 51–55 | 0.0619 |
| CEP-ITS4- | TCCTCYGCTKAYTGATATGC | 56–64 | 94.8111 |
| CEP-ITS4-CCT | CCTYCSGCTTATTGATATGC | 58–61 | 1.3655 |
| CEP-ITS4-TCT | TCTTCYGCTTATTGATATGY | 52–57 | 0.3186 |
| Kingdom | Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|---|
| Viridiplantae | Anthophyta | Eudicotyledonae | Fabales | Fabaceae | Maackia | amurensis c, w |
| Eudicotyledonae | Fagales | Juglandacea | Juglans | hopeiensis c | ||
| Eudicotyledonae | Malpighiale | Salicaceae | Salix | heteromera c | ||
| Eudicotyledonae | Malvales | Malvaceae | Tilia | sp. c | ||
| Eudicotyledonae | Malvales | Malvaceae | Tilia | paucicostata 1 c | ||
| Eudicotyledonae | Malvales | Malvaceae | Tilia | paucicostata 2 c | ||
| Eudicotyledonae | Rosales | Moraceae | Morus | Morus alba 1 c, w | ||
| Eudicotyledonae | Rosales | Moraceae | Morus | Morus alba 2 c, w | ||
| Eudicotyledonae | Rosales | Rosaceae | Prunus | sp. 1 c, w | ||
| Eudicotyledonae | Rosales | Rosaceae | Prunus | sp. 2 c, w | ||
| Eudicotyledonae | Rosales | Rosaceae | Prunus | serrulata c | ||
| Eudicotyledonae | Rosales | Rosaceae | Rubus | microphyllus w | ||
| Chlorophyta | Chlorophycea | Sphaeropleales | Radiococcaceae | Coenochloris | sp. w | |
| Trebouxiophyceae | Prasiolales | Stichococcaceae | Pseudostichococcus | monallantoides w | ||
| Trebouxiophyceae | Prasiolales | Stichococcaceae | Pseudostichococcus | sp. w |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, C.-E.; Cho, B.-J.; Kim, M.-J.; Kim, M.-C.; Park, M.-K.; Son, J.-I.; Park, H.-C.; Shin, J.-H. Ecoinformatic Analysis of the Gut Ecological Diversity of Wild and Captive Long-Tailed Gorals Using Improved ITS2 Region Primers to Support Their Conservation. Microorganisms 2023, 11, 1368. https://doi.org/10.3390/microorganisms11061368
Park C-E, Cho B-J, Kim M-J, Kim M-C, Park M-K, Son J-I, Park H-C, Shin J-H. Ecoinformatic Analysis of the Gut Ecological Diversity of Wild and Captive Long-Tailed Gorals Using Improved ITS2 Region Primers to Support Their Conservation. Microorganisms. 2023; 11(6):1368. https://doi.org/10.3390/microorganisms11061368
Chicago/Turabian StylePark, Chang-Eon, Bum-Joon Cho, Min-Ji Kim, Min-Chul Kim, Min-Kyu Park, Jang-Ick Son, Hee-Cheon Park, and Jae-Ho Shin. 2023. "Ecoinformatic Analysis of the Gut Ecological Diversity of Wild and Captive Long-Tailed Gorals Using Improved ITS2 Region Primers to Support Their Conservation" Microorganisms 11, no. 6: 1368. https://doi.org/10.3390/microorganisms11061368
APA StylePark, C.-E., Cho, B.-J., Kim, M.-J., Kim, M.-C., Park, M.-K., Son, J.-I., Park, H.-C., & Shin, J.-H. (2023). Ecoinformatic Analysis of the Gut Ecological Diversity of Wild and Captive Long-Tailed Gorals Using Improved ITS2 Region Primers to Support Their Conservation. Microorganisms, 11(6), 1368. https://doi.org/10.3390/microorganisms11061368






