Fecal Microbiome Analysis Distinguishes Bacterial Taxa Biomarkers Associated with Red Fillet Color in Rainbow Trout
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Rainbow Trout Population, Experimental Design, Treatments, and Sampling
2.3. DNA Extraction
2.4. Library Preparation
2.5. Bioinformatics and Statistical Analyses
3. Results
3.1. Mean Saturation Index (S.I) Values between the Red versus White Fillet Group
3.2. Fecal Microbiome Overall Assessment
3.3. Fecal Microbiota Composition in Red and White Fillet Fish
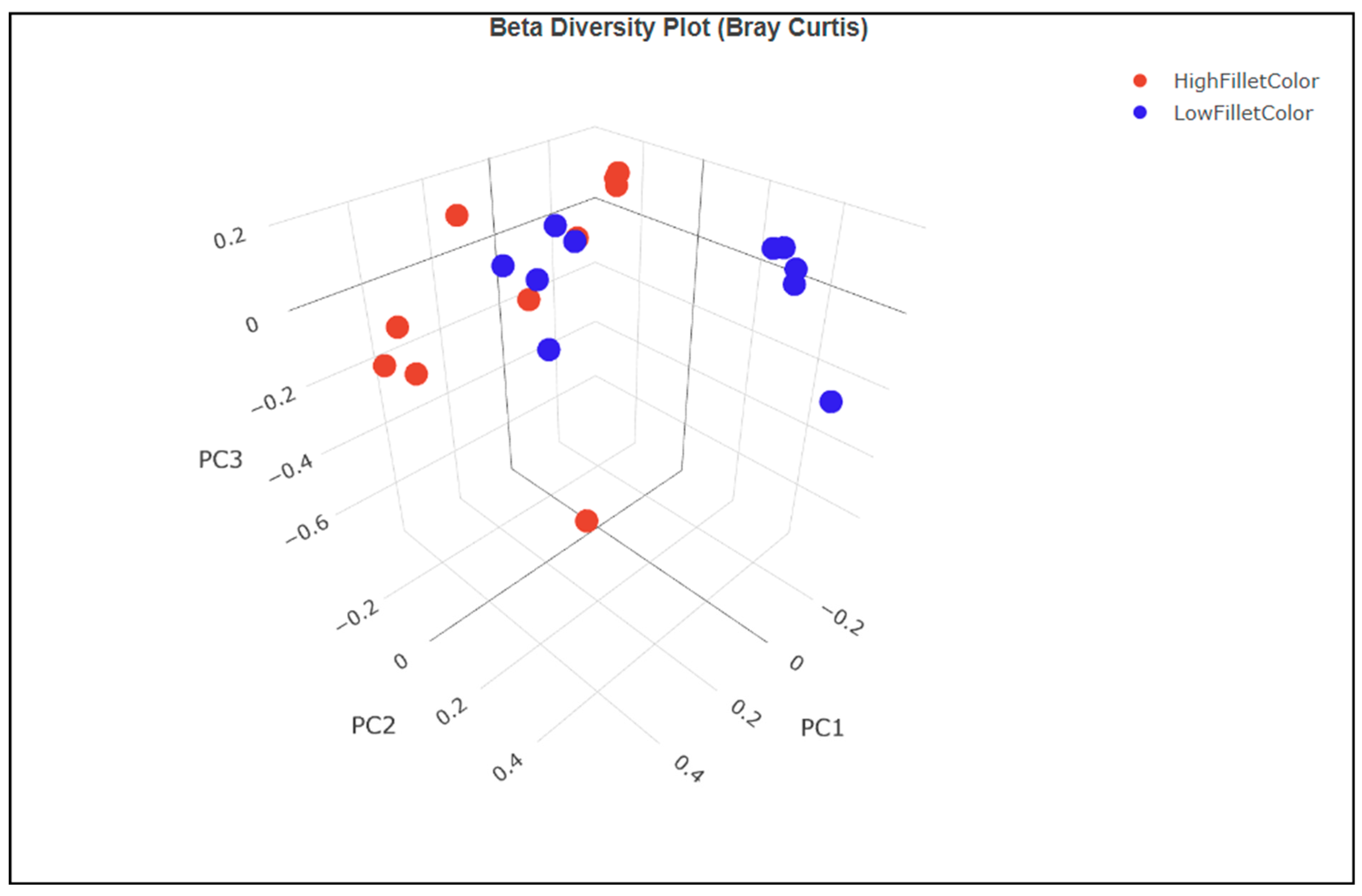
3.3.1. Alpha and Beta Diversity
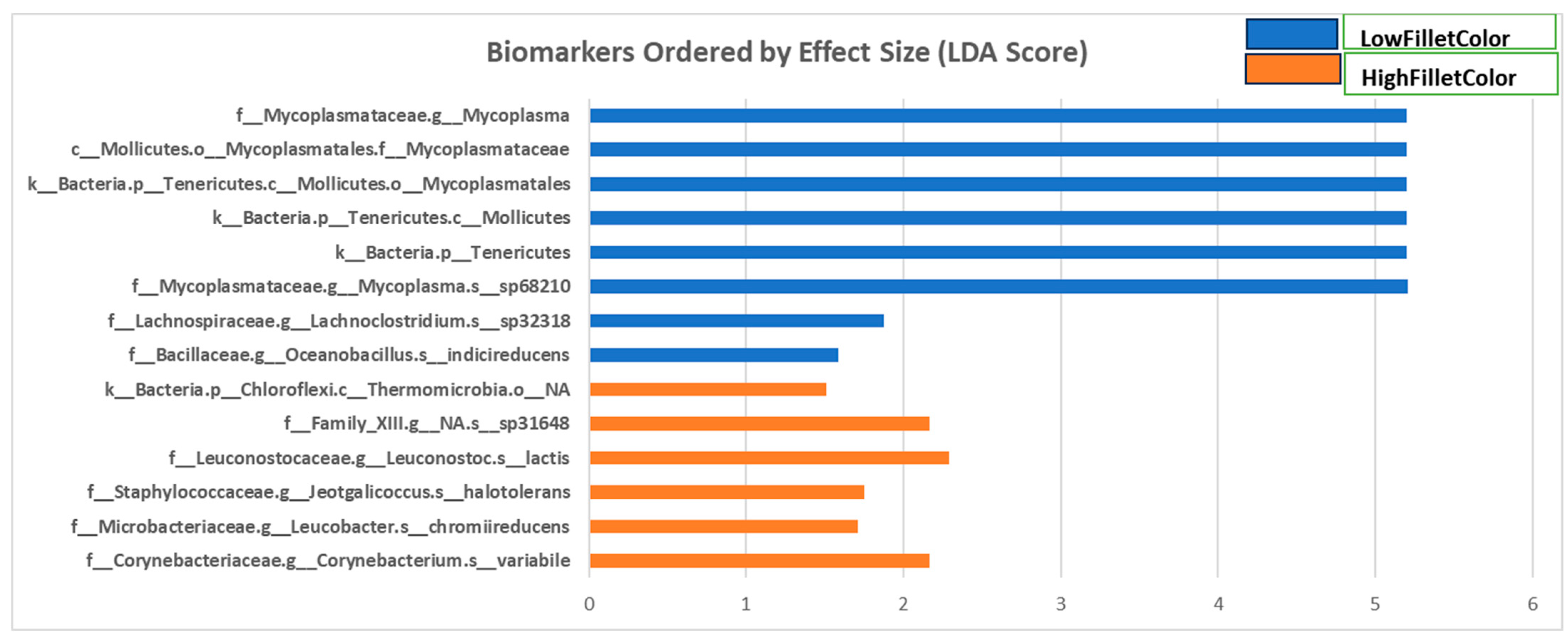
3.3.2. Linear Discriminant Analysis for Effect Size
4. Discussion
4.1. Bacterial Taxa Enriched in the Red Fillet Fish
4.2. Bacterial Taxa Enriched in the White Fillet Fish
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Kumari, S.; Rajarani, A.; Bansal, N.; Dahuja, A.; Praveen, S.; Krishnan, V.; Kumar, S. Extraction and estimation of provitamin A carotenoids from carrot. Omics Meet Plant Biochem. Appl. Nutr. Enhanc. One Health Perspect. 2019, 221, 56–60. [Google Scholar]
- Torrissen, O.J. Strategies for salmonid pigmentation. J. Appl. Ichthyol.-Z. Fur Angew. Ichthyol. 1995, 11, 276–281. [Google Scholar] [CrossRef]
- Torrissen, O.J. Pigmentation of Salmonids—Interactions of Astaxanthin and Canthaxanthin on Pigment Deposition in Rainbow-Trout. Aquaculture 1989, 79, 363–374. [Google Scholar] [CrossRef]
- Aas, G.H.; Bjerkeng, B.; Storebakken, T.; Ruyter, B. Blood appearance, metabolic transformation and plasma transport proteins of C-astaxanthin in Atlantic salmon. Fish Physiol. Biochem. 1999, 21, 325–334. [Google Scholar] [CrossRef]
- Baker, R.T.M.; Pfeiffer, A.M.; Schöner, F.J.; Smith-Lemmon, L. Pigmenting efficacy of astaxanthin and canthaxanthin in fresh-water reared Atlantic salmon. Anim. Feed Sci. Technol. 2002, 99, 97–106. [Google Scholar] [CrossRef]
- Hill, G.E. Is there an immunological cost to carotenoid-based ornamental coloration? Am. Nat. 1999, 154, 589–595. [Google Scholar] [CrossRef]
- Lozano, G.A. Carotenoids, Parasites, and Sexual Selection. Oikos 1994, 70, 309–311. [Google Scholar] [CrossRef]
- Nayak, S.K. Role of gastrointestinal microbiota in fish. Aquac. Res. 2010, 41, 1553–1573. [Google Scholar] [CrossRef]
- Wang, A.R.; Ran, C.; Ringo, E.; Zhou, Z.G. Progress in fish gastrointestinal microbiota research. Rev. Aquac. 2018, 10, 626–640. [Google Scholar] [CrossRef]
- Brugman, S.; Nieuwenhuis, E.E. Mucosal control of the intestinal microbial community. J. Mol. Med. 2010, 88, 881–888. [Google Scholar] [CrossRef]
- Cerf-Bensussan, N.; Gaboriau-Routhiau, V. The immune system and the gut microbiota: Friends or foes? Nat. Rev. Immunol. 2010, 10, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Viney, M.E.; Riley, E.M. From immunology to eco-immunology: More than a new name. In Eco-Immunology: Evolutive Aspects and Future Perspectives; Springer: Dordrecht, The Netherlands, 2014; pp. 1–19. [Google Scholar]
- Ringø, E.; Strøm, E.; Tabachek, J.A. Intestinal microflora of salmonids: A review. Aquac. Res. 1995, 26, 773–789. [Google Scholar] [CrossRef]
- Navarrete, P.; Magne, F.; Araneda, C.; Fuentes, P.; Barros, L.; Opazo, R.; Espejo, R.; Romero, J. PCR-TTGE analysis of 16S rRNA from rainbow trout (Oncorhynchus mykiss) gut microbiota reveals host-specific communities of active bacteria. PLoS ONE 2012, 7, e31335. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.R.; Links, M.G.; Collins, S.A.; Mansfield, G.S.; Drew, M.D.; Van Kessel, A.G.; Hill, J.E. Effects of plant-based diets on the distal gut microbiome of rainbow trout (Oncorhynchus mykiss). Aquaculture 2012, 350, 134–142. [Google Scholar] [CrossRef]
- Mansfield, G.S.; Desai, A.R.; Nilson, S.A.; Van Kessel, A.G.; Drew, M.D.; Hill, J.E. Characterization of rainbow trout (Oncorhynchus mykiss) intestinal microbiota and inflammatory marker gene expression in a recirculating aquaculture system. Aquaculture 2010, 307, 95–104. [Google Scholar] [CrossRef]
- Ray, A.K.; Ghosh, K.; Ringo, E. Enzyme-producing bacteria isolated from fish gut: A review. Aquac. Nutr. 2012, 18, 465–492. [Google Scholar] [CrossRef]
- Rawls, J.F.; Samuel, B.S.; Gordon, J.I. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. USA 2004, 101, 4596–4601. [Google Scholar] [CrossRef]
- Semova, I.; Carten, J.D.; Stombaugh, J.; Mackey, L.C.; Knight, R.; Farber, S.A.; Rawls, J.F. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 2012, 12, 277–288. [Google Scholar] [CrossRef]
- Camp, J.G.; Jazwa, A.L.; Trent, C.M.; Rawls, J.F. Intronic cis-regulatory modules mediate tissue-specific and microbial control of angptl4/fiaf transcription. PLoS Genet 2012, 8, e1002585. [Google Scholar] [CrossRef]
- Bates, J.M.; Mittge, E.; Kuhlman, J.; Baden, K.N.; Cheesman, S.E.; Guillemin, K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 2006, 297, 374–386. [Google Scholar] [CrossRef]
- Bates, J.M.; Akerlund, J.; Mittge, E.; Guillemin, K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2007, 2, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Chapagain, P.; Arivett, B.; Cleveland, B.M.; Walker, D.M.; Salem, M. Analysis of the fecal microbiota of fast- and slow-growing rainbow trout (Oncorhynchus mykiss). BMC Genom. 2019, 20, 788. [Google Scholar] [CrossRef] [PubMed]
- Gildberg, A.; Johansen, A.; Bogwald, J. Growth and survival of Atlantic salmon (Salmo salar) fry given diets supplemented with fish protein hydrolysate and lactic acid bacteria during a challenge trial with Aeromonas salmonicida. Aquaculture 1995, 138, 23–34. [Google Scholar] [CrossRef]
- Ringo, E.; Salinas, I.; Olsen, R.E.; Nyhaug, A.; Myklebust, R.; Mayhew, T.M. Histological changes in intestine of Atlantic salmon (Salmo salar L.) following in vitro exposure to pathogenic and probiotic bacterial strains. Cell Tissue Res. 2007, 328, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.B.; Medeiros, D.M. The microbiome as a major function of the gastrointestinal tract and its implication in micronutrient metabolism and chronic diseases. Nutr. Res. 2023, 112, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Hua, Y. Carotenoid biosynthesis in extremophilic Deinococcus-Thermus bacteria. Trends Microbiol. 2010, 18, 512–520. [Google Scholar] [CrossRef]
- Misawa, N.; Maoka, T.; Takemura, M. Carotenoids: Carotenoid and apocarotenoid analysis—Use of E. coli to produce carotenoid standards. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2022; Volume 670, pp. 87–137. [Google Scholar]
- Kanamoto, H.; Nakamura, K.; Misawa, N. Carotenoid Production in Oleaginous Yeasts. Adv. Exp. Med. Biol. 2021, 1261, 153–163. [Google Scholar] [CrossRef]
- Shindo, K.; Misawa, N. New and Rare Carotenoids Isolated from Marine Bacteria and Their Antioxidant Activities. Mar. Drugs 2014, 12, 1690–1698. [Google Scholar] [CrossRef]
- Ram, S.; Mitra, M.; Shah, F.; Tirkey, S.R.; Mishra, S. Bacteria as an alternate biofactory for carotenoid production: A review of its applications, opportunities and challenges. J. Funct. Foods 2020, 67, 103867. [Google Scholar] [CrossRef]
- Umeno, D.; Tobias, A.V.; Arnold, F.H. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. Rev. 2005, 69, 51–78. [Google Scholar] [CrossRef]
- Nguyen, C.D.H.; Amoroso, G.; Ventura, T.; Minich, J.J.; Elizur, A. Atlantic Salmon (Salmo salar L., 1758) Gut Microbiota Profile Correlates with Flesh Pigmentation: Cause or Effect? Mar. Biotechnol. 2020, 22, 786–804. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.D.H.; Amoroso, G.; Ventura, T.; Elizur, A. Assessing the Pyloric Caeca and Distal Gut Microbiota Correlation with Flesh Color in Atlantic Salmon (Salmo salar L., 1758). Microorganisms 2020, 8, 1244. [Google Scholar] [CrossRef] [PubMed]
- Bonder, M.J.; Kurilshikov, A.; Tigchelaar, E.F.; Mujagic, Z.; Imhann, F.; Vila, A.V.; Deelen, P.; Vatanen, T.; Schirmer, M.; Smeekens, S.P.; et al. The effect of host genetics on the gut microbiome. Nat. Genet. 2016, 48, 1407–1412. [Google Scholar] [CrossRef] [PubMed]
- Awany, D.; Allali, I.; Dalvie, S.; Hemmings, S.; Mwaikono, K.S.; Thomford, N.E.; Gomez, A.; Mulder, N.; Chimusa, E.R. Host and Microbiome Genome-Wide Association Studies: Current State and Challenges. Front. Genet. 2018, 9, 637. [Google Scholar] [CrossRef]
- Buitenhuis, B.; Lassen, J.; Noel, S.J.; Plichta, D.R.; Sorensen, P.; Difford, G.F.; Poulsen, N.A. Impact of the rumen microbiome on milk fatty acid composition of Holstein cattle. Genet. Sel. Evol. 2019, 51, 23. [Google Scholar] [CrossRef]
- Percie du Sert, N.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. J. Cereb. Blood Flow Metab. 2020, 40, 1769–1777. [Google Scholar] [CrossRef]
- Leeds, T.D.; Vallejo, R.L.; Weber, G.M.; Gonzalez-Pena, D.; Silverstein, J.T. Response to five generations of selection for growth performance traits in rainbow trout (Oncorhynchus mykiss). Aquaculture 2016, 465, 341–351. [Google Scholar] [CrossRef]
- Cleveland, B.M.; Radler, L.M.; Leeds, T.D. Growth, fillet yield, and muscle quality traits are not affected by a genotype by diet interaction in rainbow trout consuming diets that differ in lipid content. J. World Aquac. Soc. 2023. [Google Scholar] [CrossRef]
- Garcia, A.; Tsuruta, S.; Gao, G.; Palti, Y.; Lourenco, D.; Leeds, T. Genomic selection models substantially improve the accuracy of genetic merit predictions for fillet yield and body weight in rainbow trout using a multi-trait model and multi-generation progeny testing. Genet. Sel. Evol. 2023, 55, 11. [Google Scholar] [CrossRef]
- AMSA. Guidelines for meat color evaluation. In Proceedings of the Proceedings: The 44th Annual Reciprocal Meat Conference, Chicago, KS, USA, 9–12 June 1991; pp. 3–17. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Park, J.; Jang, J.K.; Lee, B.H.; Park, Y.S. Structural Analysis of Gluco-Oligosaccharides Produced by and Their Prebiotic Effect. Molecules 2019, 24, 3998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, M.; Dai, X. Biological characteristics and probiotic effect of Leuconostoc lactis strain isolated from the intestine of black porgy fish. Braz. J. Microbiol. 2013, 44, 685–691. [Google Scholar] [CrossRef] [PubMed]
- He, S.X.; Ran, C.; Qin, C.B.; Li, S.N.; Zhang, H.L.; de Vos, W.M.; Ringo, E.; Zhou, Z.G. Anti-Infective Effect of Adhesive Probiotic in Fish is Correlated with Their Spatial Distribution in the Intestinal Tissue. Sci. Rep. 2017, 7, 13195. [Google Scholar] [CrossRef]
- Robertson, P.A.W.; O’Dowd, C.; Burrells, C.; Williams, P.; Austin, B. Use of Carnobacterium sp. as a probiotic for Atlantic salmon (Salmo salar L.) and rainbow trout (Oncorhynchus mykiss, Walbaum). Aquaculture 2000, 185, 235–243. [Google Scholar] [CrossRef]
- Brennan, N.M.; Ward, A.C.; Beresford, T.P.; Fox, P.F.; Goodfellow, M.; Cogan, T.M. Biodiversity of the bacterial flora on the surface of a smear cheese. Appl. Environ. Microbiol 2002, 68, 820–830. [Google Scholar] [CrossRef]
- Schröder, J.; Maus, I.; Trost, E.; Tauch, A. Complete genome sequence of Corynebacterium variabile DSM 44702 isolated from the surface of smear-ripened cheeses and insights into cheese ripening and flavor generation. BMC Genom. 2011, 12, 545. [Google Scholar] [CrossRef]
- Mounier, J.; Irlinger, F.; Leclercq-Perlat, M.N.; Sarthou, A.S.; Spinnler, H.E.; Fitzgerald, G.F.; Cogan, T.M. Growth and colour development of some surface ripening bacteria with on aseptic cheese curd. J. Dairy Res. 2006, 73, 441–448. [Google Scholar] [CrossRef]
- Starr, M.P.; Saperstein, S. Thiamine and the carotenoid pigments of Corynebacterium poinsettiae. Arch. Biochem. Biophys. 1953, 43, 157–168. [Google Scholar] [CrossRef]
- Heider, S.A.; Peters-Wendisch, P.; Wendisch, V.F. Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum. BMC Microbiol. 2012, 12, 198. [Google Scholar] [CrossRef]
- Wariso, B.A.; Kester, A.S.; Thomas, R.D. Isolation and partial characterization of carotenoid mutants of Corynebacterium poinsettiae ATCC 9682. Glob. J. Pure Appl. Sci. 2006, 12, 203–210. [Google Scholar] [CrossRef]
- Shahidi, F.; Brown, J.A. Carotenoid pigments in seafoods and aquaculture. Crit. Rev. Food Sci. Nutr. 1998, 38, 1–67. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Lee, K.C.; Weiss, N.; Kang, K.H.; Park, Y.H. Jeotgalicoccus halotolerans gen. nov., sp. nov. and Jeotgalicoccus psychrophilus sp. nov., isolated from the traditional Korean fermented seafood jeotgal. Int. J. Syst. Evol. Microbiol. 2003, 53, 595–602. [Google Scholar] [CrossRef] [PubMed]
- de Lourdes Moreno, M.; Sanchez-Porro, C.; Garcia, M.T.; Mellado, E. Carotenoids’ production from halophilic bacteria. Methods Mol. Biol. 2012, 892, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Calegari-Santos, R.; Diogo, R.A.; Fontana, J.D.; Bonfim, T.M. Carotenoid Production by Halophilic Archaea Under Different Culture Conditions. Curr. Microbiol. 2016, 72, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Industrial and environmental applications of halophilic microorganisms. Environ. Technol. 2010, 31, 825–834. [Google Scholar] [CrossRef]
- Asker, D.; Ohta, Y. Production of canthaxanthin by extremely halophilic bacteria. J. Biosci. Bioeng. 1999, 88, 617–621. [Google Scholar] [CrossRef]
- Yatsunami, R.; Ando, A.; Yang, Y.; Takaichi, S.; Kohno, M.; Matsumura, Y.; Ikeda, H.; Fukui, T.; Nakasone, K.; Fujita, N.; et al. Identification of carotenoids from the extremely halophilic archaeon Haloarcula japonica. Front. Microbiol. 2014, 5, 100. [Google Scholar] [CrossRef]
- Morais, P.V.; Francisco, R.; Branco, R.; Chung, A.P.; da Costa, M.S. Leucobacter chromiireducens sp. nov, and Leucobacter aridicollis sp. nov., two new species isolated from a chromium contaminated environment. Syst. Appl. Microbiol. 2004, 27, 646–652. [Google Scholar] [CrossRef]
- Zhu, W.J.; Yang, Z.H.; Ma, Z.M.; Chai, L.Y. Reduction of high concentrations of chromate by Leucobacter sp. CRB1 isolated from Changsha, China. World J. Microbiol. Biotechnol. 2008, 24, 991–996. [Google Scholar] [CrossRef]
- Bharti, M.; Nagar, S.; Khurana, H.; Negi, R.K. Metagenomic insights to understand the role of polluted river Yamuna in shaping the gut microbial communities of two invasive fish species. Arch. Microbiol. 2022, 204, 509. [Google Scholar] [CrossRef] [PubMed]
- Sturm, G.; Brunner, S.; Suvorova, E.; Dempwolff, F.; Reiner, J.; Graumann, P.; Bernier-Latmani, R.; Majzlan, J.; Gescher, J. Chromate Resistance Mechanisms in Leucobacter chromiiresistens. Appl. Environ. Microbiol. 2018, 84, e02208–e02218. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tian, Y. Hexavalent chromium reducing bacteria: Mechanism of reduction and characteristics. Environ. Sci. Pollut. Res. Int. 2021, 28, 20981–20997. [Google Scholar] [CrossRef] [PubMed]
- Skalli, A.; Firmino, J.P.; Andree, K.B.; Salomon, R.; Estevez, A.; Puig, P.; Sabater-Martinez, M.; Hechavarria, T.; Gisbert, E. The Inclusion of the Microalga Scenedesmus sp. in Diets for Rainbow Trout, Onchorhynchus mykiss, Juveniles. Animal 2020, 10, 1656. [Google Scholar] [CrossRef] [PubMed]
- Tomás-Almenar, C.; Larrán, A.; de Mercado, E.; Sanz-Calvo, M.; Hernández, D.; Riaño, B.; García-González, M. Scenedesmus almeriensis from an integrated system waste-nutrient, as sustainable protein source for feed to rainbow trout (Oncorhynchus mykiss). Aquaculture 2018, 497, 422–430. [Google Scholar] [CrossRef]
- Sánchez, J.; Fernández, J.; Acién, F.; Rueda, A.; Pérez-Parra, J.; Molina, E. Influence of culture conditions on the productivity and lutein content of the new strain Scenedesmus almeriensis. Process Biochem. 2008, 43, 398–405. [Google Scholar] [CrossRef]
- Soler-Vila, A.; Coughlan, S.; Guiry, M.D.; Kraan, S. The red alga Porphyra dioica as a fish-feed ingredient for rainbow trout (Oncorhynchus mykiss): Effects on growth, feed efficiency, and carcass composition. J. Appl. Phycol. 2009, 21, 617–624. [Google Scholar] [CrossRef]
- Teimouri, M.; Amirkolaie, A.K.; Yeganeh, S. The effects of Spirulina platensis meal as a feed supplement on growth performance and pigmentation of rainbow trout (Oncorhynchus mykiss). Aquaculture 2013, 396, 14–19. [Google Scholar] [CrossRef]
- Zheng, A.; Luo, J.; Meng, K.; Li, J.; Zhang, S.; Li, K.; Liu, G.; Cai, H.; Bryden, W.L.; Yao, B. Proteome changes underpin improved meat quality and yield of chickens (Gallus gallus) fed the probiotic Enterococcus faecium. BMC Genom. 2014, 15, 1167. [Google Scholar] [CrossRef]
- Meng, Q.W.; Yan, L.; Ao, X.; Zhou, T.X.; Wang, J.P.; Lee, J.H.; Kim, I.H. Influence of probiotics in different energy and nutrient density diets on growth performance, nutrient digestibility, meat quality, and blood characteristics in growing-finishing pigs. J. Anim. Sci. 2010, 88, 3320–3326. [Google Scholar] [CrossRef]
- Zhang, X.; Han, L.; Gui, L.; Raza, S.H.A.; Hou, S.; Yang, B.; Wang, Z.; Ma, Y.; Makhlof, R.T.M.; Alhuwaymil, Z.; et al. Metabolome and microbiome analysis revealed the effect mechanism of different feeding modes on the meat quality of Black Tibetan sheep. Front. Microbiol. 2022, 13, 1076675. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.; Hayflick, L. Highlights of mycoplasma research—An historical perspective. Biologicals 2010, 38, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Aino, K.; Nodasaka, Y.; Yumoto, I. Oceanobacillus indicireducens sp. nov., a facultative alkaliphile that reduces an indigo dye. Int. J. Syst. Evol. Microbiol. 2013, 63, 1437–1442. [Google Scholar] [CrossRef]
- Chavan, R. Indigo dye and reduction techniques. In Denim; Elsevier: Amsterdam, The Netherlands, 2015; pp. 37–67. [Google Scholar]
- Dandachi, I.; Anani, H.; Hadjadj, L.; Brahimi, S.; Lagier, J.C.; Daoud, Z.; Rolain, J.M. Genome analysis of Lachnoclostridium phocaeense isolated from a patient after kidney transplantation in Marseille. New Microbes New Infect. 2021, 41, 100863. [Google Scholar] [CrossRef]
- Liang, J.Q.; Li, T.; Nakatsu, G.; Chen, Y.X.; Yau, T.O.; Chu, E.; Wong, S.; Szeto, C.H.; Ng, S.C.; Chan, F.K.L.; et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut 2020, 69, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.-Y.; Huang, F.-Q.; Lao, X.; Lu, Y.; Gao, X.; Alolga, R.N.; Yin, K.; Zhou, X.; Wang, Y.; Liu, B. Integrated metagenomics identifies a crucial role for trimethylamine-producing Lachnoclostridium in promoting atherosclerosis. NPJ Biofilms Microbiomes 2022, 8, 11. [Google Scholar] [CrossRef]
- Nogal, A.; Louca, P.; Zhang, X.; Wells, P.M.; Steves, C.J.; Spector, T.D.; Falchi, M.; Valdes, A.M.; Menni, C. Circulating Levels of the Short-Chain Fatty Acid Acetate Mediate the Effect of the Gut Microbiome on Visceral Fat. Front. Microbiol. 2021, 12, 711359. [Google Scholar] [CrossRef]
- Chen, Y.X.; Xie, Y.N.; Zhong, R.Q.; Liu, L.; Lin, C.G.; Xiao, L.; Chen, L.; Zhang, H.F.; Beckers, Y.; Everaert, N. Effects of Xylo-Oligosaccharides on Growth and Gut Microbiota as Potential Replacements for Antibiotic in Weaning Piglets. Front. Microbiol. 2021, 12, 641172. [Google Scholar] [CrossRef]
- Cheng, M.P.; Domingo, M.C.; Levesque, S.; Yansouni, C.P. A case report of a deep surgical site infection with Terrisporobacter glycolicus/T. Mayombei and review of the literature. BMC Infect. Dis. 2016, 16, 529. [Google Scholar] [CrossRef]
- Guo, G.; Wu, Y.; Liu, Y.; Wang, Z.; Xu, G.; Wang, X.; Liang, F.; Lai, W.; Xiao, X.; Zhu, Q.; et al. Exploring the causal effects of the gut microbiome on serum lipid levels: A two-sample Mendelian randomization analysis. Front. Microbiol. 2023, 14, 1113334. [Google Scholar] [CrossRef]
- Jones, J.; Reinke, S.N.; Ali, A.; Palmer, D.J.; Christophersen, C.T. Fecal sample collection methods and time of day impact microbiome composition and short chain fatty acid concentrations. Sci. Rep. 2021, 11, 13964. [Google Scholar] [CrossRef] [PubMed]
- Taft, D.H.; Ambalavanan, N.; Schibler, K.R.; Yu, Z.; Newburg, D.S.; Ward, D.V.; Morrow, A.L. Intestinal microbiota of preterm infants differ over time and between hospitals. Microbiome 2014, 2, 36. [Google Scholar] [CrossRef] [PubMed]


| Phyla | Red Fillet Group | White Fillet Group | Total |
|---|---|---|---|
| Firmicutes | 22.73 | 23.73 | 46.46 |
| Fusobacteria | 16.31 | 6.52 | 22.83 |
| Proteobacteria | 2.41 | 2.71 | 5.12 |
| Tenericutes | 7.58 | 15.10 | 22.68 |
| Others | 1.10 | 1.81 | 2.91 |
| Genera | |||
| Enterococcus | 4.81 | 0.35 | 5.17 |
| Lactobacillus | 2.71 | 3.61 | 6.32 |
| Peptostreptococcus | 1.71 | 6.32 | 8.02 |
| Romboutsia | 4.86 | 0.40 | 5.27 |
| Cetobacterium | 16.30 | 6.52 | 22.82 |
| Mycoplasma | 7.57 | 15.10 | 22.67 |
| Others | 12.19 | 17.55 | 29.74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, R.O.; Ali, A.; Leeds, T.; Salem, M. Fecal Microbiome Analysis Distinguishes Bacterial Taxa Biomarkers Associated with Red Fillet Color in Rainbow Trout. Microorganisms 2023, 11, 2704. https://doi.org/10.3390/microorganisms11112704
Ahmed RO, Ali A, Leeds T, Salem M. Fecal Microbiome Analysis Distinguishes Bacterial Taxa Biomarkers Associated with Red Fillet Color in Rainbow Trout. Microorganisms. 2023; 11(11):2704. https://doi.org/10.3390/microorganisms11112704
Chicago/Turabian StyleAhmed, Ridwan O., Ali Ali, Tim Leeds, and Mohamed Salem. 2023. "Fecal Microbiome Analysis Distinguishes Bacterial Taxa Biomarkers Associated with Red Fillet Color in Rainbow Trout" Microorganisms 11, no. 11: 2704. https://doi.org/10.3390/microorganisms11112704
APA StyleAhmed, R. O., Ali, A., Leeds, T., & Salem, M. (2023). Fecal Microbiome Analysis Distinguishes Bacterial Taxa Biomarkers Associated with Red Fillet Color in Rainbow Trout. Microorganisms, 11(11), 2704. https://doi.org/10.3390/microorganisms11112704






