Antimicrobial Resistance in Bacteria from Meat and Meat Products: A One Health Perspective
Abstract
:1. Introduction
2. The One Health Approach
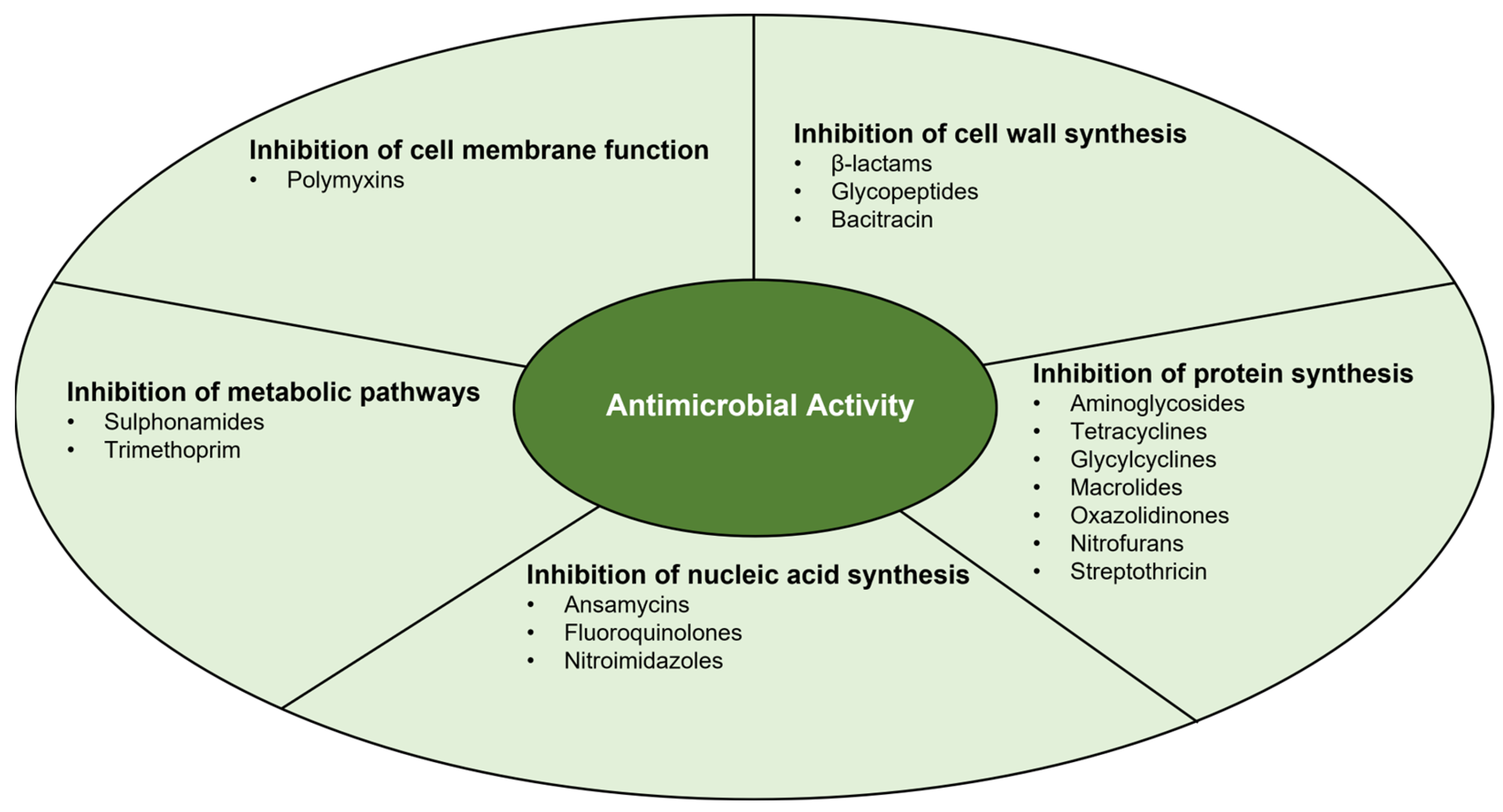
3. Antimicrobial Activity
3.1. Inhibition of Cell Wall Synthesis
3.2. Inhibition of Protein Synthesis
3.3. Inhibition of Nucleic Acid Synthesis
3.4. Inhibition of Metabolic Pathways
3.5. Inhibition of Cell Membrane Function
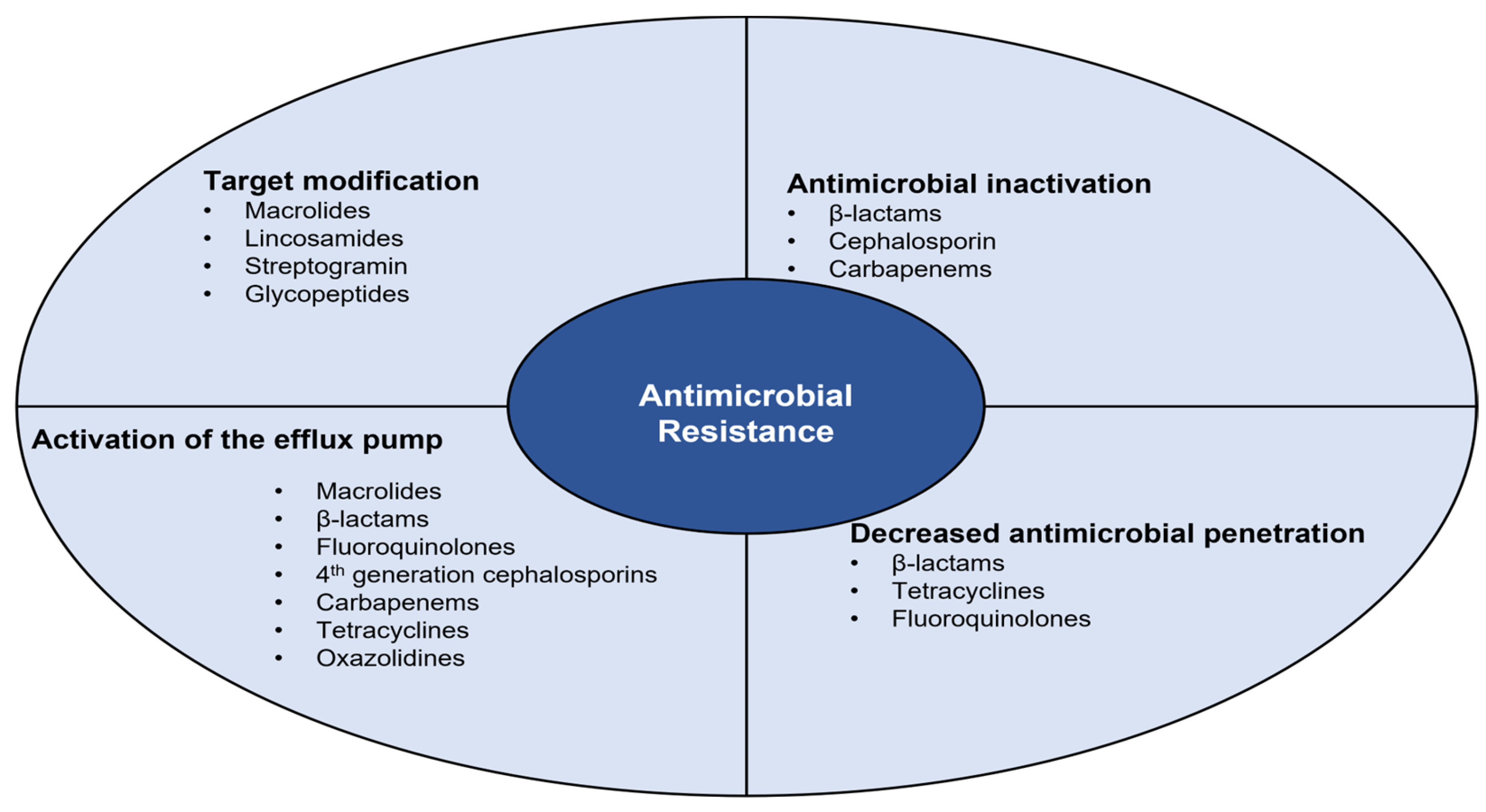
4. Antimicrobial Resistance
4.1. Antimicrobial Inactivation
4.2. Decreased Antimicrobial Penetration
4.3. Activation of the Efflux Pump
4.4. Target Modification
5. Antimicrobial Resistance and Farm-to-Fork Transmission
5.1. The Role of Meat in the Transmission of Antimicrobial Resistance
5.2. Antimicrobial Resistance in Staphylococcus aureus
5.3. Antimicrobial Resistance in ESBL-Producing Enterobacteriaceae
5.4. Antimicrobial Resistance in Vancomycin-Resistant Enterococcus spp.
5.5. Antimicrobial Resistance in ESKAPE Bacteria
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Scott, H.M.; Acuff, G.; Bergeron, G.; Bourassa, M.W.; Gill, J.; Graham, D.W.; Kahn, L.H.; Morley, P.S.; Salois, M.J.; Simjee, S.; et al. Critically important antibiotics: Criteria and approaches for measuring and reducing their use in food animal agriculture. Ann. N. Y. Acad. Sci. 2019, 1441, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Gargiullo, L.; Del Chierico, F.; D’Argenio, P.; Putignani, L. Gut microbiota modulation for multidrug-resistant organism decolonization: Present and future perspectives. Front. Microbiol. 2019, 10, 1704. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Zhang, Z.; Wu, Q.; Huang, Z.; Malakar, P.K.; Chen, L.; Liu, H.; Pan, Y.; Zhao, Y. Antibacterial peptides from seafood: A promising weapon to combat bacterial hazards in food. Food Control 2021, 125, 108004. [Google Scholar] [CrossRef]
- Queiroga, C.; Andrade, N.; Laranjo, M. Antimicrobial action of propolis extracts against staphylococci. In Understanding Microbial Pathogens Current Knowledge and Educational Ideas on Antimicrobial Research; Formatex Research Center: Badajoz, Spain, 2018; pp. 1–8. [Google Scholar]
- Queiroga, M.C.; Laranjo, M.; Andrade, N.; Marques, M.; Costa, A.R.; Antunes, C.M. Antimicrobial, Antibiofilm and Toxicological Assessment of Propolis. Antibiotics 2023, 12, 347. [Google Scholar] [CrossRef]
- Queiroga, M.C.; Pinto Coelho, M.; Arantes, S.M.; Potes, M.E.; Martins, M.R. Antimicrobial Activity of Essential Oils of Lamiaceae Aromatic Spices Towards Sheep mastitis-Causing Staphylococcus aureus and Staphylococcus epidermidis. J. Essent. Oil Bear. Plants 2018, 21, 1155–1165. [Google Scholar] [CrossRef]
- Laranjo, M.; Fernández-León, A.M.; Agulheiro-Santos, A.C.; Potes, M.E.; Elias, M. Essential oils of aromatic and medicinal plants play a role in food safety. J. Food Process. Preserv. 2019, 46, e14278. [Google Scholar] [CrossRef]
- Mohammadi Gheisar, M.; Kim, I.H. Phytobiotics in poultry and swine nutrition—A review. Ital. J. Anim. Sci. 2017, 17, 92–99. [Google Scholar] [CrossRef]
- Dellit, T.H.; Owens, R.C.; McGowan, J.E.; Gerding, D.N.; Weinstein, R.A.; Burke, J.P.; Huskins, W.C.; Paterson, D.L.; Fishman, N.O.; Carpenter, C.F.; et al. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America Guidelines for Developing an Institutional Program to Enhance Antimicrobial Stewardship. Clin. Infect. Dis. 2007, 44, 159–177. [Google Scholar] [CrossRef]
- Sudatip, D.; Chasiri, K.; Kritiyakan, A.; Phanprasit, W.; Thinphovong, C.; Tiengrim, S.; Thamlikitkul, V.; Abdallah, R.; Baron, S.A.; Rolain, J.M.; et al. A One Health approach to assessing occupational exposure to antimicrobial resistance in Thailand: The FarmResist project. PLoS ONE 2021, 16, e0245250. [Google Scholar] [CrossRef]
- Nations, U. Transforming Our World: The 2030 Agenda for Sustainable Development. 2015. Available online: https://sdgs.un.org/publications/transforming-our-world-2030-agenda-sustainable-development-17981 (accessed on 13 July 2023).
- Huan, Y.; Yu, Y.; Liang, T.; Burgman, M. A method for assessing the impacts of an international agreement on regional progress towards Sustainable Development Goals. Sci. Total Environ. 2021, 785, 147336. [Google Scholar] [CrossRef]
- Khaled, R.; Ali, H.; Mohamed, E.K.A. The Sustainable Development Goals and corporate sustainability performance: Mapping, extent and determinants. J. Clean. Prod. 2021, 311, 127599. [Google Scholar] [CrossRef]
- Pohlmann, C.R.; Scavarda, A.J.; Alves, M.B.; Korzenowski, A.L. The role of the focal company in sustainable development goals: A Brazilian food poultry supply chain case study. J. Clean. Prod. 2020, 245, 118798. [Google Scholar] [CrossRef]
- Hou, J.; Long, X.; Wang, X.; Li, L.; Mao, D.; Luo, Y.; Ren, H. Global trend of antimicrobial resistance in common bacterial pathogens in response to antibiotic consumption. J. Hazard. Mater. 2023, 442, 130042. [Google Scholar] [CrossRef]
- Michael, A.; Kelman, T.; Pitesky, M. Overview of quantitative methodologies to understand antimicrobial resistance via minimum inhibitory concentration. Animals 2020, 10, 1405. [Google Scholar] [CrossRef] [PubMed]
- Mouiche, M.M.M.; Moffo, F.; Akoachere, J.F.T.K.; Okah-Nnane, N.H.; Mapiefou, N.P.; Ndze, V.N.; Wade, A.; Djuikwo-Teukeng, F.F.; Toghoua, D.G.T.; Zambou, H.R.; et al. Antimicrobial resistance from a one health perspective in Cameroon: A systematic review and meta-analysis. BMC Public Health 2019, 19, 1135. [Google Scholar] [CrossRef]
- Bordier, M.; Binot, A.; Pauchard, Q.; Nguyen, D.T.; Trung, T.N.; Fortané, N.; Goutard, F.L. Antibiotic resistance in Vietnam: Moving towards a One Health surveillance system. BMC Public Health 2018, 18, 1136. [Google Scholar] [CrossRef]
- Sun, Q.; Wang, Y.; Hulth, A.; Xiao, Y.; Nilsson, L.E.; Li, X.; Bi, Z.; Liu, Y.; Yin, H.; Luo, Y.; et al. Study protocol for One Health data collections, analyses and intervention of the Sino-Swedish integrated multisectoral partnership for antibiotic resistance containment (IMPACT). BMJ Open 2018, 8, e017832. [Google Scholar] [CrossRef]
- Collineau, L.; Boerlin, P.; Carson, C.A.; Chapman, B.; Fazil, A.; Hetman, B.; McEwen, S.A.; Jane Parmley, E.; Reid-Smith, R.J.; Taboada, E.N.; et al. Integrating whole-genome sequencing data into quantitative risk assessment of foodborne antimicrobial resistance: A review of opportunities and challenges. Front. Microbiol. 2019, 10, 1107. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, F.; Mercanoglu Taban, B. A State-of-Art Review on Multi-Drug Resistant Pathogens in Foods of Animal Origin: Risk Factors and Mitigation Strategies. Front. Microbiol. 2019, 10, 2091. [Google Scholar] [CrossRef]
- Zhuo, A.; Labbate, M.; Norris, J.M.; Gilbert, G.L.; Ward, M.P.; Bajorek, B.V.; Degeling, C.; Rowbotham, S.J.; Dawson, A.; Nguyen, K.A.; et al. Opportunities and challenges to improving antibiotic prescribing practices through a One Health approach: Results of a comparative survey of doctors, dentists and veterinarians in Australia. BMJ Open 2018, 8, e020439. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Cui, X.; Xu, H.; Liu, W.; Tao, F.; Shao, T.; Pan, X.; Zheng, B. Whole genome sequencing of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from a wastewater treatment plant in China. Front. Microbiol. 2019, 10, 1797. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Andersen, P.S.; Stegger, M.; Sieber, R.N.; Ingmer, H.; Staubrand, N.; Dalsgaard, A.; Leisner, J.J. Antimicrobial Resistance and Virulence Gene Profiles of Methicillin-Resistant and -Susceptible Staphylococcus aureus From Food Products in Denmark. Front. Microbiol. 2019, 10, 2681. [Google Scholar] [CrossRef]
- Wen, Z.; Shang, Y.; Xu, G.; Pu, Z.; Lin, Z.; Bai, B.; Chen, Z.; Zheng, J.; Deng, Q.; Yu, Z. Mechanism of Eravacycline Resistance in Clinical Enterococcus faecalis Isolates from China. Front. Microbiol. 2020, 11, 916. [Google Scholar] [CrossRef] [PubMed]
- Cieślik, M.; Bagińska, N.; Górski, A.; Jończyk-Matysiak, E. Animal models in the evaluation of the effectiveness of phage therapy for infections caused by gram-negative bacteria from the ESKAPE group and the reliability of its use in humans. Microorganisms 2021, 9, 206. [Google Scholar] [CrossRef] [PubMed]
- Collis, R.M.; Burgess, S.A.; Biggs, P.J.; Midwinter, A.C.; French, N.P.; Toombs-Ruane, L.; Cookson, A.L. Extended-spectrum beta-lactamase-producing enterobacteriaceae in dairy farm environments: A New Zealand perspective. Foodborne Pathog. Dis. 2019, 16, 5–22. [Google Scholar] [CrossRef]
- Igbinosa, E.O.; Beshiru, A. Antimicrobial resistance, virulence determinants, and biofilm formation of Enterococcus species from ready-to-eat seafood. Front. Microbiol. 2019, 10, 728. [Google Scholar] [CrossRef]
- Lopes, E.; Conceição, T.; Poirel, L.; de Lencastre, H.; Aires-De-Sousa, M. Epidemiology and antimicrobial resistance of methicillin-resistant Staphylococcus aureus isolates colonizing pigs with different exposure to antibiotics. PLoS ONE 2019, 14, e0225497. [Google Scholar] [CrossRef]
- Aaliya, B.; Valiyapeediyekkal Sunooj, K.; Navaf, M.; Parambil Akhila, P.; Sudheesh, C.; Ahmed Mir, S.; Sabu, S.; Sasidharan, A.; Theingi Hlaing, M.; George, J. Recent trends in bacterial decontamination of food products by hurdle technology: A synergistic approach using thermal and non-thermal processing techniques. Food Res. Int. 2021, 147, 110514. [Google Scholar] [CrossRef]
- Cooper, A.L.; Low, A.J.; Koziol, A.G.; Thomas, M.C.; Leclair, D.; Tamber, S.; Wong, A.; Blais, B.W.; Carrillo, C.D. Systematic Evaluation of Whole Genome Sequence-Based Predictions of Salmonella Serotype and Antimicrobial Resistance. Front. Microbiol. 2020, 11, 549. [Google Scholar] [CrossRef]
- Thapa, S.P.; Shrestha, S.; Anal, A.K. Addressing the antibiotic resistance and improving the food safety in food supply chain (farm-to-fork) in Southeast Asia. Food Control 2020, 108, 106809. [Google Scholar] [CrossRef]
- Adisasmito, W.B.; Almuhairi, S.; Behravesh, C.B.; Bilivogui, P.; Bukachi, S.A.; Casas, N.; Becerra, N.C.; Charron, D.F.; Chaudhary, A.; Ciacci Zanella, J.R.; et al. One Health: A new definition for a sustainable and healthy future. PLoS Pathog. 2022, 18, e1010537. [Google Scholar] [CrossRef]
- What Is One Health? Available online: https://www.onehealthcommission.org/en/why_one_health/what_is_one_health/ (accessed on 6 June 2023).
- Mitchell, M.E.V.; Alders, R.; Unger, F.; Nguyen-Viet, H.; Le, T.T.H.; Toribio, J.A. The challenges of investigating antimicrobial resistance in Vietnam—What benefits does a One Health approach offer the animal and human health sectors? BMC Public Health 2020, 20, 213. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Coque, T.M.; Martínez, J.L.; Aracil-Gisbert, S.; Lanza, V.F. Gene Transmission in the One Health Microbiosphere and the Channels of Antimicrobial Resistance. Front. Microbiol. 2019, 10, 2892. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Wu, Z.; Zhu, Y.; Liu, C. One Health: A holistic approach for food safety in livestock. Sci. One Health 2022, 1, 100015. [Google Scholar] [CrossRef]
- Netea, M.G.; Dominguez-Andres, J.; Barreiro, L.B.; Chavakis, T.; Divangahi, M.; Fuchs, E.; Joosten, L.A.B.; van der Meer, J.W.M.; Mhlanga, M.M.; Mulder, W.J.M.; et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020, 20, 375–388. [Google Scholar] [CrossRef]
- Byrne, K.A.; Tuggle, C.K.; Loving, C.L. Differential induction of innate memory in porcine monocytes by beta-glucan or bacillus Calmette-Guerin. Innate Immun. 2021, 27, 448–460. [Google Scholar] [CrossRef]
- Campos, M.; Capilla, R.; Naya, F.; Futami, R.; Coque, T.; Moya, A.; Fernandez-Lanza, V.; Cantón, R.; Sempere, J.M.; Llorens, C.; et al. Simulating Multilevel Dynamics of Antimicrobial Resistance in a Membrane Computing Model. mBio 2019, 10, e02460-18. [Google Scholar] [CrossRef]
- Bizzaro, G.; Vatland, A.K.; Pampanin, D.M. The One-Health approach in seaweed food production. Environ. Int. 2022, 158, 106948. [Google Scholar] [CrossRef]
- Jeleff, M.; Lehner, L.; Giles-Vernick, T.; Dückers, M.L.A.; Napier, D.; Jirovsky-Platter, E.; Kutalek, R. Vulnerability and One Health assessment approaches for infectious threats from a social science perspective: A systematic scoping review. Lancet Planet. Health 2022, 6, e682–e693. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.M.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Pulingam, T.; Parumasivam, T.; Gazzali, A.M.; Sulaiman, A.M.; Chee, J.Y.; Lakshmanan, M.; Chin, C.F.; Sudesh, K. Antimicrobial resistance: Prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur. J. Pharm. Sci. 2022, 170, 106103. [Google Scholar] [CrossRef] [PubMed]
- Upmanyu, N.; Malviya, V.N. Antibiotics: Mechanisms of action and modern challenges. In Microorganisms for Sustainable Environment and Health; Elsevier: Amsterdam, The Netherlands, 2020; pp. 367–382. [Google Scholar]
- Singh, S.P.; Qureshi, A.; Hassan, W. Mechanisms of action by antimicrobial agents: A review. McGill J. Med. 2021, 19. [Google Scholar] [CrossRef]
- Lin, J.; Zhou, D.; Steitz, T.A.; Polikanov, Y.S.; Gagnon, M.G. Ribosome-Targeting Antibiotics: Modes of Action, Mechanisms of Resistance, and Implications for Drug Design. Annu. Rev. Biochem. 2018, 87, 451–478. [Google Scholar] [CrossRef]
- Johnston, N.; Mukhtar, T.; Wright, G. Streptogramin Antibiotics: Mode of Action and Resistance. Curr. Drug Targets 2002, 3, 335–344. [Google Scholar] [CrossRef]
- Zuckerman, J.M.; Qamar, F.; Bono, B.R. Review of Macrolides (Azithromycin, Clarithromycin), Ketolids (Telithromycin) and Glycylcyclines (Tigecycline). Med. Clin. N. Am. 2011, 95, 761–791. [Google Scholar] [CrossRef]
- Wijma, R.A.; Huttner, A.; Koch, B.C.P.; Mouton, J.W.; Muller, A.E. Review of the pharmacokinetic properties of nitrofurantoin and nitroxoline. J. Antimicrob. Chemother. 2018, 73, 2916–2926. [Google Scholar] [CrossRef]
- Vallée, M.; Harding, C.; Hall, J.; Aldridge, P.D.; Tan, A. Exploring the in situ evolution of nitrofurantoin resistance in clinically derived uropathogenic Escherichia coli isolates. J. Antimicrob. Chemother. 2022, 78, 373–379. [Google Scholar] [CrossRef]
- Morgan, C.E.; Kang, Y.-S.; Green, A.B.; Smith, K.P.; Dowgiallo, M.G.; Miller, B.C.; Chiaraviglio, L.; Truelson, K.A.; Zulauf, K.E.; Rodriguez, S.; et al. Streptothricin F is a bactericidal antibiotic effective against highly drug-resistant gram-negative bacteria that interacts with the 30S subunit of the 70S ribosome. PLoS Biol. 2023, 21, e3002091. [Google Scholar] [CrossRef]
- Edwards, D.I. Nitroimidazole drugs-action and resistance mechanisms I. Mechanisms of action. J. Antimicrob. Chemother. 1993, 31, 9–20. [Google Scholar] [CrossRef]
- De Lucia, A.; Card, R.M.; Duggett, N.; Smith, R.P.; Davies, R.; Cawthraw, S.A.; Anjum, M.F.; Rambaldi, M.; Ostanello, F.; Martelli, F. Reduction in antimicrobial resistance prevalence in Escherichia coli from a pig farm following withdrawal of group antimicrobial treatment. Vet. Microbiol. 2021, 258, 109125. [Google Scholar] [CrossRef] [PubMed]
- Mellor, K.C.; Petrovska, L.; Thomson, N.R.; Harris, K.; Reid, S.W.J.; Mather, A.E. Antimicrobial resistance diversity suggestive of distinct salmonella typhimurium sources or selective pressures in food-production animals. Front. Microbiol. 2019, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- Tsigalou, C.; Konstantinidis, T.; Stavropoulou, E.; Bezirtzoglou, E.E.; Tsakris, A. Potential Elimination of Human Gut Resistome by Exploiting the Benefits of Functional Foods. Front. Microbiol. 2020, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Rafailidis, P.I.; Kofteridis, D. Proposed amendments regarding the definitions of multidrug-resistant and extensively drug-resistant bacteria. Expert Rev. Anti-Infect. Ther. 2021, 20, 139–146. [Google Scholar] [CrossRef]
- Hickman, R.A.; Leangapichart, T.; Lunha, K.; Jiwakanon, J.; Angkititrakul, S.; Magnusson, U.; Sunde, M.; Järhult, J.D. Exploring the Antibiotic Resistance Burden in Livestock, Livestock Handlers and Their Non-Livestock Handling Contacts: A One Health Perspective. Front. Microbiol. 2021, 12, 651461. [Google Scholar] [CrossRef]
- Lai, C.K.C.; Ng, R.W.Y.; Leung, S.S.Y.; Hui, M.; Ip, M. Overcoming the rising incidence and evolving mechanisms of antibiotic resistance by novel drug delivery approaches—An overview. Adv. Drug Deliv. Rev. 2022, 181, 114078. [Google Scholar] [CrossRef]
- Li, L.; Xiao, Y.; Wang, C.; Olsen, R.H.; Meng, H.; Shi, L. Exploring the resistome, virulome, mobilome and microbiome along pork production chain using metagenomics. Int. J. Food Microbiol. 2022, 371, 109674. [Google Scholar] [CrossRef]
- Jans, C.; Sarno, E.; Collineau, L.; Meile, L.; Stärk, K.D.C.; Stephan, R. Consumer exposure to antimicrobial resistant bacteria from food at Swiss retail level. Front. Microbiol. 2018, 9, 362. [Google Scholar] [CrossRef]
- Jiang, Z.; Anwar, T.M.; Peng, X.; Biswas, S.; Elbediwi, M.; Li, Y.; Fang, W.; Yue, M. Prevalence and antimicrobial resistance of Salmonella recovered from pig-borne food products in Henan, China. Food Control 2021, 121, 107535. [Google Scholar] [CrossRef]
- Reid, C.J.; Blau, K.; Jechalke, S.; Smalla, K.; Djordjevic, S.P. Whole Genome Sequencing of Escherichia coli From Store-Bought Produce. Front. Microbiol. 2020, 10, 3050. [Google Scholar] [CrossRef]
- Kanokudom, S.; Assawakongkarat, T.; Akeda, Y.; Ratthawongjirakul, P.; Chuanchuen, R.; Chaichanawongsaroj, N. Rapid detection of extended spectrum β-lactamase producing Escherichia coli isolated from fresh pork meat and pig cecum samples using multiplex recombinase polymerase amplification and lateral flow strip analysis. PLoS ONE 2021, 16, e0248536. [Google Scholar] [CrossRef] [PubMed]
- Ferrocino, I.; Rantsiou, K.; McClure, R.; Kostic, T.; de Souza, R.S.C.; Lange, L.; FitzGerald, J.; Kriaa, A.; Cotter, P.; Maguin, E.; et al. The need for an integrated multi-OMICs approach in microbiome science in the food system. Compr. Rev. Food Sci. Food Saf. 2023, 22, 1082–1103. [Google Scholar] [CrossRef] [PubMed]
- Farm to Fork Strategy. Available online: https://food.ec.europa.eu/horizontal-topics/farm-fork-strategy_en (accessed on 6 June 2023).
- Pennone, V.; Cobo-Díaz, J.F.; Prieto, M.; Alvarez-Ordóñez, A. Application of genomics and metagenomics to improve food safety based on an enhanced characterisation of antimicrobial resistance. Curr. Opin. Food Sci. 2022, 43, 183–188. [Google Scholar] [CrossRef]
- Rocchetti, G.; Ferronato, G.; Sarv, V.; Kerner, K.; Venskutonis, P.R.; Lucini, L. Meat extenders from different sources as protein-rich alternatives to improve the technological properties and functional quality of meat products. Curr. Opin. Food Sci. 2023, 49, 100967. [Google Scholar] [CrossRef]
- Lu, N.; Ma, J.; Sun, D.W. Enhancing physical and chemical quality attributes of frozen meat and meat products: Mechanisms, techniques and applications. Trends Food Sci. Technol. 2022, 124, 63–85. [Google Scholar] [CrossRef]
- de Araújo, P.D.; Araújo, W.M.C.; Patarata, L.; Fraqueza, M.J. Understanding the main factors that influence consumer quality perception and attitude towards meat and processed meat products. Meat Sci. 2022, 193, 108952. [Google Scholar] [CrossRef]
- Van Reckem, E.; De Vuyst, L.; Weckx, S.; Leroy, F. Next-generation sequencing to enhance the taxonomic resolution of the microbiological analysis of meat and meat-derived products. Curr. Opin. Food Sci. 2021, 37, 58–65. [Google Scholar] [CrossRef]
- Chaudhary, P.; Kumar, Y. Recent advances in multiplex molecular techniques for meat species identification. J. Food Compos. Anal. 2022, 110, 104581. [Google Scholar] [CrossRef]
- World Consumption of Meat. Available online: https://www.theworldcounts.com/challenges/foods-and-beverages/world-consumption-of-meat (accessed on 6 June 2023).
- Brinck, J.E.; Lassen, S.B.; Forouzandeh, A.; Pan, T.; Wang, Y.-Z.; Monteiro, A.; Blavi, L.; Solà-Oriol, D.; Stein, H.H.; Su, J.-Q.; et al. Impacts of dietary copper on the swine gut microbiome and antibiotic resistome. Sci. Total Environ. 2023, 857, 159609. [Google Scholar] [CrossRef]
- Li, X.; Rensing, C.; Vestergaard, G.; Arumugam, M.; Nesme, J.; Gupta, S.; Brejnrod, A.D.; Sørensen, S.J. Metagenomic evidence for co-occurrence of antibiotic, biocide and metal resistance genes in pigs. Environ. Int. 2022, 158, 106899. [Google Scholar] [CrossRef]
- Luu, Q.H.; Nguyen, T.L.A.; Pham, T.N.; Vo, N.G.; Padungtod, P. Antimicrobial use in household, semi-industrialized, and industrialized pig and poultry farms in Viet Nam. Prev. Vet. Med. 2021, 189, 105292. [Google Scholar] [CrossRef] [PubMed]
- Pholwat, S.; Pongpan, T.; Chinli, R.; Rogawski McQuade, E.T.; Thaipisuttikul, I.; Ratanakorn, P.; Liu, J.; Taniuchi, M.; Houpt, E.R.; Foongladda, S. Antimicrobial Resistance in Swine Fecal Specimens Across Different Farm Management Systems. Front. Microbiol. 2020, 11, 1238. [Google Scholar] [CrossRef] [PubMed]
- Sirichokchatchawan, W.; Apiwatsiri, P.; Pupa, P.; Saenkankam, I.; Khine, N.O.; Lekagul, A.; Lugsomya, K.; Hampson, D.J.; Prapasarakul, N. Reducing the Risk of Transmission of Critical Antimicrobial Resistance Determinants from Contaminated Pork Products to Humans in South-East Asia. Front. Microbiol. 2021, 12, 689015. [Google Scholar] [CrossRef]
- Wang, Y.; Sutton, N.B.; Zheng, Y.; Dong, H.; Rijnaarts, H.H.M. Seasonal variation in antibiotic resistance genes and bacterial phenotypes in swine wastewater during three-chamber anaerobic pond treatment. Environ. Res. 2023, 216, 114495. [Google Scholar] [CrossRef] [PubMed]
- Calero, G.C.; Gomez, N.C.; Benomar, N.; Montoro, B.P.; Knapp, C.W.; Galvez, A.; Abriouel, H. Deciphering Resistome and Virulome Diversity in a Porcine Slaughterhouse and Pork Products Through Its Production Chain. Front. Microbiol. 2018, 9, 2099. [Google Scholar] [CrossRef]
- Oswaldi, V.; Luth, S.; Dzierzon, J.; Meemken, D.; Schwarz, S.; Fessler, A.T.; Felix, B.; Langforth, S. Distribution and Characteristics of Listeria spp. in Pigs and Pork Production Chains in Germany. Microorganisms 2022, 10, 512. [Google Scholar] [CrossRef]
- Díaz-Jiménez, D.; García-Meniño, I.; Herrera, A.; Lestón, L.; Mora, A. Microbiological risk assessment of Turkey and chicken meat for consumer: Significant differences regarding multidrug resistance, mcr or presence of hybrid aEPEC/ExPEC pathotypes of E. coli. Food Control 2021, 123, 107713. [Google Scholar] [CrossRef]
- Das, T.; Paino, D.; Manoharan, A.; Farrell, J.; Whiteley, G.; Kriel, F.H.; Glasbey, T.; Manos, J. Conditions under which glutathione disrupts the biofilms and improves antibiotic efficacy of both ESKAPE and NON-ESKAPE species. Front. Microbiol. 2019, 10, 2000. [Google Scholar] [CrossRef]
- Zohra, T.; Numan, M.; Ikram, A.; Salman, M.; Khan, T.; Din, M.; Salman, M.; Farooq, A.; Amir, A.; Ali, M. Cracking the Challenge of Antimicrobial Drug Resistance with CRISPR/Cas9, Nanotechnology and Other Strategies in ESKAPE Pathogens. Microorganisms 2021, 9, 954. [Google Scholar] [CrossRef]
- Klimienė, I.; Virgailis, M.; Kerzienė, S.; Šiugždinienė, R.; Mockeliūnas, R.; Ružauskas, M. Evaluation of genotypical antimicrobial resistance in ESBL producing Escherichia coli phylogenetic groups isolated from retail poultry meat. J. Food Saf. 2018, 38, e12370. [Google Scholar] [CrossRef]
- Correia Santos, S.; Fraqueza, M.J.; Elias, M.; Salvador Barreto, A.; Semedo-Lemsaddek, T. Traditional dry smoked fermented meat sausages: Characterization of autochthonous enterococci. LWT-Food Sci. Technol. 2017, 79, 410–415. [Google Scholar] [CrossRef]
- Yu, T.; Jiang, X.; Liang, Y.; Zhu, Y.; Tian, J.; Ying, H.; Wang, X.; Shi, L. Characterization and horizontal transfer of antimicrobial resistance genes and integrons in bacteria isolated from cooked meat products in China. J. Food Prot. 2017, 80, 2048–2055. [Google Scholar] [CrossRef] [PubMed]
- Bouchami, O.; Fraqueza, M.J.; Faria, N.A.; Alves, V.; Lawal, O.U.; de Lencastre, H.; Miragaia, M. Evidence for the dissemination to humans of methicillin-resistant Staphylococcus aureus ST398 through the pork production chain: A study in a portuguese slaughterhouse. Microorganisms 2020, 8, 1892. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Bougouffa, S.; Park, T.-J.; Lau, A.; Tong, M.-K.; Chow, K.-H.; Ho, P.-L. Sharing of Antimicrobial Resistance Genes between Humans and Food Animals. mSystems 2022, 7, e00775-22. [Google Scholar] [CrossRef]
- Lauteri, C.; Festino, A.R.; Conter, M.; Vergara, A. Prevalence and antimicrobial resistance profile in Salmonella spp. isolates from swine food chain. Ital. J. Food Saf. 2022, 11, 9980. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Chan, E.W.-C.; Chen, S. Complete Genetic Analysis of Plasmids Carrying Multiple Resistance, Virulence, and Phage-Like Genes in Foodborne Escherichia coli Isolate. Microbiol. Spectr. 2023, 11, e02820-22. [Google Scholar] [CrossRef]
- Sin, M.; Yoon, S.; Kim, Y.B.; Noh, E.B.; Seo, K.W.; Lee, Y.J. Molecular characteristics of antimicrobial resistance determinants and integrons in Salmonella isolated from chicken meat in Korea. J. Appl. Poult. Res. 2020, 29, 502–514. [Google Scholar] [CrossRef]
- Wang, W.; Chen, J.; Shao, X.; Huang, P.; Zha, J.; Ye, Y. Occurrence and antimicrobial resistance of Salmonella isolated from retail meats in Anhui, China. Food Sci. Nutr. 2021, 9, 4701–4710. [Google Scholar] [CrossRef]
- Wang, Y.T.; Lin, Y.T.; Wan, T.W.; Wang, D.Y.; Lin, H.Y.; Lin, C.Y.; Chen, Y.C.; Teng, L.J. Distribution of antibiotic resistance genes among Staphylococcus species isolated from ready-to-eat foods. J. Food Drug Anal. 2019, 27, 841–848. [Google Scholar] [CrossRef]
- Guerrero-Ramos, E.; Molina-González, D.; Blanco-Morán, S.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C.; Capita, R. Prevalence, antimicrobial resistance, and genotypic characterization of vancomycin-resistant enterococci in meat preparations. J. Food Prot. 2016, 79, 748–756. [Google Scholar] [CrossRef]
- Belloso Daza, M.V.; Milani, G.; Cortimiglia, C.; Pietta, E.; Bassi, D.; Cocconcelli, P.S. Genomic Insights of Enterococcus faecium UC7251, a Multi-Drug Resistant Strain from Ready-to-Eat Food, Highlight the Risk of Antimicrobial Resistance in the Food Chain. Front. Microbiol. 2022, 13, 894241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fu, Y.; Xiong, Z.; Ma, Y.; Wei, Y.; Qu, X.; Zhang, H.; Zhang, J.; Liao, M. Highly Prevalent Multidrug-Resistant Salmonella from Chicken and Pork Meat at Retail Markets in Guangdong, China. Front. Microbiol. 2018, 9, 2104. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wei, X.; Fan, M. Assessment of Antibiotic Susceptibility within Lactic Acid Bacteria and Coagulase-Negative Staphylococci Isolated from Hunan Smoked Pork, a Naturally Fermented Meat Product in China. J. Food Sci. 2018, 83, 1707–1715. [Google Scholar] [CrossRef] [PubMed]
- Heo, E.J.; Ko, E.K.; Kang, H.J.; Kim, Y.J.; Park, H.J.; Wee, S.H.; Moon, J.S. Prevalence and Antimicrobial Characteristics of Shiga Toxin-Producing Escherichia coli Isolates from Pork in Korea. Foodborne Pathog. Dis. 2020, 17, 602–607. [Google Scholar] [CrossRef]
- Li, L.; Heidemann Olsen, R.; Ye, L.; Yan, H.; Nie, Q.; Meng, H.; Shi, L. Antimicrobial Resistance and Resistance Genes in Aerobic Bacteria Isolated from Pork at Slaughter. J. Food Prot. 2016, 79, 589–597. [Google Scholar] [CrossRef]
- Liu, Z.; Klümper, U.; Shi, L.; Ye, L.; Li, M. From pig breeding environment to subsequently produced pork: Comparative analysis of antibiotic resistance genes and bacterial community composition. Front. Microbiol. 2019, 10, 43. [Google Scholar] [CrossRef]
- Li, L.; Ye, L.; Yu, L.; Zhou, C.; Meng, H. Characterization of Extended Spectrum Beta-Lactamase Producing Enterobacteria and Methicillin-Resistant Staphylococcus aureus Isolated from Raw Pork and Cooked Pork Products in South China. J. Food Sci. 2016, 81, M1773–M1777. [Google Scholar] [CrossRef]
- Bacci, C.; Lanzoni, E.; Vismarra, A.; Alpigiani, I.; Nuvoloni, R.; Bonardi, S.; Brindani, F. Antibiotic resistance and resistance genes in Salmonella enterica isolated from pork meat and pig carcasses in Northern Italy. Large Anim. Rev. 2014, 20, 201–207. [Google Scholar]
- Chajecka-Wierzchowska, W.; Zadernowska, A.; Nalepa, B.; Sierpinska, M.; Laniewska-Trokenheim, L. Coagulase-negative staphylococci (CoNS) isolated from ready-to-eat food of animal origin—Phenotypic and genotypic antibiotic resistance. Food Microbiol. 2015, 46, 222–226. [Google Scholar] [CrossRef]
- Lawal, O.U.; Fraqueza, M.J.; Bouchami, O.; Worning, P.; Bartels, M.D.; Gonçalves, M.L.; Paixao, P.; Gonçalves, E.; Toscano, C.; Empel, J.; et al. Foodborne origin and local and global spread of staphylococcus saprophyticus causing human urinary tract infections. Emerg. Infect. Dis. 2021, 27, 880–893. [Google Scholar] [CrossRef]
- Henriques, A.R.; Melo Cristino, J.; Fraqueza, M.J. Genetic characterization of listeria monocytogenes isolates from industrial and retail ready-to-eat meat-based foods and their relationship with clinical strains from human listeriosis in Portugal. J. Food Prot. 2017, 80, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Rolain, J.M. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front. Microbiol. 2013, 4, 173. [Google Scholar] [CrossRef] [PubMed]
- Stanton, I.C.; Bethel, A.; Leonard, A.F.C.; Gaze, W.H.; Garside, R. Existing evidence on antibiotic resistance exposure and transmission to humans from the environment: A systematic map. Environ. Evid. 2022, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Igrejas, G.; Correia, S.; Silva, V.; Hébraud, M.; Caniça, M.; Torres, C.; Gomes, C.; Nogueira, F.; Poeta, P. Planning a one health case study to evaluate Methicillin resistant Staphylococcus aureus and its economic burden in Portugal. Front. Microbiol. 2018, 9, 2964. [Google Scholar] [CrossRef]
- Neyaz, L.; Rajagopal, N.; Wells, H.; Fakhr, M.K. Molecular Characterization of Staphylococcus aureus Plasmids Associated with Strains Isolated From Various Retail Meats. Front. Microbiol. 2020, 11, 223. [Google Scholar] [CrossRef]
- Okorie-Kanu, O.J.; Anyanwu, M.U.; Ezenduka, E.V.; Mgbeahuruike, A.C.; Thapaliya, D.; Gerbig, G.; Ugwuijem, E.E.; Okorie-Kanu, C.O.; Agbowo, P.; Olorunleke, S.; et al. Molecular epidemiology, genetic diversity and antimicrobial resistance of Staphylococcus aureus isolated from chicken and pig carcasses, and carcass handlers. PLoS ONE 2020, 15, e0232913. [Google Scholar] [CrossRef]
- Şanlıbaba, P. Prevalence, antibiotic resistance, and enterotoxin production of Staphylococcus aureus isolated from retail raw beef, sheep, and lamb meat in Turkey. Int. J. Food Microbiol. 2022, 361, 109461. [Google Scholar] [CrossRef]
- Barros, E.M.; Martin, M.J.; Selleck, E.M.; Lebreton, F.; Sampaio, J.L.M.; Gilmore, M.S. Daptomycin Resistance and Tolerance Due to Loss of Function in Staphylococcus aureus dsp1 and asp23. Antimicrob. Agents Chemother. 2019, 63, e01542-18. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, J.; Lei, H.; Liang, H.; Li, X.; Li, B. The development of variation-based rifampicin resistance in Staphylococcus aureus deciphered through genomic and transcriptomic study. J. Hazard. Mater. 2023, 442, 130112. [Google Scholar] [CrossRef]
- Fraqueza, M.J.; Laranjo, M.; Elias, M.; Patarata, L. Microbiological hazards associated with salt and nitrite reduction in cured meat products: Control strategies based on antimicrobial effect of natural ingredients and protective microbiota. Curr. Opin. Food Sci. 2021, 38, 32–39. [Google Scholar] [CrossRef]
- Vestergaard, M.; Nøhr-Meldgaard, K.; Ingmer, H. Multiple pathways towards reduced membrane potential and concomitant reduction in aminoglycoside susceptibility in Staphylococcus aureus. Int. J. Antimicrob. Agents 2018, 51, 132–135. [Google Scholar] [CrossRef]
- Correia, S.; Silva, V.; García-Díez, J.; Teixeira, P.; Pimenta, K.; Pereira, J.E.; Oliveira, S.; Rocha, J.; Manaia, C.M.; Igrejas, G.; et al. One Health Approach Reveals the Absence of Methicillin-Resistant Staphylococcus aureus in Autochthonous Cattle and Their Environments. Front. Microbiol. 2019, 10, 2735. [Google Scholar] [CrossRef]
- Hau, S.J.; Haan, J.S.; Davies, P.R.; Frana, T.; Nicholson, T.L. Antimicrobial resistance distribution differs among methicillin resistant Staphylococcus aureus sequence type (ST) 5 isolates from health care and agricultural sources. Front. Microbiol. 2018, 9, 2102. [Google Scholar] [CrossRef] [PubMed]
- Watkins, R.R.; Holubar, M.; David, M.Z. Antimicrobial Resistance in Methicillin-Resistant Staphylococcus aureus to Newer Antimicrobial Agents. Antimicrob. Agents Chemother. 2019, 63, e01216-19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Qin, L.; Jin, C.; Ju, H.; Jiang, R.; Li, L.; Zhang, H.; Gao, W.; Wei, X.; Dong, H.; et al. Molecular Characteristics and Antibiotic Resistance of Staphylococcus aureus Isolated from Patient and Food Samples in Shijiazhuang, China. Pathogens 2022, 11, 1333. [Google Scholar] [CrossRef] [PubMed]
- Lv, G.; Jiang, R.; Zhang, H.; Wang, L.; Li, L.; Gao, W.; Zhang, H.; Pei, Y.; Wei, X.; Dong, H.; et al. Molecular Characteristics of Staphylococcus aureus From Food Samples and Food Poisoning Outbreaks in Shijiazhuang, China. Front. Microbiol. 2021, 12, 652276. [Google Scholar] [CrossRef]
- Bonardi, S.; Cabassi, C.S.; Manfreda, G.; Parisi, A.; Fiaccadori, E.; Sabatino, A.; Cavirani, S.; Bacci, C.; Rega, M.; Spadini, C.; et al. Survey on Carbapenem-Resistant Bacteria in Pigs at Slaughter and Comparison with Human Clinical Isolates in Italy. Antibiotics 2022, 11, 777. [Google Scholar] [CrossRef]
- Moosavian, M.; Emam, N. The first report of emerging mobilized colistin-resistance (Mcr) genes and ERIC-PCR typing in Escherichia coli and Klebsiella pneumoniae clinical isolates in southwest Iran. Infect. Drug Resist. 2019, 12, 1001–1010. [Google Scholar] [CrossRef]
- Teklu, D.S.; Negeri, A.A.; Legese, M.H.; Bedada, T.L.; Woldemariam, H.K.; Tullu, K.D. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control 2019, 8, 39. [Google Scholar] [CrossRef]
- Alizadeh, N.; Rezaee, M.A.; Kafil, H.S.; Hasani, A.; Barhaghi, M.H.S.; Milani, M.; Sefidan, F.Y.; Memar, M.Y.; Lalehzadeh, A.; Ghotaslo, R. Evaluation of Resistance Mechanisms in Carbapenem-Resistant Enterobacteriaceae. Infect. Drug Resist. 2020, 13, 1377–1385. [Google Scholar] [CrossRef]
- Castanheira, M.; Deshpande, L.M.; Mendes, R.E.; Canton, R.; Sader, H.S.; Jones, R.N. Variations in the Occurrence of Resistance Phenotypes and Carbapenemase Genes Among Enterobacteriaceae Isolates in 20 Years of the SENTRY Antimicrobial Surveillance Program. Open Forum Infect. Dis. 2019, 6, S23–S33. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Oh, S.S.; Kim, J.; Park, S.; Shin, J. Clinically Relevant Extended-Spectrum β-Lactamase–Producing Escherichia coli Isolates from Food Animals in South Korea. Front. Microbiol. 2020, 11, 604. [Google Scholar] [CrossRef]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 6 June 2023).
- Jiang, B.; Du, P.; Jia, P.; Liu, E.; Kudinha, T.; Zhang, H.; Li, D.; Xu, Y.; Xie, L.; Yang, Q. Antimicrobial Susceptibility and Virulence of mcr-1-Positive Enterobacteriaceae in China, a Multicenter Longitudinal Epidemiological Study. Front. Microbiol. 2020, 11, 1611. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, G.; Tanzarella, E.S.; Cutuli, S.L.; De Pascale, G. Treatment of severe infections caused by ESBL or carbapenemases-producing Enterobacteriaceae. Med. Intensiv. Engl. Ed. 2022, 47, 34–44. [Google Scholar] [CrossRef]
- Richter, L.; du Plessis, E.M.; Duvenage, S.; Korsten, L. Occurrence, Phenotypic and Molecular Characterization of Extended-Spectrum- and AmpC- β-Lactamase Producing Enterobacteriaceae Isolated from Selected Commercial Spinach Supply Chains in South Africa. Front. Microbiol. 2019, 11, 638. [Google Scholar] [CrossRef]
- Ibrahim, M.E.; Abbas, M.; Al-Shahrai, A.M.; Elamin, B.K. Phenotypic Characterization and Antibiotic Resistance Patterns of Extended-Spectrum β-Lactamase- and AmpC β-Lactamase-Producing Gram-Negative Bacteria in a Referral Hospital, Saudi Arabia. Can. J. Infect. Dis. Med. Microbiol. 2019, 2019, 6054694. [Google Scholar] [CrossRef]
- Rubee Chanu, T.; K Shah, P.; Soni, S.; Ghosh, A. Phenotypic detection of extended spectrum, AmpC, Metallo beta-lactamases and their coexistence in clinical isolates of commonly isolated gram negativebacteria in GKGH hospital, Bhuj. IP Int. J. Med. Microbiol. Trop. Dis. 2019, 5, 52–56. [Google Scholar] [CrossRef]
- Ejikeugwu, C.; Nworie, O.; Saki, M.; Al-Dahmoshi, H.O.M.; Al-Khafaji, N.S.K.; Ezeador, C.; Nwakaeze, E.; Eze, P.; Oni, E.; Obi, C.; et al. Metallo-β-lactamase and AmpC genes in Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa isolates from abattoir and poultry origin in Nigeria. BMC Microbiol. 2021, 21, 124. [Google Scholar] [CrossRef]
- Sultan, I.; Siddiqui, M.T.; Gogry, F.A.; Haq, Q.M.R. Molecular characterization of resistance determinants and mobile genetic elements of ESBL producing multidrug-resistant bacteria from freshwater lakes in Kashmir, India. Sci. Total Environ. 2022, 827, 154221. [Google Scholar] [CrossRef]
- Carlet, J.; Jarlier, V.; Acar, J.; Debaere, O.; Dehaumont, P.; Grandbastien, B.; Le Coz, P.; Lina, G.; Pean, Y.; Rambaud, C.; et al. Trends in Antibiotic Consumption and Resistance in France over 20 Years: Large and Continuous Efforts but Contrasting Results. Open Forum Infect. Dis. 2020, 7, ofaa452. [Google Scholar] [CrossRef]
- Seenama, C.; Thamlikitkul, V.; Ratthawongjirakul, P. Multilocus sequence typing and bla (ESBL) characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolated from healthy humans and swine in Northern Thailand. Infect. Drug Resist. 2019, 12, 2201–2214. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.N.; Hoang, H.T.T.; Xavier, B.B.; Lammens, C.; Le, H.T.; Hoang, N.T.B.; Nguyen, S.T.; Pham, N.T.; Goossens, H.; Dang, A.D.; et al. Prospective One Health genetic surveillance in Vietnam identifies distinct bla(CTX-M)-harbouring Escherichia coli in food-chain and human-derived samples. Clin. Microbiol. Infect. 2021, 27, 1515.e1–1515.e8. [Google Scholar] [CrossRef] [PubMed]
- Ayala, D.I.; Cook, P.W.; Franco, J.G.; Bugarel, M.; Kottapalli, K.R.; Loneragan, G.H.; Brashears, M.M.; Nightingale, K.K. A systematic approach to identify and characterize the effectiveness and safety of novel probiotic strains to control foodborne pathogens. Front. Microbiol. 2019, 10, 1108. [Google Scholar] [CrossRef] [PubMed]
- Markwart, R.; Willrich, N.; Haller, S.; Noll, I.; Koppe, U.; Werner, G.; Eckmanns, T.; Reuss, A. The rise in vancomycin-resistant Enterococcus faecium in Germany: Data from the German Antimicrobial Resistance Surveillance (ARS). Antimicrob. Resist. Infect. Control 2019, 8, 147. [Google Scholar] [CrossRef]
- Chilambi, G.S.; Hinks, J.; Matysik, A.; Zhu, X.; Choo, P.Y.; Liu, X.; Chan-Park, M.B.; Bazan, G.C.; Kline, K.A.; Rice, S.A. Enterococcus faecalis Adapts to Antimicrobial Conjugated Oligoelectrolytes by Lipid Rearrangement and Differential Expression of Membrane Stress Response Genes. Front. Microbiol. 2020, 11, 155. [Google Scholar] [CrossRef]
- Di Cesare, A.; Frangipani, E.; Citterio, B.; Sabatino, R.; Corno, G.; Fontaneto, D.; Mangiaterra, G.; Bencardino, D.; Zoppi, S.; Di Blasio, A.; et al. Class 1 integron and Enterococcus spp. abundances in swine farms from the “Suckling piglets” to the “Fatteners” production category. Vet. Microbiol. 2022, 274, 109576. [Google Scholar] [CrossRef]
- Liu, M.; Kemper, N.; Volkmann, N.; Schulz, J. Resistance of enterococcus spp. in dust from farm animal houses: A retrospective study. Front. Microbiol. 2018, 9, 3074. [Google Scholar] [CrossRef]
- Mahony, A.A.; Buultjens, A.H.; Ballard, S.A.; Grabsch, E.A.; Xie, S.; Seemann, T.; Stuart, R.L.; Kotsanas, D.; Cheng, A.; Heffernan, H.; et al. Vancomycin-resistant Enterococcus faecium sequence type 796-rapid international dissemination of a new epidemic clone. Antimicrob. Resist. Infect. Control 2018, 7, 44. [Google Scholar] [CrossRef]
- Abbo, L.; Shukla, B.S.; Giles, A.; Aragon, L.; Jimenez, A.; Camargo, J.F.; Simkins, J.; Sposato, K.; Tran, T.T.; Diaz, L.; et al. Linezolid-and vancomycin-resistant Enterococcus faecium in solid organ transplant recipients: Infection control and antimicrobial stewardship using whole genome sequencing. Clin. Infect. Dis. 2019, 69, 259–265. [Google Scholar] [CrossRef]
- Farman, M.; Yasir, M.; Al-Hindi, R.R.; Farraj, S.A.; Jiman-Fatani, A.A.; Alawi, M.; Azhar, E.I. Genomic analysis of multidrug-resistant clinical Enterococcus faecalis isolates for antimicrobial resistance genes and virulence factors from the western region of Saudi Arabia. Antimicrob. Resist. Infect. Control 2019, 8, 55. [Google Scholar] [CrossRef]
- Kang, X.; Wei, Y.; Fan, X.; Luo, S.; Luo, X.; Zhao, S.; Wang, G. Analysis of virulence genes, drug resistance detection, and pathogenicity in Enterococcus from farm animals. Microb. Pathog. 2022, 171, 105745. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, L.M.; Castanheira, M.; Flamm, R.K.; Mendes, R.E. Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: Results from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 2018, 73, 2314–2322. [Google Scholar] [CrossRef] [PubMed]
- Sacramento, A.G.; Andrade, A.C.D.; Teotonio, B.N.; de Oliveira Santos, L.M.; da Silva, L.C.B.A.; Lincopan, N.; Sellera, F.P. WHO critical priority van-type vancomycin-resistant Enterococcus in dogs and cats. Prev. Vet. Med. 2022, 202, 105614. [Google Scholar] [CrossRef] [PubMed]
- Bender, J.K.; Cattoir, V.; Hegstad, K.; Sadowy, E.; Coque, T.M.; Westh, H.; Hammerum, A.M.; Schaffer, K.; Burns, K.; Murchan, S.; et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature. Drug Resist. Updates 2018, 40, 25–39. [Google Scholar] [CrossRef]
- Larsen, J.; Schonheyder, H.C.; Lester, C.H.; Olsen, S.S.; Porsbo, L.J.; Garcia-Migura, L.; Jensen, L.B.; Bisgaard, M.; Hammerum, A.M. Porcine-origin gentamicin-resistant Enterococcus faecalis in humans, Denmark. Emerg. Infect. Dis. 2010, 16, 682–684. [Google Scholar] [CrossRef]
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE pathogens in the environment: Antibiotic resistance status, community-acquired infection and risk to human health. Int. J. Hyg. Environ. Health 2022, 244, 114006. [Google Scholar] [CrossRef]
- Ma, Y.X.; Wang, C.Y.; Li, Y.Y.; Li, J.; Wan, Q.Q.; Chen, J.H.; Tay, F.R.; Niu, L.N. Considerations and Caveats in Combating ESKAPE Pathogens against Nosocomial Infections. Adv. Sci. 2020, 7, 1901872. [Google Scholar] [CrossRef]
- de Aledo, M.G.; González-Bardanca, M.; Blasco, L.; Pacios, O.; Bleriot, I.; Fernández-García, L.; Fernández-Quejo, M.; López, M.; Bou, G.; Tomás, M. Crispr-cas, a revolution in the treatment and study of eskape infections: Pre-clinical studies. Antibiotics 2021, 10, 756. [Google Scholar] [CrossRef]
- Intra, J.; Carcione, D.; Sala, R.M.; Siracusa, C.; Brambilla, P.; Leoni, V. Antimicrobial Resistance Patterns of Enterobacter cloacae and Klebsiella aerogenes Strains Isolated from Clinical Specimens: A Twenty-Year Surveillance Study. Antibiotics 2023, 12, 775. [Google Scholar] [CrossRef]

| Type of Food | Microbiota | Antimicrobial Resistance Genes | Reference |
|---|---|---|---|
| Raw poultry, pork and beef | Enterococcus spp. | Vancomycin: vanA, vanB and vanC1,2,3 | [96] |
| Tetracycline: tetM, tetL | |||
| Erythromycin: ermA and ermB | |||
| Quinupristin-dalfopristin: vat[D] and vat[E] | |||
| Retail poultry meat | Escherichia coli | β-lactam: blaTEM, blaSHV, blaCMY-2 and blaCTX-M | [86] |
| Sulphamethoxazole: sul2 | |||
| Tetracycline: tetA and tetB | |||
| Chloramphenicol: cmlA | |||
| Aminoglycoside: aphA1 and aadA | |||
| Trimethoprim: dfrA1 | |||
| Bull-cooked meat products | Enterobacter spp. Escherichia coli Citrobacter spp. Pseudomonas spp. | β-lactam: blaTEM-1 and blaCTX-M-14 | [88] |
| Gentamicin: aac(3)-IIa | |||
| Streptomycin: strA and strB | |||
| Quinolone: qnrB and qnrS | |||
| Sulphamethoxazole: sul1, sul2 and sul3 | |||
| Chloramphenicol: cat1 and cat3 | |||
| Tetracycline: tetM. tetA and tetB | |||
| Animal-based products (ready-to-eat food) | Staphylococcus saprophyticus Staphylococcus sciuri Staphylococcus xylosus. | Oxacillin: mecA | [95] |
| β-lactam: blaZ | |||
| Tetracycline: tetK | |||
| Erythromycin: msrA, msrB, ermA | |||
| Gentamycin: aacA-aphD | |||
| Fusidic acid: fusD | |||
| Trimethoprim/sulfamethoxazole: dfrG | |||
| Chicken meat | Salmonella Albany Salmonella Virchow Salmonella Enteritidis Salmonella Infantis | β-lactam: blaCTZ-M-15, blaCTX-M-79 and blaCMY-2 | [93] |
| Tetracycline: tetA and tetB | |||
| Sulfonamide: sul1 and sul2 | |||
| Chloramphenicol: catA1 and cmlA | |||
| Retail meat (pork, chicken and duck) | Salmonella Enteritidis Salmonella Typhimurium Salmonella Typhi Salmonella Goldcoast Salmonella Ouakam Salmonella Paratyphi | Tetracycline: tetA | [94] |
| β-lactam: blaTEM | |||
| Aminoglycoside: aadA1 and aadA2 | |||
| Sulfonamide: sul1 and sul2 | |||
| Dry fermented Italian salami | Enterococcus faecium UC7251 | Ampicillin: pbp5-S1/R20 | [97] |
| Gentamycin: aac(6′)-li | |||
| Kanamycin: aph(3′)-lll | |||
| Streptomycin: aad6 and aadE | |||
| Erythromycin: ermB, mrsC and sat4 | |||
| Clindamycin: ermB, lnuB and lsaE | |||
| Tylosine: ermB | |||
| Tetracycline: tetL and tetM | |||
| Traditional pork dry sausages | Salmonella Enteritidis Salmonella Typhi Salmonella Typhimurium | Quinolone: gyrA and parC | [91] |
| Chloramphenicol: catA1 | |||
| Trimethoprim: drfA | |||
| Tetracycline: tetA and tetB | |||
| Nitrofurantoin: nfsA and nfsB | |||
| Ampicillin: blaTEM | |||
| Chicken meat | Escherichia coli isolate 1108 | β-lactam: blaNDM-1, blaTEM-1, blaCTZX-M-64 and blaCMY-2 | [92] |
| Bleomycin: bleMBL | |||
| Sulfonamide: sul1 and sul2 | |||
| Tetracycline: tetA and tetR | |||
| Aminoglycosides: strA | |||
| Quinolone: oqxA and oqxB | |||
| Phenicol: floR | |||
| Streptomycin: aadA2 | |||
| Trimethoprim: dfrA12 | |||
| Retail meat (chicken and pork) | Salmonella Kentucky Salmonella Indiana Salmonella Derby Salmonella Typhimurium Salmonella Litchfield Salmonella Schwarzengrun | β-lactam: blaCTX-M-55, blaTEM-206, blaTEM-214, blaOXA-1, blaCTX-M-123, blaTEM-1, blaCTX-M-64 and blaCTX-M-15 | [98] |
| Naturally fermented smoked pork | Staphylococcus carnosus Lactobacillus plantarum Labctobacillus brevis Lactobacillus sakei Weissella confusa Weissella cibaria | Tetracycline: tetO and tetM Erythromycin: ereA Chloramphenicol: catA Streptomycin: strA and strB | [99] |
| Pork meat | Aeromonas aquariorum Aeromonas hydrophila Aeromonas jandaei Aeromonas veronii Acinetobacter baumannii Acinetobacter bereziniae Acinetobacter johnsonii Acinetobacter septicus Acinetobacter ursingii Citrobacter sp. Citrobacter freundii Citrobacter murliniae Enterobacteriaceae Enterobacter sp. Enterobacter asburiae Enterobacter cloacae Enterobacter hormaechei Enterobacter ludwigii Escherichia coli Klebsiella sp. Klebsiella oxytoca Klebsiella terrigena Lactobacillus casei Leclercia sp. Lactococcus garvieae Lactococcus lactis Micrococcus caseolyticus Myroides phaeus Myroides marinus Myroides odoratimimus Oceanobacillus Pantoea sp. Pantoea dispersa Pantoea agglomerans Proteus penneri Providencia alcalifaciens Pseudomonas sp. Raoultella sp. Raoultella terrigena Serratia sp. Serratia marcescens Sphingobacterium Staphylococcus sp. Staphylococcus sciuri Staphylococcus epidermidis Vibrio cincinnatiensis Wautersiella falsenii genomovar 1 Kurthia sp. Bacillus sp. Morganella sp. Micrococcus caseolyticus Vagococcus sp. Raoultella ornithinolytica Comamonas sp. Budvicia sp. Aeromonas sp. Klebsiella sp. | β-lactam: blaTEM, blaCTX-M, blaCMY-2 Tetracycline: tetA, tetC, tetE, tetK, tetL, tetM and tetS Sulfonamide: sul1 and sul2 Aminoglycoside: aadA and aphA-1 Chloramphenicol: cmlA Macrolide: ermB Florfenicol: floR | [100,101,102] |
| Raw and cooked pork | Citrobacter freundii Serratia marcescens Escherichia coli | β-lactams: blaTEM, blaCTX-M-1, blaSHV and blaCTX-M-9 | [103] |
| Pork meat and pork meat preparations (cotechino, hamburger, sausages and Zuccotto of Bismantova) | Salmonella Derby Salmonella Typhimurium Salmonella Bredeney Salmonella London Salmonella Anatum Salmonella Agona Salmonella Virchow Salmonella Senftenberg Salmonella Livingstone Salmonella India Salmonella Heidelberg Salmonella Bovis-morbificans Salmonella Coeln | Ampicillin: blaPSE-1 Gentamicin: ant (2″)-Ia Sulfamethoxazole: sul1 Tetracycline: tetA, tetB, tetG and marRAB | [104] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conceição, S.; Queiroga, M.C.; Laranjo, M. Antimicrobial Resistance in Bacteria from Meat and Meat Products: A One Health Perspective. Microorganisms 2023, 11, 2581. https://doi.org/10.3390/microorganisms11102581
Conceição S, Queiroga MC, Laranjo M. Antimicrobial Resistance in Bacteria from Meat and Meat Products: A One Health Perspective. Microorganisms. 2023; 11(10):2581. https://doi.org/10.3390/microorganisms11102581
Chicago/Turabian StyleConceição, Sara, Maria Cristina Queiroga, and Marta Laranjo. 2023. "Antimicrobial Resistance in Bacteria from Meat and Meat Products: A One Health Perspective" Microorganisms 11, no. 10: 2581. https://doi.org/10.3390/microorganisms11102581
APA StyleConceição, S., Queiroga, M. C., & Laranjo, M. (2023). Antimicrobial Resistance in Bacteria from Meat and Meat Products: A One Health Perspective. Microorganisms, 11(10), 2581. https://doi.org/10.3390/microorganisms11102581







