Mechanism of the Change in the Intestinal Microbiota of C-Strain Spodoptera frugiperda (Lepidoptera: Noctuidae) after an Interspecific Transference between Rice and Corn
Abstract
:1. Introduction
2. Materials and Methods
2.1. Host Plants and Insects
2.2. Strain Analysis for S. frugiperda Individuals Feeding on Rice and Corn
2.3. Intestinal Bacteria of S. frugiperda Fed on Rice and Corn
2.4. Enzyme Activity
2.5. Statistical Analysis
3. Results
3.1. Host Strain Analysis
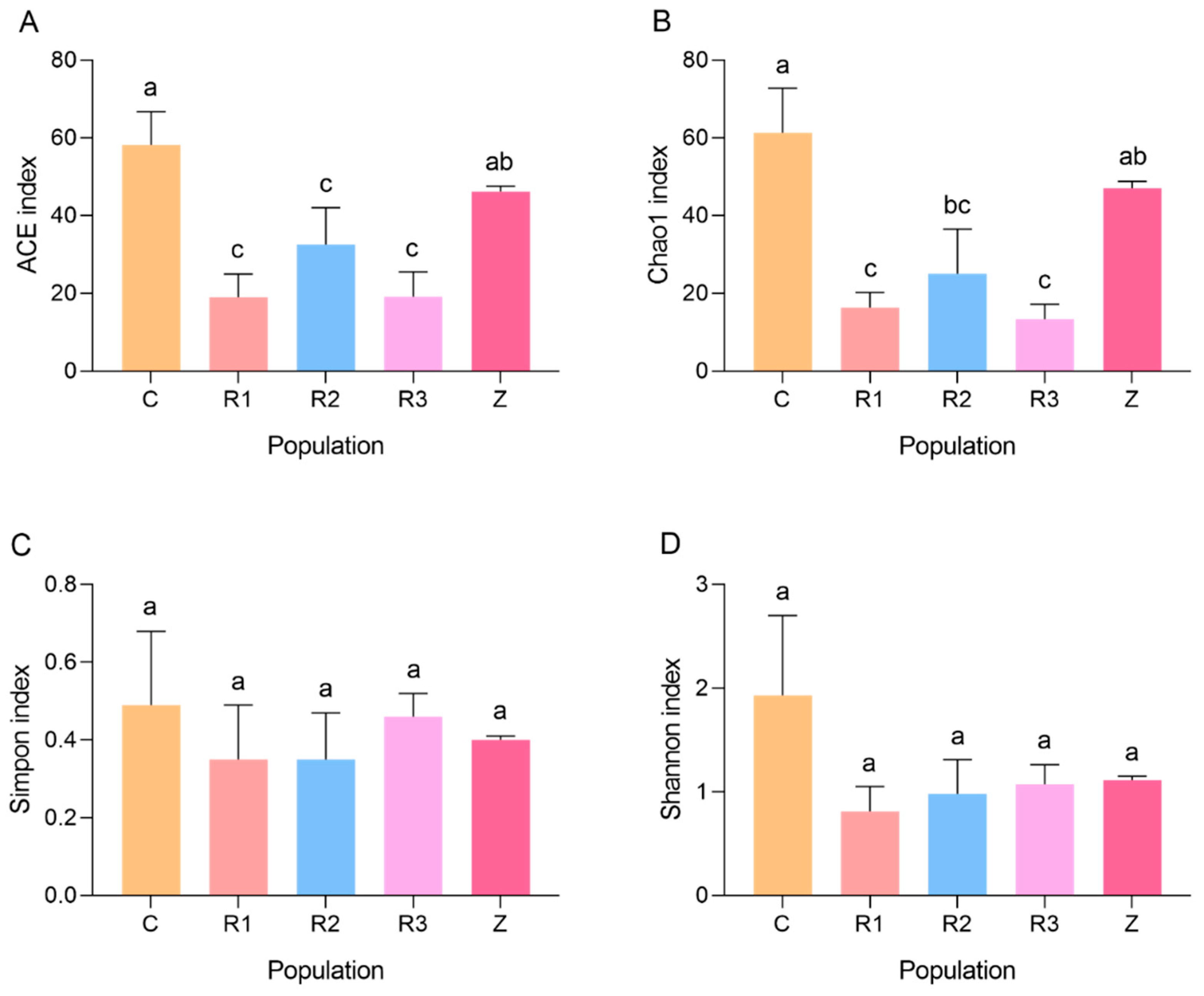
3.2. Influence of Host-Plant-Switching on Intestinal Bacteria
3.3. Enzyme Activity of S. frugiperda Fed on Corn and Rice
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| C-strain | Corn strain |
| R-strain | Rice strain |
| DNA | Deoxyribo Nucleic Acid |
| CTAB | Cetyltrimethyl Ammonium Bromide |
| Tpi | Triosephosphate isomerase |
| OTU | Operational Taxonomic Units |
| RDP | The Ribosomal Database Project |
| CAT | Catalase |
| POD | Peroxidase |
| SOD | Superoxide dismutase |
| GST | Glutathione S-transferase |
| CarE | Carboxylesterase |
References
- Todd, E.L.; Poole, R.W. Keys and illustrations for the armyworm moths of the Noctuid genus Spodoptera Guenée from the western hemisphere. Ann. Entomol. Soc. Am. 1980, 73, 722–738. [Google Scholar] [CrossRef]
- Sparks, A.N. A review of the biology of the fall armyworm. Fla. Entomol. Soc. 1979, 62, 82–87. [Google Scholar] [CrossRef]
- Montezano, D.; Spechtk, A.; Sosa-Gómez, D.; Roque-Specht, V.; Sousa-Silva, J.; Paula-Moraes, S.; Peterson, J.; Hunt, T. Host plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr. Entomol. 2018, 26, 286–300. [Google Scholar] [CrossRef]
- Song, Y.; Yang, X.; Zhang, H.; Zhang, D.; He, W.; Wyckhuys, K.A.G.; Wu, K. Interference competition and predation between invasive and native herbivores in maize. J. Pest. Sci. 2021, 94, 1053–1063. [Google Scholar] [CrossRef]
- Pashley, D. Host-associated genetic differentiation in fall armyworm (Lepidoptera: Noctuidae): A sibling species complex? Ann. Entomol. Soc. Am. 1986, 79, 898–904. [Google Scholar] [CrossRef]
- Nagoshi, R.N.; Meagher, R.L. Behavior and distribution of the two fall armyworm host strains in Florida. Fla. Entomol. 2004, 87, 440–449. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, B.; Jiang, Y.Y.; Liu, J.; Wu, K.M.; Xiao, Y.T. Molecular characterization analysis of fall armyworm populations in China. Plant Prot. 2019, 45, 20–27. [Google Scholar]
- Guo, J.; Zhang, M.; Gao, Z.; Wang, D.; He, K.; Wang, Z. Comparison of larval performance and oviposition preference of Spodoptera frugiperda among three host plants: Potential risks to potato and tobacco crops. Insect Sci. 2020, 28, 602–610. [Google Scholar] [CrossRef]
- Guo, Z.B.; Jiang, X.R.; Tang, Y.L.; Gu, R.C.; Li, Q.Y.; Xing, T.; Xiang, L.; Wu, Y.Y.; Hu, Y.; Liu, X.; et al. Identification of new isolates of gut bacteria of Spodoptera frugiperda feeding on sorghum in Chongqing area. J. Southwest Univ. 2019, 41, 9–16. [Google Scholar]
- Blytt, H.J.; Guscar, T.K.; Butler, L.G. Antinutritional effects and ecological significance of dietary condensed tannins may not be due to binding and inhibiting digestive enzymes. J. Chem. Ecol. 1988, 14, 1455–1465. [Google Scholar] [CrossRef]
- Liu, S.; Guo, J.; Wang, Q.; He, K.; Wang, Z. Resistance evaluation of corn hybrid Jingke 968 silks to Ostrinia furnacalis and related resistance mechanism. J. Environ. Entomol. 2019, 41, 25–32. [Google Scholar]
- Simmonds, M.S.J. Flavonoid-insect interactions: Recent advances in our knowledge. Phytochemistry 2003, 64, 21–30. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defence against insect herbivores. Plant Signal Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef]
- Binyameen, M.; Hussain, A.; Yousefi, F.; Birgersson, G.; Schlyter, F. Modulation of reproductive behaviors by non-host volatiles in the polyphagous Egyptian cotton leafworm, Spodoptera littoralis. J. Chem. Ecol. 2013, 39, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Mukhopadhyay, A.; Bahadur, M. Effect of host plants on fitness traits and detoxifying enzymes activity of Helopeltis theivora, a major sucking insect pest of tea. Phytoparasitica 2012, 40, 433–444. [Google Scholar] [CrossRef]
- Huang, X.; Ma, J.; Qin, X.; Tu, X.; Cao, G.; Wang, G.; Nong, X.; Zhang, Z. Biology, physiology and gene expression of grasshopper Oedaleus asiaticus exposed to diet stress from plant secondary compounds. Sci. Rep. 2017, 7, 8655. [Google Scholar] [CrossRef]
- Su, Q.; Zhou, X.M.; Zhang, Y.J. Symbiont-mediated functions in insect hosts. Commun. Integr. Biol. 2013, 6, e23804. [Google Scholar] [CrossRef]
- Schretter, C.E.; Vielmetter, J.; Bartos, I.; Marka, Z.; Marka, S.; Argade, S.; Mazmanian, S.K. A gut microbial factor modulates locomotor behavior Drosophila. Nature 2018, 563, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, R.G.; Selvakumar, G.; Abraham, V.; Sudhagar, S.; Ravi, M. Diversity of the cultivable gut bacterial communities associated with the fruit flies Bactrocera dorsalis and Bactrocera cucurbitae (Diptera: Tephritidae). Phytoparasitica 2017, 45, 453–460. [Google Scholar]
- Strano, C.P.; Malacrinò, A.; Campolo, O.; Palmeri, V. Influence of host plant on Thaumetopoea piyocampa gut bacterial community. Microb. Ecol. 2017, 75, 487–494. [Google Scholar] [CrossRef]
- de Almeida, L.G.; de Moraes, L.A.B.; Trigo, J.R.; Omoto, C.; Cônsoli, F.L. The gut microbiota of insecticide-resistant insects houses insecticide-degrading bacteria: A potential source for biotechnological exploitantion. PLoS ONE 2017, 12, e0174754. [Google Scholar]
- Koch, H.; Abrol, D.P.; Li, J.L.; Schmid-Hempel, P. Diversity and evolutional patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 2013, 22, 2028–2044. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.W.; Zhang, H.Y. The impact of environmental heterogeneity and life stage on the hindgut microbiota of Holotrichia parallela larvae (Coleoptera: Scarabaeidae). PLoS ONE 2017, 8, e57169. [Google Scholar] [CrossRef] [PubMed]
- Gichuhi, J.; Sevgan, S.; Khamis, F.; Van den Berg, J.; Plessis, H.; Ekesi, S.; Herren, J.K. Diversity of fall armyworm, Spodoptera frugiperda and their gut bacterial community in Kenya. PeerJ 2020, 8, e8701. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Y.; Tang, Y.L.; Jiang, R.X.; Zhang, Y.H.; Zhu, F.; Bai, X.R.; Gu, R.C.; Wu, Y.Y.; Wu, Y.J.; Chen, J.; et al. Isolation and identification of gut bacteria of Spodoptera frugiperda feeding on maize in Yunnan, China. J. Southwest Univ. (Nat. Sci. Ed.) 2020, 42, 1–8. [Google Scholar]
- Li, Y.P.; Du, G.Z.; Xu, T.M.; Yang, H.; Yang, J.B.; Zhang, T.F.; Yi, X.G.; Chen, B. Diversity of intestinal and fecal bacteria in fall armyworm Spodoptera frugiperda larvae from different altitudes of Yunnan Province. J. Plant Prot. 2022, 49, 559–573. [Google Scholar]
- Chang, H.; Hao, D.; Yang, X.; Xiao, R.; Liu, Y.; Qian, L.; An, Y. Comparison of four genomic DNA extraction methods with Scolytidae beetles. J. Beijing For. Univ. 2013, 35, 75–79. [Google Scholar]
- Nagoshi, R. Improvements in the identification of strains facilitate population studies of fall armyworm subgroups. Entomol. Soc. Am. 2012, 105, 351–358. [Google Scholar] [CrossRef]
- He, Y.Y.; Du, G.Z.; Xie, S.X.; Long, X.M.; He, X.H.; Zhu, Y.Y.; Chen, B. The acaricidal potential of a new agent GC16 for Tetranychus pueraricola (acari: Tetranychidae) based on developmental performance and physiological enzyme activity. J. Econ. Entomol. 2022, 115, 814–825. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, J.; Xie, M.; Li, Y.; Yang, J.; Zhang, M.; Qiu, K. Observation on law of diffusion damage of Spodoptera frugiperda in China in 2019. Plant Prot. 2019, 45, 10–19. [Google Scholar]
- He, Y.; Wang, K.; Du, G.; Zhang, Q.; Li, B.; Zhao, L.; He, P.; Chen, B. Temporal and spatial distribution patterns of Spodoptera frugiperda in mountain maize fields in China. Insects 2022, 13, 938. [Google Scholar] [CrossRef] [PubMed]
- Acharya, R.; Malekera, M.J.; Dhungana, S.K.; Sharma, S.R.; Lee, K. Impact of rice and potato host plants is higher on the reproduction than growth of corn strain fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects 2022, 13, 256. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Q.; Zhou, H.C.; Lai, Y.S.; Chen, Q.; Yu, X.Q.; Wang, X.Y. Gut microbiota dysbiosis influences metabolic homeostasis in Spodoptera frugiperda. Frot. Microbiol. 2021, 12, 727434. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.J.; Xiang, H.; Feng, Q.L.; Cheng, J. Preliminary metagenomic analysis of gut microorganisms in fall armyworm Spodoptera frugiperda larvae. J. Plant Prot. 2021, 48, 1254–1261. [Google Scholar]
- Piskorski, R.; Dorn, S. How the oligophage codling moth Cydia pomonella survives on walnut despite its secondary metabolite juglone. J. Insect Physiol. 2011, 57, 744–750. [Google Scholar] [CrossRef]
- Zámocky, M.; Koller, F. Understanding the structure and function of catalases: Clues from molecular evolution and in vitro mutagenesis. Prog. Biophys. Mol. Biol. 1999, 72, 19–66. [Google Scholar] [CrossRef]
- Lu, Z.H.; Chen, Y.P.; Zhou, A.C.; He, S.Q.; Li, H.; Bao, Y.Y.; Gui, F.R. Effects of multigeneration feeding with different host plants on activities of enzyme in Spodoptera frugiperda larvae. J. Environ. Entomol. 2020, 42, 1361–1368. [Google Scholar]



| Group Information | Sample ID | Effective CCS | AvgLen (bp) | Tags Num | Goods Coverage |
|---|---|---|---|---|---|
| C | C01 | 12,913 | 1484 | 12,339 | 0.9993 |
| C02 | 10,909 | 1483 | 9274 | 0.9987 | |
| C03 | 10,626 | 1474 | 10,000 | 0.9992 | |
| average | 11,483 | 1480 | 10,538 | 0.9991 | |
| R1 | R101 | 12,991 | 1483 | 12,930 | 0.9998 |
| R102 | 12,844 | 1471 | 12,784 | 0.9995 | |
| R103 | 12,880 | 1473 | 12,821 | 1.0000 | |
| average | 12,905 | 1476 | 12,845 | 0.9998 | |
| R2 | R201 | 12,833 | 1461 | 12,641 | 0.9999 |
| R202 | 12,755 | 1464 | 12,466 | 0.9994 | |
| R203 | 12,768 | 1467 | 12,689 | 0.9997 | |
| average | 12,785 | 1464 | 12,599 | 0.9997 | |
| R3 | R301 | 12,879 | 1470 | 12,779 | 0.9998 |
| R302 | 13,011 | 1472 | 12,879 | 0.9998 | |
| R303 | 12,817 | 1463 | 12,605 | 0.9995 | |
| average | 12,902 | 1468 | 12,754 | 0.9997 | |
| Z | Z01 | 12,311 | 1463 | 11,894 | 0.9991 |
| Z02 | 12,893 | 1464 | 12,525 | 0.9992 | |
| Z03 | 11,383 | 1464 | 10,906 | 0.9990 | |
| average | 12,196 | 1464 | 11,775 | 0.9991 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di, T.; Li, Y.; Du, G.; He, Y.; Wang, W.; Shen, Y.; Meng, J.; Xiao, W.; Xiao, G.; Chen, B. Mechanism of the Change in the Intestinal Microbiota of C-Strain Spodoptera frugiperda (Lepidoptera: Noctuidae) after an Interspecific Transference between Rice and Corn. Microorganisms 2023, 11, 2514. https://doi.org/10.3390/microorganisms11102514
Di T, Li Y, Du G, He Y, Wang W, Shen Y, Meng J, Xiao W, Xiao G, Chen B. Mechanism of the Change in the Intestinal Microbiota of C-Strain Spodoptera frugiperda (Lepidoptera: Noctuidae) after an Interspecific Transference between Rice and Corn. Microorganisms. 2023; 11(10):2514. https://doi.org/10.3390/microorganisms11102514
Chicago/Turabian StyleDi, Teng, Yongping Li, Guangzu Du, Yanyan He, Wenqian Wang, Yunfeng Shen, Jizhi Meng, Wenxiang Xiao, Guanli Xiao, and Bin Chen. 2023. "Mechanism of the Change in the Intestinal Microbiota of C-Strain Spodoptera frugiperda (Lepidoptera: Noctuidae) after an Interspecific Transference between Rice and Corn" Microorganisms 11, no. 10: 2514. https://doi.org/10.3390/microorganisms11102514
APA StyleDi, T., Li, Y., Du, G., He, Y., Wang, W., Shen, Y., Meng, J., Xiao, W., Xiao, G., & Chen, B. (2023). Mechanism of the Change in the Intestinal Microbiota of C-Strain Spodoptera frugiperda (Lepidoptera: Noctuidae) after an Interspecific Transference between Rice and Corn. Microorganisms, 11(10), 2514. https://doi.org/10.3390/microorganisms11102514







