A New Kayfunavirus-like Escherichia Phage vB_EcoP-Ro45lw with Antimicrobial Potential of Shiga Toxin-Producing Escherichia coli O45 Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. Phage Isolation
2.2. Bacterial Culture
2.3. Genomic Analysis
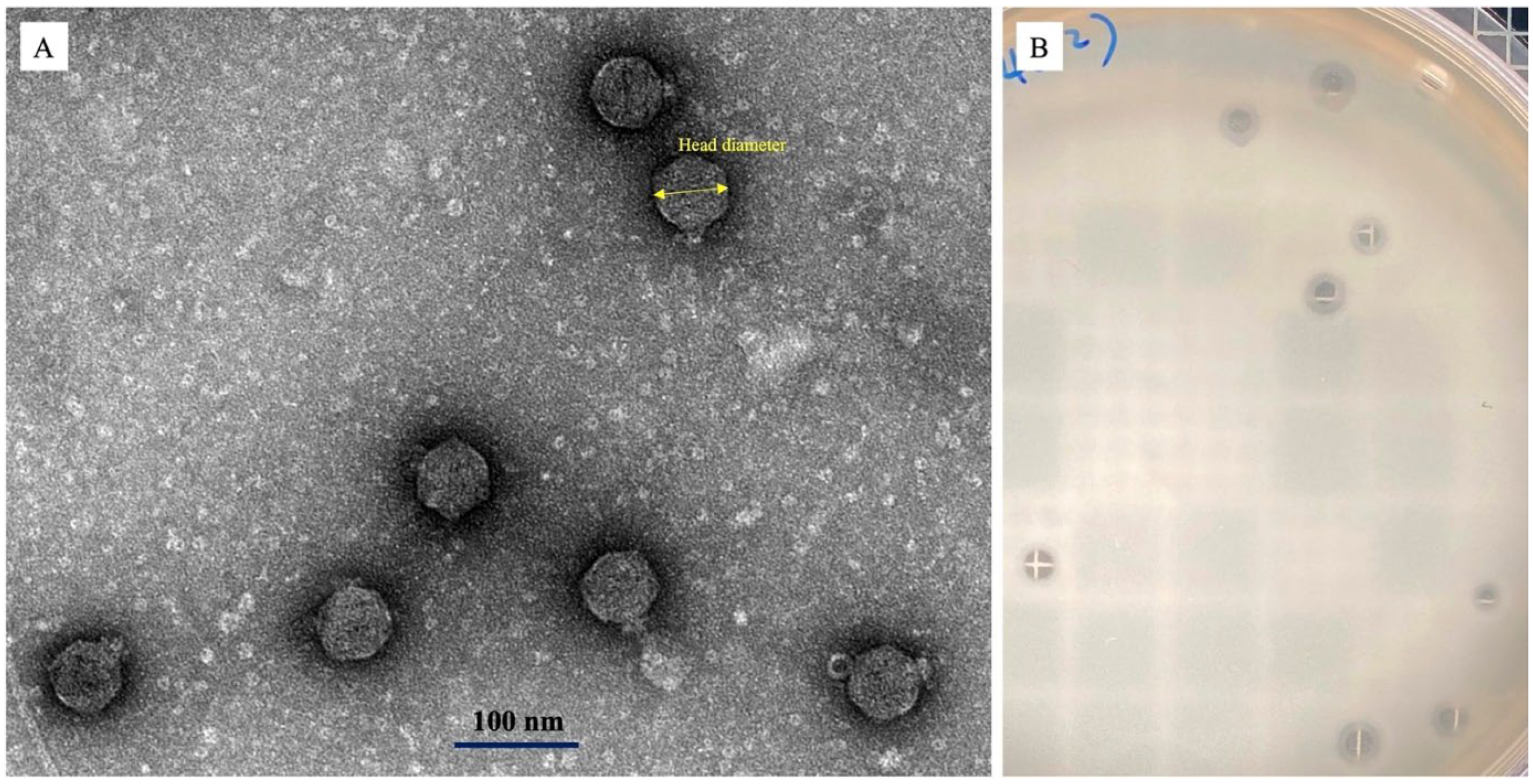
2.4. Transmission Electron Microscopy (TEM)
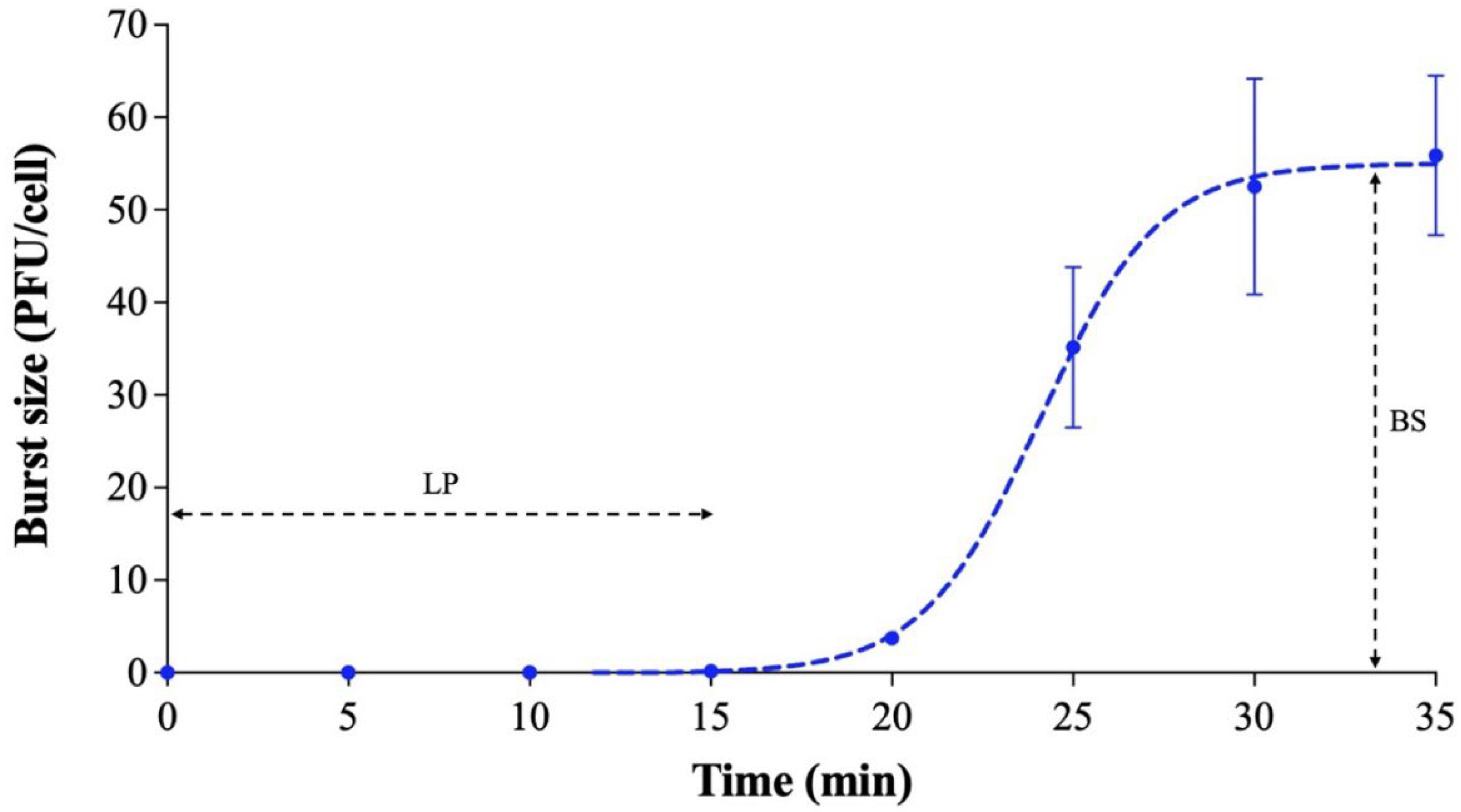
2.5. One-Step Growth Curve
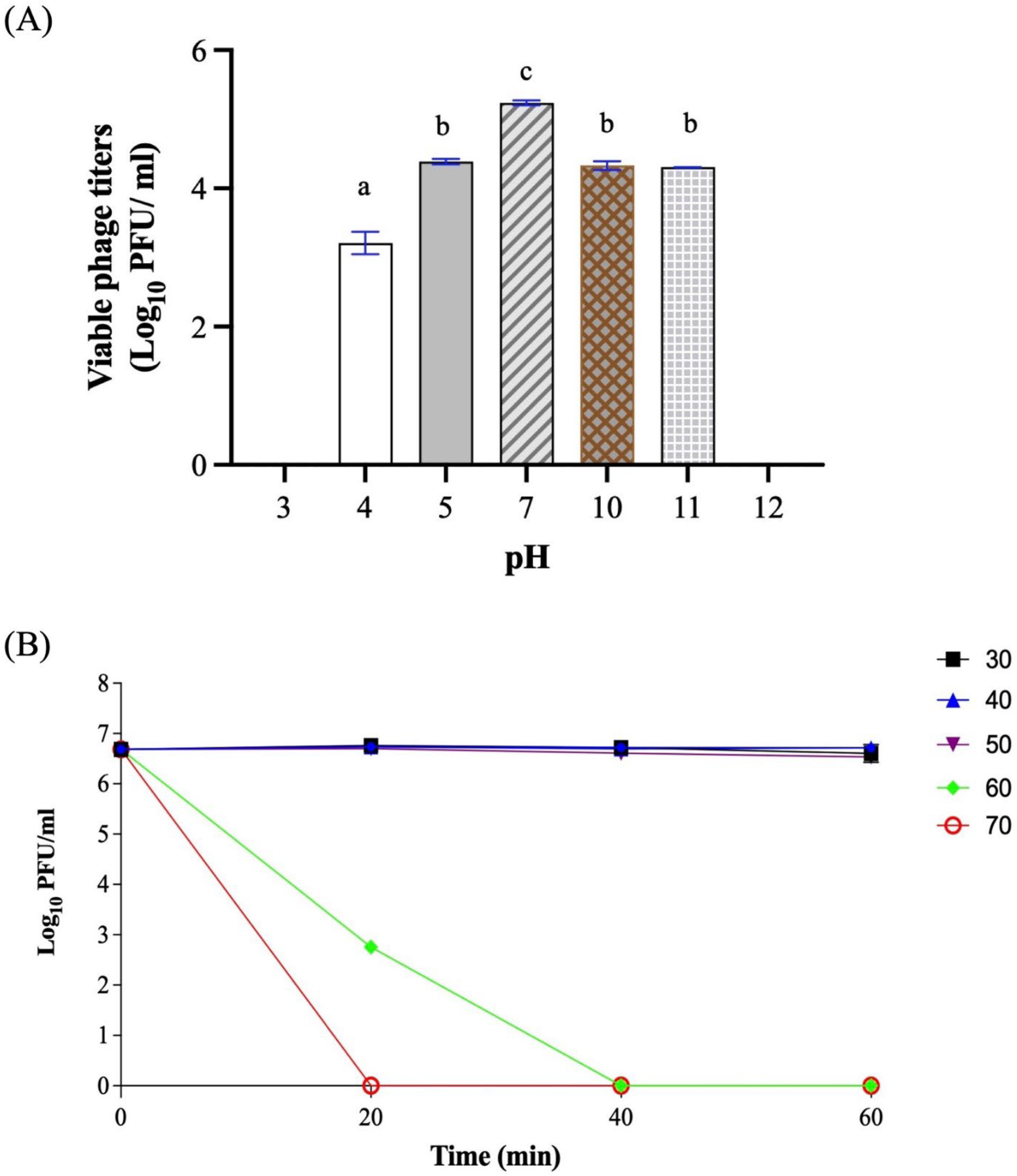
2.6. pH and Temperature Stability
2.7. Host Range and Efficiency of Plating (EOP)
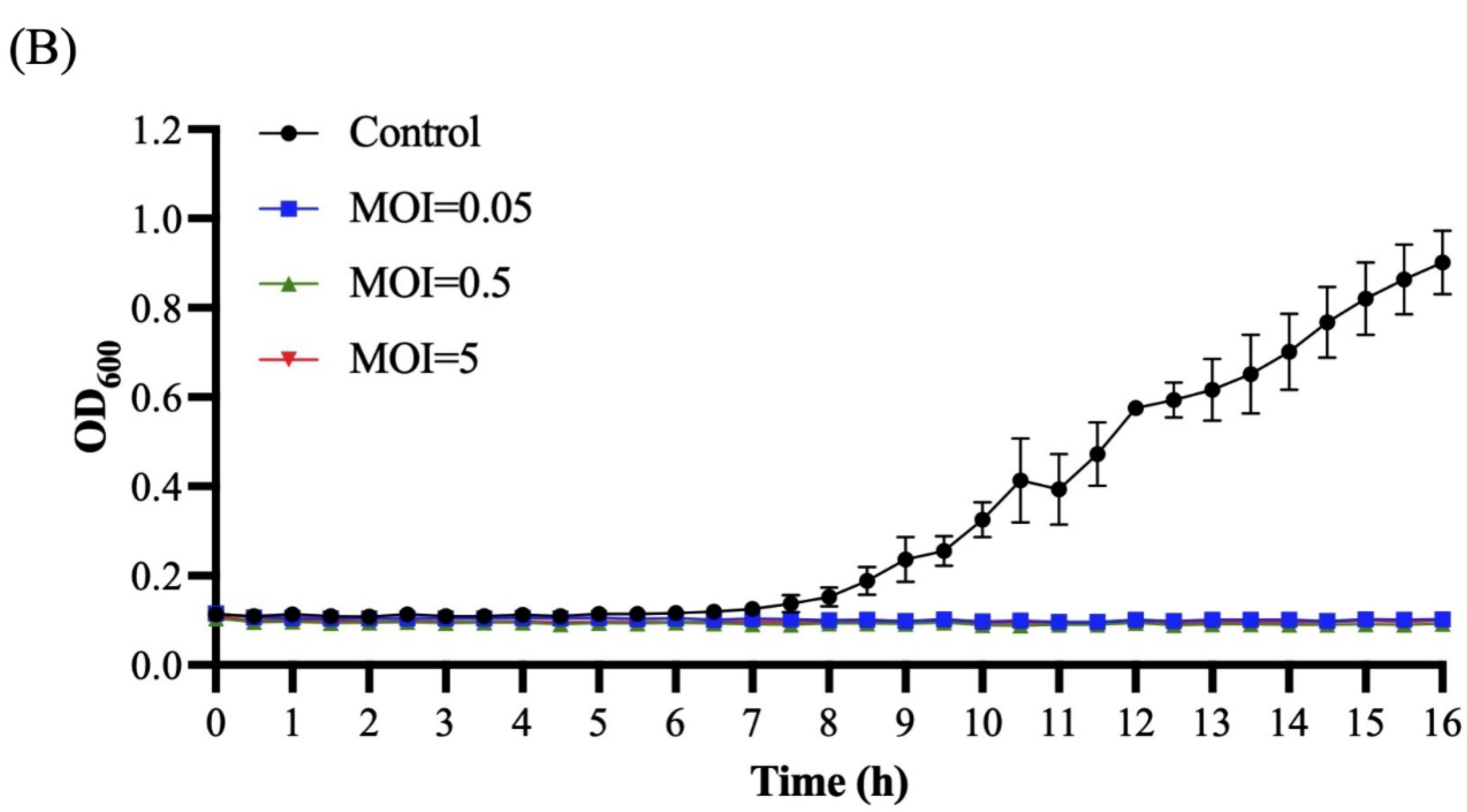
2.8. Bacterial Challenge Assay
2.9. Statistical Analysis
3. Results
3.1. Genomic Features of Ro45lw
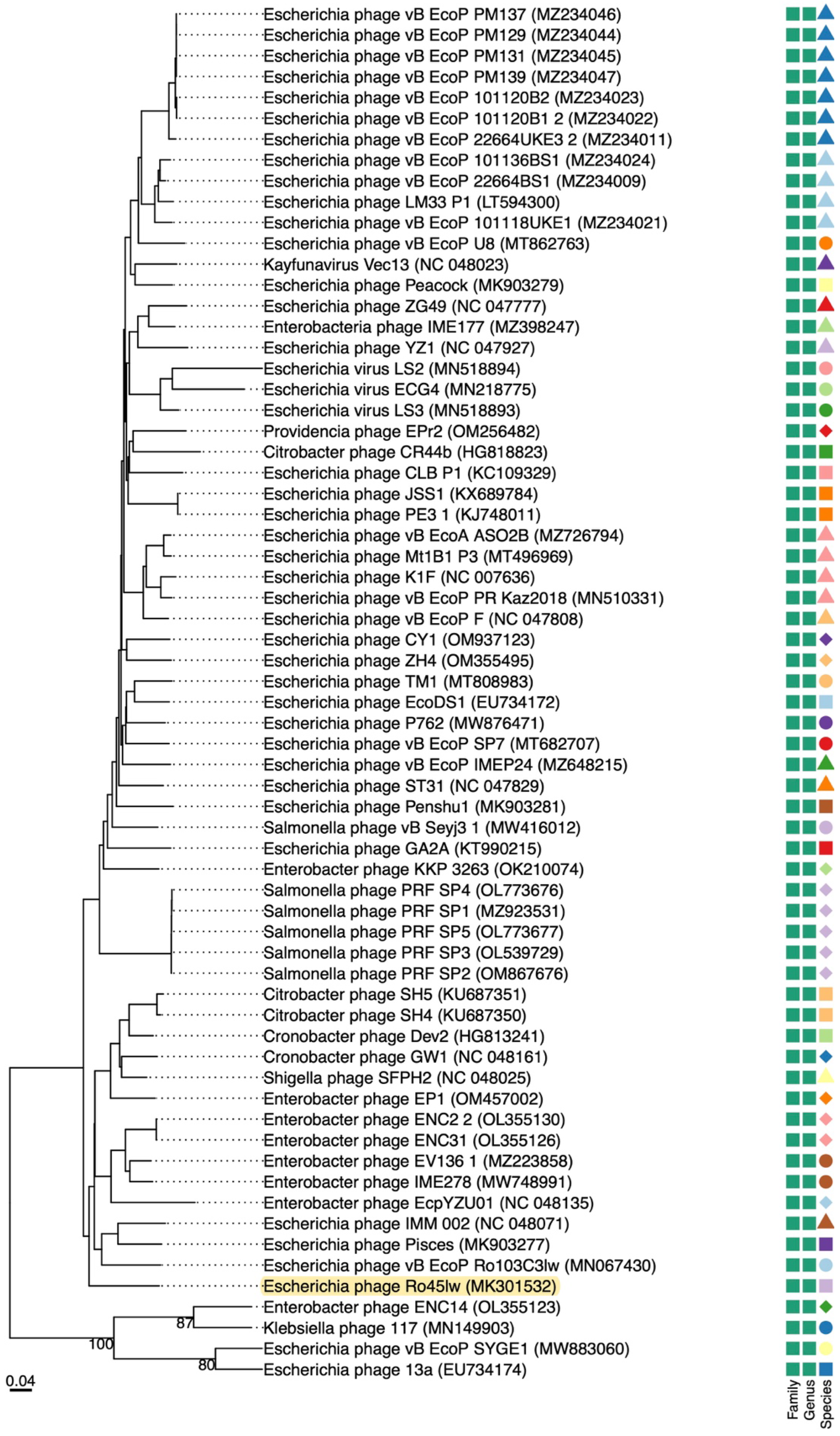
Phylogenetic Analysis
3.2. Phage Morphology
3.3. One-Step Growth Curve and Stability Test
3.4. Host Range and Productive Infection
3.5. Bacterial Challenge Assay of Ro45lw
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karmali, M.A. Emerging Public Health Challenges of Shiga Toxin-Producing Escherichia coli Related to Changes in the Pathogen, the Population, and the Environment. Clin. Infect. Dis. 2017, 64, 371–376. [Google Scholar] [CrossRef] [PubMed]
- CDC. National Shiga Toxin-Producing Escherichia coli (STEC) Surveillance Overview; US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2012.
- CDC. National Enteric Disease Surveillance: Shiga Toxin-Producing E. coli (STEC) Annual Report. 2017. Available online: https://www.cdc.gov/ecoli/surv2017/index.html (accessed on 30 September 2022).
- Luna-Gierke, R.E.; Griffin, P.M.; Gould, L.H.; Herman, K.; Bopp, C.A.; Strockbine, N.; Mody, R.K. Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol. Infect. 2014, 142, 2270–2280. [Google Scholar] [CrossRef] [PubMed]
- Institutional Meat Purchase Specifications for Fresh Beef Products; Agricultural Marketing Service, U.S. Department of Agriculture: Washington, DC, USA, 2011.
- Schaffzin, J.K.; Coronado, F.; Dumas, N.B.; Root, T.P.; Halse, T.A.; Schoonmaker-Bopp, D.J.; Lurie, M.M.; Nicholas, D.; Gerzonich, B.; Johnson, G.S.; et al. Public health approach to detection of non-O157 Shiga toxin-producing Escherichia coli: Summary of two outbreaks and laboratory procedures. Epidemiol. Infect. 2011, 40, 283–289. [Google Scholar] [CrossRef]
- CDC. Importance of culture confirmation of shiga toxin-producing Escherichia coli infection as illustrated by outbreaks of gastroenteritis—New York and North Carolina, 2005. MMWR Morbility Mortal. Wkly. Rep. 2006, 55, 1042–1045. [Google Scholar]
- Zhang, Y.; Liao, Y.-T.; Sun, X.; Wu, V.C.H. Is Shiga Toxin-Producing Escherichia coli O45 No Longer a Food Safety Threat? The Danger is Still Out There. Microorganisms 2020, 8, 782. [Google Scholar] [CrossRef]
- Bielaszewska, M.; Mellmann, A.; Zhang, W.; Köck, R.; Fruth, A.; Bauwens, A.; Peters, G.; Karch, H. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: A microbiological study. Lancet Infect. Dis. 2011, 11, 671–676. [Google Scholar] [CrossRef]
- Liao, Y.-T.; Brook, J.C.; Martin, J.N.; Echeverry, A.; Loneragan, G.H.; Brashears, M.M. Antimicrobial interventions for O157:H7 and non-O157 Shiga toxin-producing Escherichia coli on beef subprimal and mechanically tenderized steaks. J. Food Prot. 2015, 78, 511–517. [Google Scholar] [CrossRef][Green Version]
- O’Sullivan, L.; Bolton, D.; McAuliffe, O.; Coffey, A. Bacteriophages in Food Applications: From Foe to Friend. Annu. Rev. Food Sci. Technol. 2019, 10, 151–172. [Google Scholar] [CrossRef]
- Milho, C.; Silva, M.D.; Alves, D.; Oliveira, H.; Sousa, C.; Pastrana, L.M.; Azeredo, J.; Sillankorva, S. Escherichia coli and Salmonella Enteritidis dual-species biofilms: Interspecies interactions and antibiofilm efficacy of phages. Sci. Rep. 2019, 9, 18183. [Google Scholar] [CrossRef]
- Clokie, M.R.J.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Hassan, A.Y.; Lin, J.T.; Ricker, N.; Anany, H. The Age of Phage: Friend or Foe in the New Dawn of Therapeutic and Biocontrol Applications? Pharmaceuticals 2021, 14, 199. [Google Scholar] [CrossRef] [PubMed]
- Amarillas, L.; Chaidez, C.; Gonzalez-Robles, A.; Lugo-Melchor, Y.; Leon-Felix, J. Characterization of novel bacteriophage phiC119 capable of lysing multidrug-resistant Shiga toxin-producing Escherichia coli O157:H7. PeerJ 2016, 4, e2423. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Snyder, A.B.; Perry, J.J.; Yousef, A.E. Developing and optimizing bacteriophage treatment to control enterohemorrhagic Escherichia coli on fresh produce. Int. J. Food Microbiol. 2016, 236, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, Z.; Kalbasi-Ashtari, A.; Riskowski, G.; Castillo, A. Reduction of Salmonella and Shiga toxin-producing Escherichia coli on alfalfa seeds and sprouts using an ozone generating system. Int. J. Food Microbiol. 2019, 289, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Tokman, J.I.; Woolston, J.; Sulakvelidze, A. Phage Biocontrol Improves Food Safety by Significantly Reducing the Level and Prevalence of Escherichia coli O157:H7 in Various Foods. J. Food Prot. 2020, 83, 668–676. [Google Scholar] [CrossRef]
- Liao, Y.-T.; Liu, F.; Wu, V.C.H. Complete Genome Sequence of a Lytic T7-Like Phage, Escherichia Phage vB_EcoP-Ro45lw, Isolated from Nonfecal Compost Samples. Microbiol. Resour. Announc. 2019, 8, e00036-00019. [Google Scholar] [CrossRef]
- Liao, Y.-T.; Zhang, Y.; Salvador, A.; Harden, L.A.; Wu, V.C.H. Characterization of a T4-like Bacteriophage vB_EcoM-Sa45lw as a Potential Biocontrol Agent for Shiga Toxin-Producing Escherichia coli O45 Contaminated on Mung Bean Seeds. Microbiol. Spectr. 2022, 10, e02220–e02221. [Google Scholar] [CrossRef]
- Zhang, Y.; Liao, Y.T.; Salvador, A.; Lavenburg, V.M.; Wu, V.C.H. Characterization of Two New Shiga Toxin-Producing Escherichia coli O103-Infecting Phages Isolated from an Organic Farm. Microorganisms 2021, 9, 1527. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Bairoch, A.; Apweiler, R.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. The Universal Protein Resource (UniProt). Nucleic Acids Res. 2005, 33, D154–D159. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef]
- Liao, Y.-T.; Salvador, A.; Harden, L.A.; Liu, F.; Lavenburg, V.M.; Li, R.W.; Wu, V.C.H. Characterization of a Lytic Bacteriophage as an Antimicrobial Agent for Biocontrol of Shiga Toxin-Producing Escherichia coli O145 Strains. Antibiotics 2019, 8, 74. [Google Scholar] [CrossRef]
- Liao, W.C.; Ng, W.V.; Lin, I.H.; Syu, W., Jr.; Liu, T.-T.; Chang, C.H. T4-Like genome organization of the Escherichia coli O157:H7 lytic phage AR1. J. Virol. 2011, 85, 6567–6578. [Google Scholar] [CrossRef][Green Version]
- Heiberger, R.M.; Neuwirth, E. One-way Anova. In R through Excel; Springer: Berlin/Heidelberg, Germany, 2009; pp. 165–191. [Google Scholar]
- Bardina, C.; Colom, J.; Spricigo, D.A.; Otero, J.; Sanchez-Osuna, M.; Cortes, P.; Llagostera, M. Genomics of Three New Bacteriophages Useful in the Biocontrol of Salmonella. Front. Microbiol. 2016, 7, 545. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Cuervo, A.; Pulido-Cid, M.; Chagoyen, M.; Arranz, R.; González-García, V.A.; Garcia-Doval, C.; Castón, J.R.; Valpuesta, J.M.; van Raaij, M.J.; Martín-Benito, J.; et al. Structural characterization of the bacteriophage T7 tail machinery. J. Biol. Chem. 2013, 288, 26290–26299. [Google Scholar] [CrossRef]
- Yoshikawa, G.; Askora, A.; Blanc-Mathieu, R.; Kawasaki, T.; Li, Y.; Nakano, M.; Ogata, H.; Yamada, T. Xanthomonas citri jumbo phage XacN1 exhibits a wide host range and high complement of tRNA genes. Sci. Rep. 2018, 8, 4486. [Google Scholar] [CrossRef]
- Pyra, A.; Brzozowska, E.; Pawlik, K.; Gamian, A.; Dauter, M.; Dauter, Z. Tail tubular protein A: A dual-function tail protein of Klebsiella pneumoniae bacteriophage KP32. Sci. Rep. 2017, 7, 2223. [Google Scholar] [CrossRef]
- Pyra, A.; Urbańska, N.; Filik, K.; Tyrlik, K.; Brzozowska, E. Biochemical features of the novel Tail Tubular Protein A of Yersinia phage phiYeO3-12. Sci. Rep. 2020, 10, 4196. [Google Scholar] [CrossRef]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B. Bacteriophages and Phage-Derived Proteins—Application Approaches. Curr. Med. Chem. 2015, 22, 1757–1773. [Google Scholar] [CrossRef] [PubMed]
- Pyra, A.; Filik, K.; Szermer-Olearnik, B.; Czarny, A.; Brzozowska, E. New Insights on the Feature and Function of Tail Tubular Protein B and Tail Fiber Protein of the Lytic Bacteriophage φYeO3-12 Specific for Yersinia enterocolitica Serotype O:3. Molecules 2020, 25, 4392. [Google Scholar] [CrossRef] [PubMed]
- CDC. Cronobacter Infection and Infants. Available online: https://www.cdc.gov/cronobacter/infection-and-infants.html (accessed on 30 September 2022).
- Chakraborty, S.; von Mentzer, A.; Begum, Y.A.; Manzur, M.; Hasan, M.; Ghosh, A.N.; Hossain, M.A.; Camilli, A.; Qadri, F. Phenotypic and genomic analyses of bacteriophages targeting environmental and clinical CS3-expressing enterotoxigenic Escherichia coli (ETEC) strains. PLoS ONE 2018, 13, e0209357. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Olia, A.S.; Cingolani, G. Architecture of viral genome-delivery molecular machines. Curr. Opin. Struct. Biol. 2014, 25, 1–8. [Google Scholar] [CrossRef]
- Abedon, S.T.; Herschler, T.D.; Stopar, D. Bacteriophage latent-period evolution as a response to resource availability. Appl. Environ. Microbiol. 2001, 67, 4233–4241. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Kang, C. Lysis delay and burst shrinkage of coliphage T7 by deletion of terminator Tφ reversed by deletion of early genes. J. Virol. 2014, 88, 2107–2115. [Google Scholar] [CrossRef]
- Abedon, S.T. Selection for bacteriophage latent period length by bacterial density: A theoretical examination. Microb. Ecol. 1989, 18, 79–88. [Google Scholar] [CrossRef]
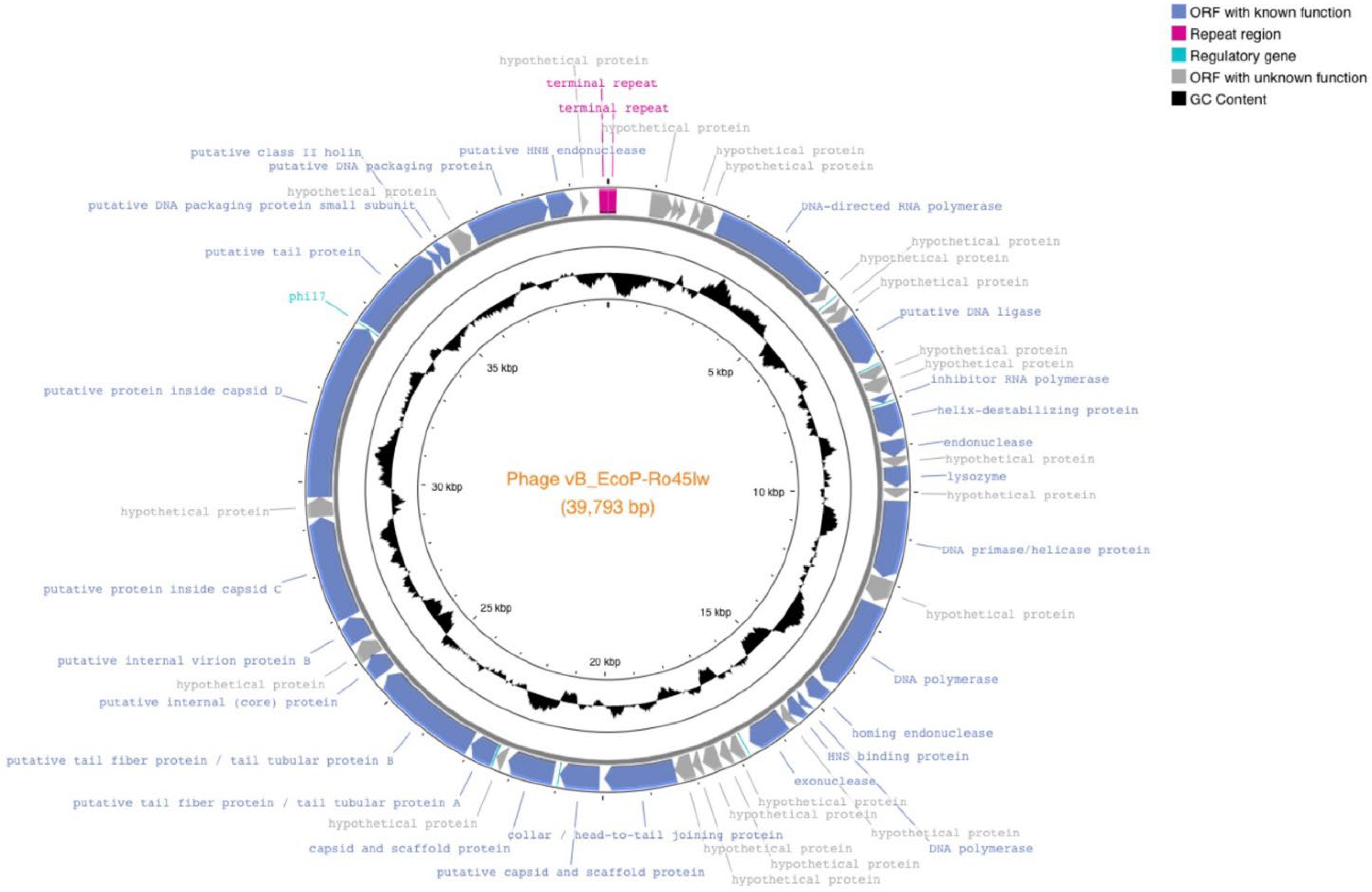


| Type | Predicted Function | Functional Category | Minimum bp | Max bp |
|---|---|---|---|---|
| Repeat region | terminal repeat region | - | 1 | 190 |
| ORF_6 | DNA-directed RNA polymerase CDS | DNA replication | 2498 | 5179 |
| regulatory | predicted promoter regulatory | - | 5483 | 5505 |
| ORF_10 | putative DNA ligase CDS | DNA replication | 6026 | 7093 |
| regulatory | phi1.6 regulatory | - | 7178 | 7200 |
| ORF_13 | inhibitor RNA polymerase CDS | DNA replication | 7864 | 8022 |
| regulatory | phi2.5 regulatory | - | 8018 | 8040 |
| ORF_14 | helix-destabilizing protein CDS | DNA replication | 8080 | 8778 |
| ORF_15 | endonuclease CDS | DNA replication | 8859 | 9233 |
| ORF_17 | lysozyme CDS | Cell lysis | 9450 | 9908 |
| ORF_19 | DNA primase/helicase protein CDS | DNA replication | 10,203 | 11,903 |
| ORF_21 | DNA polymerase CDS | DNA replication | 12,523 | 14,541 |
| ORF_22 | homing endonuclease CDS | DNA replication | 14,621 | 15,016 |
| ORF_23 | DNA polymerase CDS | DNA replication | 15,119 | 15,292 |
| ORF_24 | HNS binding protein CDS | DNA replication | 15,292 | 15,582 |
| ORF_26 | exonuclease CDS | DNA replication | 15,785 | 16,651 |
| regulatory | phi6.5 regulatory | - | 16,774 | 16,796 |
| ORF_32 | collar/head-to-tail joining protein CDS | Structural protein | 18,416 | 19,984 |
| ORF_33 | putative capsid and scaffold protein CDS | Structural protein | 20,081 | 20,962 |
| regulatory | phi regulatory | - | 20,967 | 20,989 |
| ORF_34 | capsid and scaffold protein CDS | Structural protein | 21,093 | 22,142 |
| regulatory | phiTE regulatory | - | 22,409 | 22,444 |
| ORF_36 | putative tail fiber protein/tail tubular protein A CDS | Structural protein | 22,464 | 23,030 |
| ORF_37 | putative tail fiber protein/tail tubular protein B CDS | Structural protein | 23,042 | 25,411 |
| ORF_38 | putative internal (core) protein CDS | Structural protein | 25,498 | 25,980 |
| ORF_40 | putative internal virion protein B CDS | Structural protein | 26,381 | 26,944 |
| ORF_41 | putative protein inside capsid C CDS | Structural protein | 26,956 | 29,238 |
| ORF_43 | putative protein inside capsid D CDS | Structural protein | 29,675 | 33,571 |
| regulatory | phi17 regulatory | - | 33,567 | 33,589 |
| ORF_44 | putative tail protein | Structural protein | 33,637 | 35,646 |
| ORF_45 | putative class II holin CDS | Cell lysis | 35,662 | 35,856 |
| ORF_46 | putative DNA packaging protein small subunit CDS | DNA packaging | 35,853 | 36,116 |
| ORF_48 | putative DNA packaging protein CDS | DNA packaging | 36,710 | 38,476 |
| ORF_49 | putative HNH endonuclease CDS | DNA replication | 38,486 | 39,028 |
| Repeat region | terminal repeat region | - | 39,604 | 39,793 |
| Group | Strain ID | EOP α |
|---|---|---|
| Non-O157 STEC | STEC O26, O103, O111, O121 and O145 | R * |
| E. coli O45:H- (RM10729) | H ^ | |
| E. coli O45:H2 (SJ7) | 0.10 | |
| E. coli O45:H16 (RM13752) | 0.77 | |
| E. coli O45:H16 (RM13745) | 0.84 | |
| STEC O157 | E. coli O157:H7 (RM18959, RM18974 & ATCC 43888) | R |
| Generic E. coli | ATCC 13706 | Inefficiency |
| DH5a | R | |
| Salmonella enterica | Salmonella Newport | R |
| Salmonella Enteritidis | R | |
| Salmonella Typhimurium | R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Liao, Y.-T.; Zhang, Y.; Salvador, A.; Ho, K.-J.; Wu, V.C.H. A New Kayfunavirus-like Escherichia Phage vB_EcoP-Ro45lw with Antimicrobial Potential of Shiga Toxin-Producing Escherichia coli O45 Strain. Microorganisms 2023, 11, 77. https://doi.org/10.3390/microorganisms11010077
Sun X, Liao Y-T, Zhang Y, Salvador A, Ho K-J, Wu VCH. A New Kayfunavirus-like Escherichia Phage vB_EcoP-Ro45lw with Antimicrobial Potential of Shiga Toxin-Producing Escherichia coli O45 Strain. Microorganisms. 2023; 11(1):77. https://doi.org/10.3390/microorganisms11010077
Chicago/Turabian StyleSun, Xincheng, Yen-Te Liao, Yujie Zhang, Alexandra Salvador, Kan-Ju Ho, and Vivian C. H. Wu. 2023. "A New Kayfunavirus-like Escherichia Phage vB_EcoP-Ro45lw with Antimicrobial Potential of Shiga Toxin-Producing Escherichia coli O45 Strain" Microorganisms 11, no. 1: 77. https://doi.org/10.3390/microorganisms11010077
APA StyleSun, X., Liao, Y.-T., Zhang, Y., Salvador, A., Ho, K.-J., & Wu, V. C. H. (2023). A New Kayfunavirus-like Escherichia Phage vB_EcoP-Ro45lw with Antimicrobial Potential of Shiga Toxin-Producing Escherichia coli O45 Strain. Microorganisms, 11(1), 77. https://doi.org/10.3390/microorganisms11010077






