Epidemiology and Characteristics of SARS-CoV-2 Variants of Concern: The Impacts of the Spike Mutations
Abstract
1. Introduction
1.1. Genome Organization and Proteins of SARS-CoV-2
1.2. The Viral Cycle of SARS-CoV-2
2. Genetic Variability of SARS-CoV-2
Classification and Nomenclature of the Different Variants
3. Circulation of SARS-CoV-2 Variants
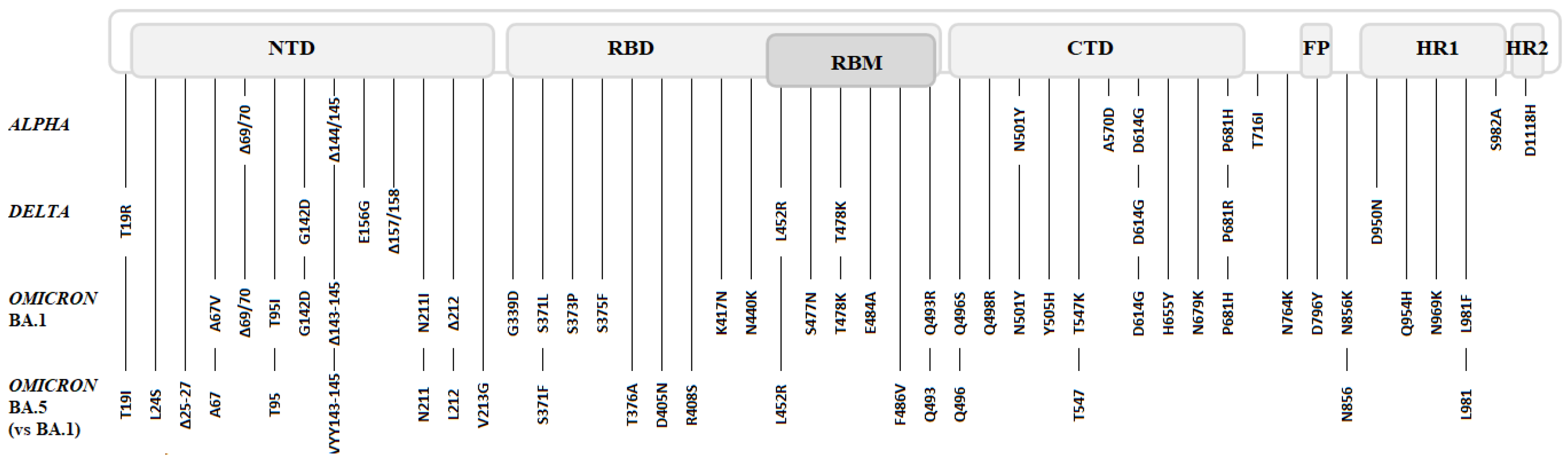
4. The Main Variants of SARS-CoV-2: Spike Mutations and Their Impacts
4.1. Clade B.1: D614G
4.2. ALPHA Variant (B.1.1.7)
4.3. BETA (B.1.351) and GAMMA (P.1) Variants
4.4. DELTA Variant (B.1.617.2 and AY Lineages)
4.5. OMICRON Variant (BA.1)
4.6. Omicron’s Sublineages (BA.2 to BA.5)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ogando, N.S.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Bredenbeek, P.J.; Posthuma, C.C.; Snijder, E.J. The Enzymatic Activity of the Nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020, 94, e01246-20. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Walls, A.C.; Tortorici, M.A.; Snijder, J.; Xiong, X.; Bosch, B.-J.; Rey, F.A.; Veesler, D. Tectonic Conformational Changes of a Coronavirus Spike Glycoprotein Promote Membrane Fusion. Proc. Natl. Acad. Sci. USA 2017, 114, 11157–11162. [Google Scholar] [CrossRef]
- Fan, X.; Cao, D.; Kong, L.; Zhang, X. Cryo-EM Analysis of the Post-Fusion Structure of the SARS-CoV Spike Glycoprotein. Nat. Commun. 2020, 11, 3618. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, Y.; Xiao, T.; Lu, J.; Peng, H.; Sterling, S.M.; Walsh, R.M.; Rits-Volloch, S.; Zhu, H.; Woosley, A.N.; et al. Structural Impact on SARS-CoV-2 Spike Protein by D614G Substitution. Science 2021, 372, 525–530. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e13. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A Neutralizing Human Antibody Binds to the N-Terminal Domain of the Spike Protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Huang, Y.; Lau, S.K.P.; Yuen, K.-Y. Coronavirus Genomics and Bioinformatics Analysis. Viruses 2010, 2, 1804–1820. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Zemelman, B.V.; McNew, J.A.; Westermann, B.; Gmachl, M.; Parlati, F.; Söllner, T.H.; Rothman, J.E. SNAREpins: Minimal Machinery for Membrane Fusion. Cell 1998, 92, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 Infects Cells after Viral Entry via Clathrin-Mediated Endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Xiong, X.; Park, Y.-J.; Tortorici, M.A.; Snijder, J.; Quispe, J.; Cameroni, E.; Gopal, R.; Dai, M.; Lanzavecchia, A.; et al. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell 2019, 176, 1026–1039.e15. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, S.G.; Sawicki, D.L. Coronaviruses Use Discontinuous Extension for Synthesis of Subgenome-Length Negative Strands. In Corona- and Related Viruses; Talbot, P.J., Levy, G.A., Eds.; Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 1995; Volume 380, pp. 499–506. ISBN 978-1-4613-5775-9. [Google Scholar]
- Sztuba-Solińska, J.; Stollar, V.; Bujarski, J.J. Subgenomic Messenger RNAs: Mastering Regulation of (+)-Strand RNA Virus Life Cycle. Virology 2011, 412, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Mistry, P.; Barmania, F.; Mellet, J.; Peta, K.; Strydom, A.; Viljoen, I.M.; James, W.; Gordon, S.; Pepper, M.S. SARS-CoV-2 Variants, Vaccines, and Host Immunity. Front. Immunol. 2022, 12, 809244. [Google Scholar] [CrossRef]
- Papanikolaou, V.; Chrysovergis, A.; Ragos, V.; Tsiambas, E.; Katsinis, S.; Manoli, A.; Papouliakos, S.; Roukas, D.; Mastronikolis, S.; Peschos, D.; et al. From Delta to Omicron: S1-RBD/S2 Mutation/Deletion Equilibrium in SARS-CoV-2 Defined Variants. Gene 2022, 814, 146134. [Google Scholar] [CrossRef]
- World Health Organization Website. Available online: www.who.int/fr (accessed on 20 November 2022).
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Johns Hopkins Coronavirus Resource Center. Available online: https://coronavirus.jhu.edu/map.html (accessed on 20 November 2022).
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Galloway, S.E.; Paul, P.; MacCannell, D.R.; Johansson, M.A.; Brooks, J.T.; MacNeil, A.; Slayton, R.B.; Tong, S.; Silk, B.J.; Armstrong, G.L.; et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage—United States, December 29, 2020–January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Weaver, S.; Tegally, H.; San, J.E.; Shank, S.D.; Wilkinson, E.; Lucaci, A.G.; Giandhari, J.; Naidoo, S.; Pillay, Y.; et al. The Emergence and Ongoing Convergent Evolution of the SARS-CoV-2 N501Y Lineages. Cell 2021, 184, 5189–5200.e7. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D. da S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and Epidemiology of the P.1 SARS-CoV-2 Lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Weekly Epidemiological Update: Omicron Variant of Concern (VOC)—Week 1 (Data as of 7 January 2022) EU/EEA. Available online: Https://Www.Ecdc.Europa.Eu/En/News-Events/Weekly-Epidemiological-Update-Omicron-Variant-Concern-Voc-Week-1-Data-7-January-2022 (accessed on 7 January 2022).
- European Centre for Disease Prevention and Control. Spread of the SARS-CoV-2 Omicron Variant Sub-Lineage BQ.1 in the EU/EEA. Available online: https://www.ecdc.europa.eu/en/publications-data/spread-sars-cov-2-omicron-variant-sub-lineage-bq1-eueea (accessed on 20 November 2022).
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Omotuyi, I.O.; Nash, O.; Ajiboye, O.B.; Iwegbulam, C.G.; Oyinloye, E.B.; Oyedeji, O.A.; Kashim, Z.A.; Okaiyeto, K. Atomistic Simulation Reveals Structural Mechanisms Underlying D614G Spike Glycoprotein-enhanced Fitness in SARS-COV-2. J. Comput. Chem. 2020, 41, 2158–2161. [Google Scholar] [CrossRef]
- Zhou, B.; Thao, T.T.N.; Hoffmann, D.; Taddeo, A.; Ebert, N.; Labroussaa, F.; Pohlmann, A.; King, J.; Portmann, J.; Halwe, N.J.; et al. SARS-CoV-2 Spike D614G Variant Confers Enhanced Replication and Transmissibility. Nature 2021, 592, 122–127. [Google Scholar] [CrossRef]
- Gobeil, S.M.-C.; Janowska, K.; McDowell, S.; Mansouri, K.; Parks, R.; Manne, K.; Stalls, V.; Kopp, M.F.; Henderson, R.; Edwards, R.J.; et al. D614G Mutation Alters SARS-CoV-2 Spike Conformation and Enhances Protease Cleavage at the S1/S2 Junction. Cell Rep. 2021, 34, 108630. [Google Scholar] [CrossRef]
- Ozono, S.; Zhang, Y.; Ode, H.; Sano, K.; Tan, T.S.; Imai, K.; Miyoshi, K.; Kishigami, S.; Ueno, T.; Iwatani, Y.; et al. SARS-CoV-2 D614G Spike Mutation Increases Entry Efficiency with Enhanced ACE2-Binding Affinity. Nat. Commun. 2021, 12, 848. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Peng, H.; Quinlan, B.D.; Rangarajan, E.S.; Pan, A.; Vanderheiden, A.; Suthar, M.S.; et al. SARS-CoV-2 Spike-Protein D614G Mutation Increases Virion Spike Density and Infectivity. Nat. Commun. 2020, 11, 6013. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike Mutation D614G Alters SARS-CoV-2 Fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef]
- Weissman, D.; Alameh, M.-G.; de Silva, T.; Collini, P.; Hornsby, H.; Brown, R.; LaBranche, C.C.; Edwards, R.J.; Sutherland, L.; Santra, S.; et al. D614G Spike Mutation Increases SARS-CoV-2 Susceptibility to Neutralization. Cell Host Microbe 2021, 29, 23–31.e4. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zheng, Y.; Niu, Z.; Jiang, X.; Sun, Q. The Virological Impacts of SARS-CoV-2 D614G Mutation. J. Mol. Cell Biol. 2021, 13, 712–720. [Google Scholar] [CrossRef]
- Meng, B.; Kemp, S.A.; Papa, G.; Datir, R.; Ferreira, I.A.T.M.; Marelli, S.; Harvey, W.T.; Lytras, S.; Mohamed, A.; Gallo, G.; et al. Recurrent Emergence of SARS-CoV-2 Spike Deletion H69/V70 and Its Role in the Alpha Variant B.1.1.7. Cell Rep. 2021, 35, 109292. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, K.R.; Rennick, L.J.; Nambulli, S.; Robinson-McCarthy, L.R.; Bain, W.G.; Haidar, G.; Duprex, W.P. Recurrent Deletions in the SARS-CoV-2 Spike Glycoprotein Drive Antibody Escape. Science 2021, 371, 1139–1142. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.; Shum, M.H.; Leung, G.M.; Lam, T.T.; Wu, J.T. Early Transmissibility Assessment of the N501Y Mutant Strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eurosurveillance 2021, 26, 2002106. [Google Scholar] [CrossRef]
- Zahradník, J.; Marciano, S.; Shemesh, M.; Zoler, E.; Harari, D.; Chiaravalli, J.; Meyer, B.; Rudich, Y.; Li, C.; Marton, I.; et al. SARS-CoV-2 Variant Prediction and Antiviral Drug Design Are Enabled by RBD in Vitro Evolution. Nat. Microbiol. 2021, 6, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Zia, T.; Suleman, M.; Khan, T.; Ali, S.S.; Abbasi, A.A.; Mohammad, A.; Wei, D. Higher Infectivity of the SARS-CoV-2 New Variants Is Associated with K417N/T, E484K, and N501Y Mutants: An Insight from Structural Data. J. Cell. Physiol. 2021, 236, 7045–7057. [Google Scholar] [CrossRef]
- Maison, D.P.; Ching, L.L.; Shikuma, C.M.; Nerurkar, V.R. Genetic Characteristics and Phylogeny of 969-bp S Gene Sequence of SARS-CoV-2 from Hawai’i Reveals the Worldwide Emerging P681H Mutation. Hawaii J. Health Soc. Welf. 2021, 80, 52–61. [Google Scholar]
- Zhu, Y.; Feng, F.; Hu, G.; Wang, Y.; Yu, Y.; Zhu, Y.; Xu, W.; Cai, X.; Sun, Z.; Han, W.; et al. A Genome-Wide CRISPR Screen Identifies Host Factors That Regulate SARS-CoV-2 Entry. Nat. Commun. 2021, 12, 961. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Lubinski, B.; Fernandes, M.H.V.; Frazier, L.; Tang, T.; Daniel, S.; Diel, D.G.; Jaimes, J.A.; Whittaker, G.R. Functional Evaluation of the P681H Mutation on the Proteolytic Activation of the SARS-CoV-2 Variant B.1.1.7 (Alpha) Spike. iScience 2022, 25, 103589. [Google Scholar] [CrossRef] [PubMed]
- Lista, M.J.; Winstone, H.; Wilson, H.D.; Dyer, A.; Pickering, S.; Galao, R.P.; De Lorenzo, G.; Cowton, V.M.; Furnon, W.; Suarez, N.; et al. The P681H Mutation in the Spike Glycoprotein of the Alpha Variant of SARS-CoV-2 Escapes IFITM Restriction and Is Necessary for Type I Interferon Resistance. J. Virol. 2022, 96, e01250-22. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Jang, U.S.; Soh, S.M.; Lee, J.-Y.; Lee, H.-R. The Impact on Infectivity and Neutralization Efficiency of SARS-CoV-2 Lineage B.1.351 Pseudovirus. Viruses 2021, 13, 633. [Google Scholar] [CrossRef]
- Fujino, T.; Nomoto, H.; Kutsuna, S.; Ujiie, M.; Suzuki, T.; Sato, R.; Fujimoto, T.; Kuroda, M.; Wakita, T.; Ohmagari, N. Novel SARS-CoV-2 Variant in Travelers from Brazil to Japan. Emerg. Infect. Dis. 2021, 27, 1243–1245. [Google Scholar] [CrossRef]
- Jawad, B.; Adhikari, P.; Podgornik, R.; Ching, W.-Y. Key Interacting Residues between RBD of SARS-CoV-2 and ACE2 Receptor: Combination of Molecular Dynamics Simulation and Density Functional Calculation. J. Chem. Inf. Model. 2021, 61, 4425–4441. [Google Scholar] [CrossRef]
- Yuan, M.; Huang, D.; Lee, C.-C.D.; Wu, N.C.; Jackson, A.M.; Zhu, X.; Liu, H.; Peng, L.; van Gils, M.J.; Sanders, R.W.; et al. Structural and Functional Ramifications of Antigenic Drift in Recent SARS-CoV-2 Variants. Science 2021, 373, 818–823. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 Escapes Neutralization by South African COVID-19 Donor Plasma. Nat. Med. 2021, 27, 622–625. [Google Scholar] [CrossRef]
- Network for Genomic Surveillance in South Africa; COMMIT-KZN Team; Cele, S.; Gazy, I.; Jackson, L.; Hwa, S.-H.; Tegally, H.; Lustig, G.; Giandhari, J.; Pillay, S.; et al. Escape of SARS-CoV-2 501Y.V2 from Neutralization by Convalescent Plasma. Nature 2021, 593, 142–146. [Google Scholar] [CrossRef]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 Variants to Neutralization by Monoclonal and Serum-Derived Polyclonal Antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, Z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D.; et al. Delta Spike P681R Mutation Enhances SARS-CoV-2 Fitness over Alpha Variant. Cell Rep. 2022, 39, 110829. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Baral, P.; Bhattarai, N.; Hossen, M.L.; Stebliankin, V.; Gerstman, B.S.; Narasimhan, G.; Chapagain, P.P. Mutation-Induced Changes in the Receptor-Binding Interface of the SARS-CoV-2 Delta Variant B.1.617.2 and Implications for Immune Evasion. Biochem. Biophys. Res. Commun. 2021, 574, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Singh, J.; Hasnain, S.E.; Sundar, D. Possible Link between Higher Transmissibility of Alpha, Kappa and Delta Variants of SARS-CoV-2 and Increased Structural Stability of Its Spike Protein and HACE2 Affinity. Int. J. Mol. Sci. 2021, 22, 9131. [Google Scholar] [CrossRef]
- McCallum, M.; Bassi, J.; De Marco, A.; Chen, A.; Walls, A.C.; Di Iulio, J.; Tortorici, M.A.; Navarro, M.-J.; Silacci-Fregni, C.; Saliba, C.; et al. SARS-CoV-2 Immune Evasion by the B.1.427/B.1.429 Variant of Concern. Science 2021, 373, 648–654. [Google Scholar] [CrossRef]
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, Infectivity, and Neutralization of a Spike L452R SARS-CoV-2 Variant. Cell 2021, 184, 3426–3437.e8. [Google Scholar] [CrossRef]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; et al. SARS-CoV-2 Spike L452R Variant Evades Cellular Immunity and Increases Infectivity. Cell Host Microbe 2021, 29, 1124–1136.e11. [Google Scholar] [CrossRef]
- Kiyotani, K.; Toyoshima, Y.; Nemoto, K.; Nakamura, Y. Bioinformatic Prediction of Potential T Cell Epitopes for SARS-CoV-2. J. Hum. Genet. 2020, 65, 569–575. [Google Scholar] [CrossRef]
- Di Giacomo, S.; Mercatelli, D.; Rakhimov, A.; Giorgi, F.M. Preliminary Report on Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Spike Mutation T478K. J. Med. Virol. 2021, 93, 5638–5643. [Google Scholar] [CrossRef]
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S.; et al. SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms 2021, 9, 1542. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced Fusogenicity and Pathogenicity of SARS-CoV-2 Delta P681R Mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 Usage by SARS-CoV-2 Omicron Impacts Infectivity and Fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef] [PubMed]
- McCallum, M.; Czudnochowski, N.; Rosen, L.E.; Zepeda, S.K.; Bowen, J.E.; Walls, A.C.; Hauser, K.; Joshi, A.; Stewart, C.; Dillen, J.R.; et al. Structural Basis of SARS-CoV-2 Omicron Immune Evasion and Receptor Engagement. Science 2022, 375, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron Escapes the Majority of Existing SARS-CoV-2 Neutralizing Antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhou, J.; Tian, M.; Huang, M.; Liu, S.; Xie, Y.; Han, P.; Bai, C.; Han, P.; Zheng, A.; et al. Omicron SARS-CoV-2 Mutations Stabilize Spike up-RBD Conformation and Lead to a Non-RBM-Binding Monoclonal Antibody Escape. Nat. Commun. 2022, 13, 4958. [Google Scholar] [CrossRef]
- Nyberg, T.; Ferguson, N.M.; Nash, S.G.; Webster, H.H.; Flaxman, S.; Andrews, N.; Hinsley, W.; Bernal, J.L.; Kall, M.; Bhatt, S.; et al. Comparative Analysis of the Risks of Hospitalisation and Death Associated with SARS-CoV-2 Omicron (B.1.1.529) and Delta (B.1.617.2) Variants in England: A Cohort Study. Lancet 2022, 399, 1303–1312. [Google Scholar] [CrossRef]
- Lubinski, B.; Jaimes, J.A.; Whittaker, G.R. Intrinsic Furin-Mediated Cleavability of the Spike S1/S2 Site from SARS-CoV-2 Variant B.1.1.529 (Omicron). BioRxiv 2022. [Google Scholar] [CrossRef]
- Wang, L.; Kainulainen, M.H.; Jiang, N.; Di, H.; Bonenfant, G.; Mills, L.; Currier, M.; Shrivastava-Ranjan, P.; Calderon, B.M.; Sheth, M.; et al. Differential Neutralization and Inhibition of SARS-CoV-2 Variants by Antibodies Elicited by COVID-19 MRNA Vaccines. Nat. Commun. 2022, 13, 4350. [Google Scholar] [CrossRef]
- Iketani, S.; Liu, L.; Guo, Y.; Liu, L.; Chan, J.F.-W.; Huang, Y.; Wang, M.; Luo, Y.; Yu, J.; Chu, H.; et al. Antibody Evasion Properties of SARS-CoV-2 Omicron Sublineages. Nature 2022, 604, 553–556. [Google Scholar] [CrossRef]
- Bruel, T.; Pinaud, L.; Tondeur, L.; Planas, D.; Staropoli, I.; Porrot, F.; Guivel-Benhassine, F.; Attia, M.; Pelleau, S.; Woudenberg, T.; et al. Neutralising Antibody Responses to SARS-CoV-2 Omicron among Elderly Nursing Home Residents Following a Booster Dose of BNT162b2 Vaccine: A Community-Based, Prospective, Longitudinal Cohort Study. eClinicalMedicine 2022, 51, 101576. [Google Scholar] [CrossRef] [PubMed]
- Takatsuki, Y.; Takahashi, Y.; Nakajima, J.; Iwasaki, Y.; Nagano, K.; Tani-Sassa, C.; Yuasa, S.; Kanehira, S.; Sonobe, K.; Nukui, Y.; et al. Viral Load of SARS-CoV-2 Omicron BA.5 Is Lower than That of BA.2 despite the Higher Infectivity of BA.5. Med. MedRxiv 2022. [Google Scholar] [CrossRef]
- Yamasoba, D.; Kosugi, Y.; Kimura, I.; Fujita, S.; Uriu, K.; Ito, J.; Sato, K. Neutralisation Sensitivity of SARS-CoV-2 Omicron Subvariants to Therapeutic Monoclonal Antibodies. Lancet Infect. Dis. 2022, 22, 942–943. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control (ECDC). SARS-CoV-2 Variants of Concern. Available online: Https://Www.Ecdc.Europa.Eu/En/Covid-19/Variants-Concern (accessed on 20 November 2022).
- Jian, F.; Yu, Y.; Song, W.; Yisimayi, A.; Yu, L.; Gao, Y.; Zhang, N.; Wang, Y.; Shao, F.; Hao, X.; et al. Further Humoral Immunity Evasion of Emerging SARS-CoV-2 BA.4 and BA.5 Subvariants. Lancet Infect. Dis. 2022, 22, 1535–1537. [Google Scholar] [CrossRef]
- Kurhade, C.; Zou, J.; Xia, H.; Liu, M.; Chang, H.C.; Ren, P.; Xie, X.; Shi, P.-Y. Low Neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1, and XBB.1 by 4 Doses of Parental MRNA Vaccine or a BA.5-Bivalent Booster. Nat. Med. 2022. [Google Scholar] [CrossRef]
- Davis-Gardner, M.E.; Lai, L.; Wali, B.; Samaha, H.; Solis, D.; Lee, M.; Porter-Morrison, A.; Hentenaar, I.T.; Yamamoto, F.; Godbole, S.; et al. MRNA Bivalent Booster Enhances Neutralization against BA.2.75.2 and BQ.1.1. BioRxiv 2022. [Google Scholar] [CrossRef]
- Cao, Y.; Jian, F.; Wang, J.; Yu, Y.; Song, W.; Yisimayi, A.; Wang, J.; An, R.; Chen, X.; Zhang, N.; et al. Imprinted SARS-CoV-2 Humoral Immunity Induces Convergent Omicron RBD Evolution. BioRxiv 2022. [CrossRef]
| WHO | Mutations of Interest | ||
|---|---|---|---|
| Classification | Transmissibility * | Virulence *,** | Immune Escape * |
| + | + | ||
| Alpha | |||
| N501Y | Δ69/70, P681H | ||
| + | + | ||
| Beta | |||
| N501Y, E484K, K417N | K417N, E484K | ||
| + | + | ||
| Gamma | |||
| N501Y, E484K, K417T | K417T, E484K | ||
| ++ | +++ | ++ | |
| Delta | |||
| L452R, T478K, P681R | P681R | L452R, T478K, E484Q | |
| +++ | - | +++ | |
| Omicron | S477N, Q493R, | Δ69/70, S371L, S373P, | |
| BA.1 | Q498R, N501Y | S375F, K417N, N440K, | |
| S477N, E484A, Q493R | |||
| Omicron BA.5 | +++ | - | +++ |
| L452R, Q493 | L452R | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cocherie, T.; Zafilaza, K.; Leducq, V.; Marot, S.; Calvez, V.; Marcelin, A.-G.; Todesco, E. Epidemiology and Characteristics of SARS-CoV-2 Variants of Concern: The Impacts of the Spike Mutations. Microorganisms 2023, 11, 30. https://doi.org/10.3390/microorganisms11010030
Cocherie T, Zafilaza K, Leducq V, Marot S, Calvez V, Marcelin A-G, Todesco E. Epidemiology and Characteristics of SARS-CoV-2 Variants of Concern: The Impacts of the Spike Mutations. Microorganisms. 2023; 11(1):30. https://doi.org/10.3390/microorganisms11010030
Chicago/Turabian StyleCocherie, Théophile, Karen Zafilaza, Valentin Leducq, Stéphane Marot, Vincent Calvez, Anne-Geneviève Marcelin, and Eve Todesco. 2023. "Epidemiology and Characteristics of SARS-CoV-2 Variants of Concern: The Impacts of the Spike Mutations" Microorganisms 11, no. 1: 30. https://doi.org/10.3390/microorganisms11010030
APA StyleCocherie, T., Zafilaza, K., Leducq, V., Marot, S., Calvez, V., Marcelin, A.-G., & Todesco, E. (2023). Epidemiology and Characteristics of SARS-CoV-2 Variants of Concern: The Impacts of the Spike Mutations. Microorganisms, 11(1), 30. https://doi.org/10.3390/microorganisms11010030






