Indigenous Yeasts from Rose Oil Distillation Wastewater and Their Capacity for Biotransformation of Phenolics
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of Rose Oil Distillation Wastes
2.2. Collection of Indigenous Yeast and Mold Isolates from RODW and DLRFR Samples
2.3. Isolation of Yeast and Mold Rose Flower Endophytes from Flower Buds of R. damascena Mill
2.4. Common Yeast Strains Used for Fermentation of RODW
2.5. Molecular Identification of Yeast and Mold Isolates from RODW, DLRFR and RFE Samples
2.6. Fermentation of RODW with Indigenous Yeast Isolates and Common Yeast Strains
2.7. Phenolics Extraction from RODW Samples
2.8. Analytical Methods
2.8.1. Total Phenolic Content
2.8.2. Reducing Sugar Content
2.8.3. HPLC/UV Analysis
2.8.4. Statistical Analysis
3. Results
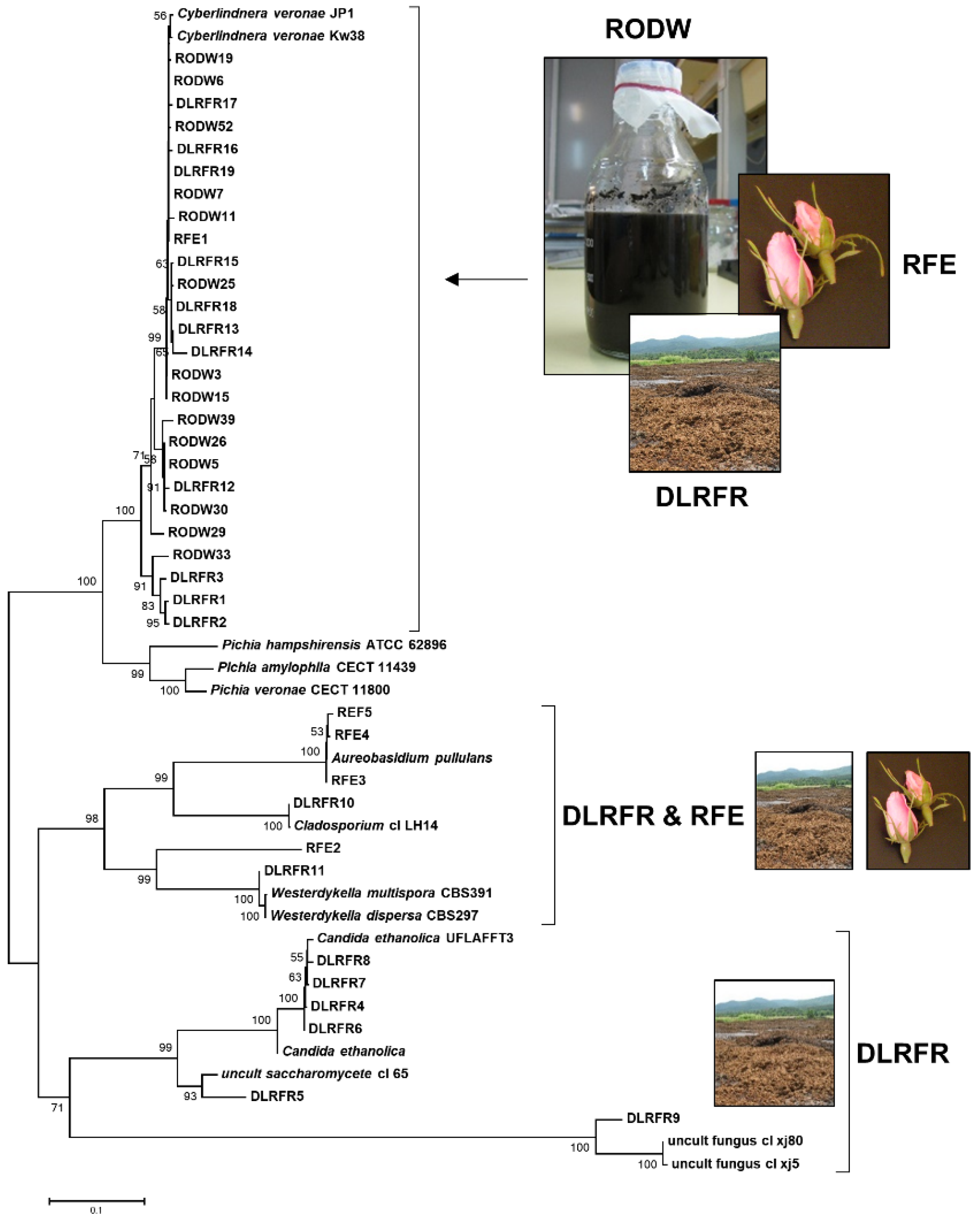
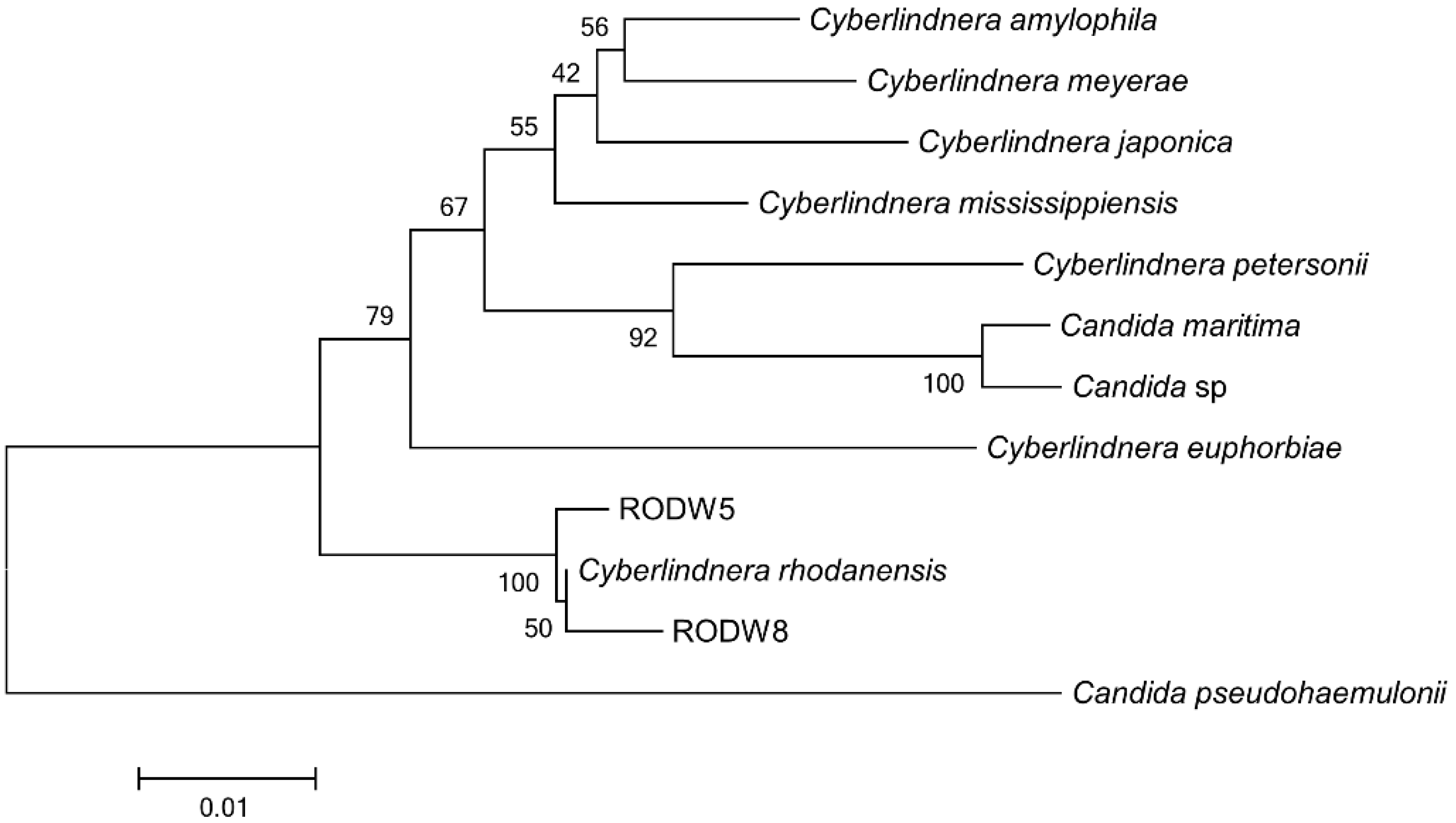
3.1. Molecular Characterization of Yeast and Mold Diversity of Rose Oil Distillation Wastes and Rose Flower Endophytes
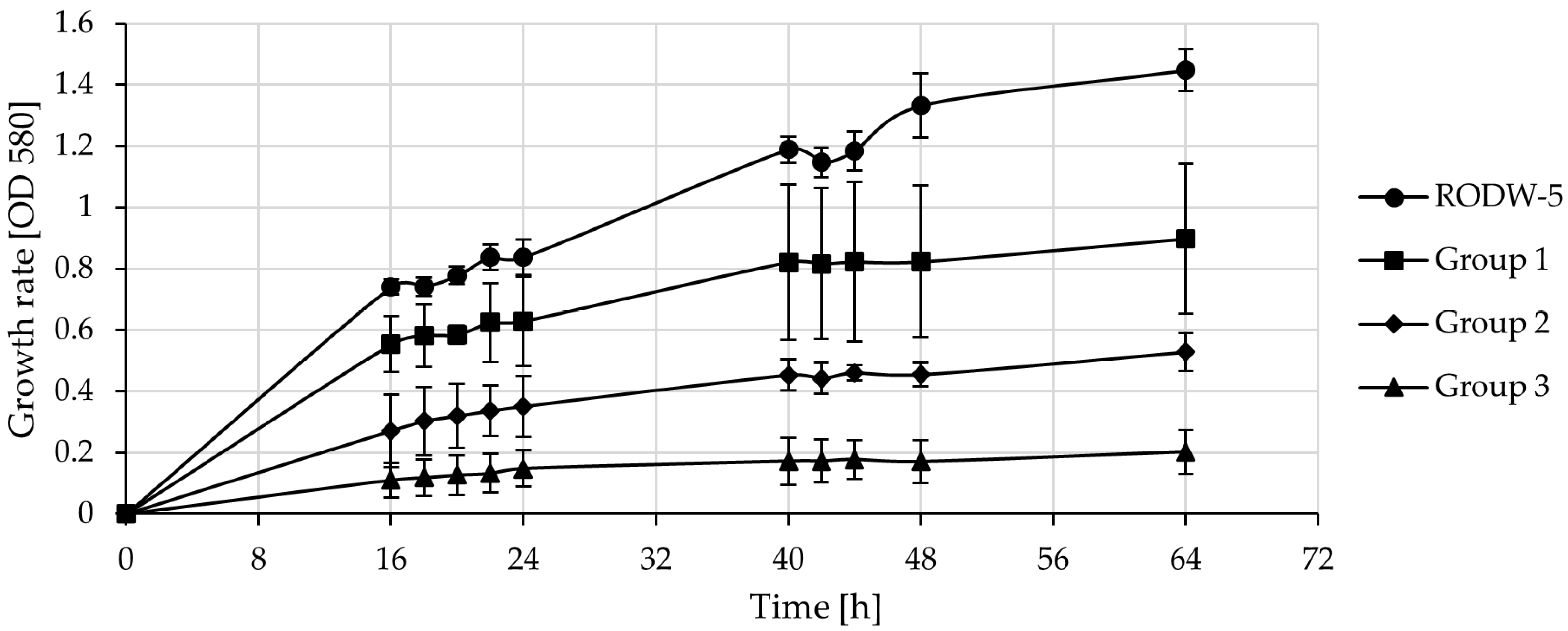
3.2. Cultivation of Indigenous Yeast RODW Isolates in RODW Media
3.3. Cultivation of Indigenous RODW Isolates and Common Yeast Strains in RODW Media
3.4. RODW Phenolic Biotransformation after Fermentation by Indigenous Rodw Yeast Isolates and Common Yeast Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kovacheva, N.; Rusanov, K.; Atanassov, I. Industrial cultivation of oil bearing rose and rose oil production in Bulgaria during 21st century, directions and challenges. Biotechnol. Biotechnol. Equip. 2010, 24, 1793–1798. [Google Scholar] [CrossRef]
- Rusanov, K.; Garo, E.; Rusanova, M.; Fertig, O.; Hamburger, M.; Atanassov, I.; Butterweck, V. Recovery of polyphenols from rose oil distillation wastewater using adsorption resins—A pilot study. Planta Med. 2014, 80, 1657–1664. [Google Scholar] [CrossRef] [PubMed]
- Slavov, A.; Vasileva, I.; Stefanov, L.; Stoyanova, A. Valorization of wastes from the rose oil industry. Rev. Environ. Sci. Bio/Technol. 2017, 16, 309–325. [Google Scholar] [CrossRef]
- Ilieva, Y.; Dimitrova, L.; Georgieva, A.; Vilhelmova-Ilieva, N.; Zaharieva, M.M.; Kokanova-Nedialkova, Z.; Dobreva, A.; Nedialkov, P.; Kussovski, V.; Kroumov, A.D. In Vitro Study of the Biological Potential of Wastewater Obtained after the Distillation of Four Bulgarian Oil-Bearing Roses. Plants 2022, 11, 1073. [Google Scholar] [CrossRef]
- Solimine, J.; Garo, E.; Wedler, J.; Rusanov, K.; Fertig, O.; Hamburger, M.; Atanassov, I.; Butterweck, V. Tyrosinase inhibitory constituents from a polyphenol enriched fraction of rose oil distillation wastewater. Fitoterapia 2016, 108, 13–19. [Google Scholar] [CrossRef]
- Wedler, J.; Rusanov, K.; Atanassov, I.; Butterweck, V. A polyphenol-enriched fraction of rose oil distillation wastewater inhibits cell proliferation, migration and TNF-α-induced VEGF secretion in human immortalized keratinocytes. Planta Med. 2016, 82, 1000–1008. [Google Scholar] [CrossRef]
- Amaral, C.; Anjos, R.; Pais, C.; Musculo, A. Bioremediation of olive mill wastewaters with fungi. Terr. Aquat. Environ. Toxicol. 2010, 4, 45–56. [Google Scholar]
- Hamimed, S.; Landoulsi, A.; Chatti, A. The bright side of olive mill wastewater: Valuables bioproducts after bioremediation. Int. J. Environ. Sci. Technol. 2021, 18, 4053–4074. [Google Scholar] [CrossRef]
- Hladnik, L.; Vicente, F.A.; Grilc, M.; Likozar, B. β-Carotene production and extraction: A case study of olive mill wastewater bioremediation by Rhodotorula glutinis with simultaneous carotenoid production. Biomass Convers. Biorefin. 2022, 1–9. [Google Scholar] [CrossRef]
- Iwissat, S.; Rahil, M.; Shahin, N.; Abuamsha, R.; Hilal, H.S.; Zyoud, A.; Nassar, I.; Voogt, W.; Kujawa, K.; Salman, M. Fungus-based bioremediation of olive mill wastewater and potential use in horticulture. Water Environ. J. 2022, 36, 380–386. [Google Scholar] [CrossRef]
- Martínez-Gallardo, M.R.; López, M.J.; López-González, J.A.; Jurado, M.M.; Suárez-Estrella, F.; Pérez-Murcia, M.D.; Sáez, J.A.; Moral, R.; Moreno, J. Microbial communities of the olive mill wastewater sludge stored in evaporation ponds: The resource for sustainable bioremediation. J. Environ. Manag. 2021, 279, 111810. [Google Scholar] [CrossRef] [PubMed]
- Di Maio, S.; Polizzotto, G.; Di Gangi, E.; Foresta, G.; Genna, G.; Verzera, A.; Scacco, A.; Amore, G.; Oliva, D. Biodiversity of indigenous Saccharomyces populations from old wineries of south-eastern Sicily (Italy): Preservation and economic potential. PLoS ONE 2012, 7, e30428. [Google Scholar] [CrossRef]
- McCarthy, G.C.; Morgan, S.C.; Martiniuk, J.T.; Newman, B.L.; McCann, S.E.; Measday, V.; Durall, D.M. An indigenous Saccharomyces uvarum population with high genetic diversity dominates uninoculated Chardonnay fermentations at a Canadian winery. PLoS ONE 2021, 16, e0225615. [Google Scholar] [CrossRef] [PubMed]
- Meersman, E.; Steensels, J.; Mathawan, M.; Wittocx, P.-J.; Saels, V.; Struyf, N.; Bernaert, H.; Vrancken, G.; Verstrepen, K.J. Detailed analysis of the microbial population in Malaysian spontaneous cocoa pulp fermentations reveals a core and variable microbiota. PLoS ONE 2013, 8, e81559. [Google Scholar] [CrossRef]
- Nardi, T. Microbial resources as a tool for enhancing sustainability in winemaking. Microorganisms 2020, 8, 507. [Google Scholar] [CrossRef]
- Steensels, J.; Verstrepen, K.J. Taming wild yeast: Potential of conventional and nonconventional yeasts in industrial fermentations. Annu. Rev. Microbiol. 2014, 68, 61–80. [Google Scholar] [CrossRef]
- Rusanov, K.; Kovacheva, N.; Rusanova, M.; Atanassov, I. Low variability of flower volatiles of Rosa damascena Mill. plants from rose plantations along the Rose Valley, Bulgaria. Ind. Crops Prod. 2012, 37, 6–10. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 2003, 3, 417–432. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventós, R.M. [14] Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1999; Volume 299, pp. 152–178. [Google Scholar]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Bougreau, M.; Ascencio, K.; Bugarel, M.; Nightingale, K.; Loneragan, G. Yeast species isolated from Texas High Plains vineyards and dynamics during spontaneous fermentations of Tempranillo grapes. PLoS ONE 2019, 14, e0216246. [Google Scholar] [CrossRef] [PubMed]
- Montoya, A.C.V.; da Silva Mazareli, R.C.; Delforno, T.P.; Centurion, V.B.; Sakamoto, I.K.; de Oliveira, V.M.; Silva, E.L.; Varesche, M.B.A. Hydrogen, alcohols and volatile fatty acids from the co-digestion of coffee waste (coffee pulp, husk, and processing wastewater) by applying autochthonous microorganisms. Int. J. Hydrog. Energy 2019, 44, 21434–21450. [Google Scholar] [CrossRef]
- Li, Q.; Huang, J.; Li, Y.; Zhang, Y.; Luo, Y.; Chen, Y.; Lin, H.; Wang, K.; Liu, Z. Fungal community succession and major components change during manufacturing process of Fu brick tea. Sci. Rep. 2017, 7, 6947. [Google Scholar] [CrossRef] [PubMed]
- Chuengcharoenphanich, N.; Watsuntorn, W.; Qi, W.; Wang, Z.; Hu, Y.; Chulalaksananukul, W. The potential of biodiesel production from grasses in Thailand through consolidated bioprocessing using a cellulolytic oleaginous yeast, Cyberlindnera rhodanensis CU-CV7. Energy 2023, 263, 125759. [Google Scholar] [CrossRef]
- Danouche, M.; Ferioun, M.; Bahafid, W.; El Ghachtouli, N. Mycoremediation of azo dyes using Cyberlindnera fabianii yeast strain: Application of designs of experiments for decolorization optimization. Water Environ. Res. 2021, 93, 1402–1416. [Google Scholar] [CrossRef]
- Kanpiengjai, A.; Chui-Chai, N.; Chaikaew, S.; Khanongnuch, C. Distribution of tannin-’tolerant yeasts isolated from Miang, a traditional fermented tea leaf (Camellia sinensis var. assamica) in northern Thailand. Int. J. Food Microbiol. 2016, 238, 121–131. [Google Scholar] [CrossRef]
- Murali, D.; Dhandayuthapani, K. Optimization of fermentation of ulva sp. hydrolysate BY novel yeast Cyberlindnera jadinii MMS7 for enhancement of polyphenol content and antioxidant activity. J. Drug Deliv. Ther. 2019, 9, 1–7. [Google Scholar]
- Rodríguez Madrera, R.; Pando Bedriñana, R.; Suárez Valles, B. Evaluation of indigenous non-Saccharomyces cider yeasts for use in brewing. Eur. Food Res. Technol. 2021, 247, 819–828. [Google Scholar] [CrossRef]
- Shi, Q.; Tang, X.; Liu, B.Q.; Liu, W.H.; Li, H.; Luo, Y.Y. Correlation between microbial communities and key odourants in fermented capsicum inoculated with Pediococcus pentosaceus and Cyberlindnera rhodanensis. J. Sci. Food Agric. 2022. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Song, L.; Shao, Y.; Tan, L. Degradation and detoxification of azo dyes by a salt-tolerant yeast Cyberlindnera samutprakarnensis S4 under high-salt conditions. World J. Microbiol. Biotechnol. 2018, 34, 131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Zhou, T.; Zhou, Y. Fungal flora and mycotoxin contamination in tea: Current status, detection methods and dietary risk assessment-A comprehensive review. Trends Food Sci. Technol. 2022, 127, 207–220. [Google Scholar] [CrossRef]

| № | Compound | RODW-5 | Pichia angusta ATCC 34438 | Pichia bispora ATCC 10627 | Pichia guilliermondi ATCC 9058 | Geotrichum fermentans ATCC 10675 | Pichia anomala ATCC 16763 | Zygoascus hellenicus ATCC 15542 |
|---|---|---|---|---|---|---|---|---|
| 1 | Elagic acid | 29 ± 3 ab | 102 ± 26 a | 81 ± 8 ab | 93 ± 8 a | 39 ± 5 a | 92 ± 13 abc | 81 ± 36 abc |
| 2 | Hyperoside | 237 ± 31 d | 131 ± 17 ab | 149 ± 11 ab | 130 ± 10 a | 150 ± 33 bcd | 104 ± 12 abc | 99 ± 27 abcd |
| 3 | Isoquercitrin | 0 ± 0 a | 127 ± 16 ab | 5 ± 1 a | 117 ± 9 a | 86 ± 25 ab | 95 ± 10 abc | 99 ± 32 abcd |
| 4 | Kaempferol-3-O-rutinoside | 225 ± 29 d | 133 ± 18 ab | 155 ± 7 abc | 135 ± 10 ab | 171 ± 19 cd | 108 ± 12 abc | 122 ± 21 bcd |
| 5 | Kaempferol 3-O-galactoside | 269 ± 35 d | 145 ± 26 ab | 171 ± 8 abc | 140 ± 9 ab | 177 ± 19 cd | 106 ± 12 abc | 130 ± 23 cd |
| 6 | Quercetin-O-methyl disaccharide | 211 ± 32 cd | 115 ± 8 ab | 79 ± 4 ab | 128 ± 9 a | 121 ± 8 abc | 132 ± 12 c | 30 ± 5 a |
| 7 | Astragalin (kaempferol 3-O-glucoside) | 17 ± 3 a | 130 ± 16 ab | 29 ± 9 ab | 109 ± 8 a | 136 ± 13 bc | 101 ± 10 abc | 119 ± 19 bcd |
| 8 | Quercitrin | 264 ± 36 d | 128 ± 17 ab | 184 ± 47 bc | 146 ± 11 ab | 175 ± 20 cd | 98 ± 12 abc | 185 ± 33 d |
| 9 | Kaempferol-3-O-xyloside | 213 ± 29 cd | 125 ± 13 ab | 67 ± 4 ab | 134 ± 10 ab | 131 ± 10 bc | 137 ± 14 c | 40 ± 6 ab |
| 10 | Multiflorin B | 119 ± 13 bc | 135 ± 16 ab | 157 ± 15 abc | 127 ± 9 a | 173 ± 20 cd | 103 ± 11 abc | 122 ± 26 bcd |
| 11 | Kaempferol-3-O-arabinoside | 545 ± 70 e | 128 ± 15 ab | 316 ± 24 c | 139 ± 10 ab | 231 ± 28 d | 111 ± 11 abc | 275 ± 54 e |
| 12 | Kaempferol-3-O-rhamnoside | 0 ± 0 a | 125 ± 14 ab | 149 ± 15 ab | 124 ± 10 a | 166 ± 20 bcd | 61 ± 6 ab | 112 ± 16 abcd |
| 13 | Multiflorin A | 0 ± 0 a | 111 ± 16 ab | 127 ± 9 ab | 113 ± 9 a | 141 ± 15 bc | 48 ± 6 a | 137 ± 23 cd |
| 14 | Quercetin | 174 ± 27 cd | 144 ± 17 ab | 626 ± 72 d | 188 ± 21 b | 354 ± 47 e | 124 ± 32 abc | 116 ± 10 abcd |
| 15 | Kaempferol | 470 ± 62 e | 155 ± 20 b | 1182 ± 192 e | 406 ± 58 c | 472 ± 66 f | 136 ± 70 c | 135 ± 55 cd |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rusanova, M.; Rusanov, K.; Butterweck, V.; Atanassov, I. Indigenous Yeasts from Rose Oil Distillation Wastewater and Their Capacity for Biotransformation of Phenolics. Microorganisms 2023, 11, 201. https://doi.org/10.3390/microorganisms11010201
Rusanova M, Rusanov K, Butterweck V, Atanassov I. Indigenous Yeasts from Rose Oil Distillation Wastewater and Their Capacity for Biotransformation of Phenolics. Microorganisms. 2023; 11(1):201. https://doi.org/10.3390/microorganisms11010201
Chicago/Turabian StyleRusanova, Mila, Krasimir Rusanov, Veronika Butterweck, and Ivan Atanassov. 2023. "Indigenous Yeasts from Rose Oil Distillation Wastewater and Their Capacity for Biotransformation of Phenolics" Microorganisms 11, no. 1: 201. https://doi.org/10.3390/microorganisms11010201
APA StyleRusanova, M., Rusanov, K., Butterweck, V., & Atanassov, I. (2023). Indigenous Yeasts from Rose Oil Distillation Wastewater and Their Capacity for Biotransformation of Phenolics. Microorganisms, 11(1), 201. https://doi.org/10.3390/microorganisms11010201







