Characterization of Two Novel Predatory Bacteria, Bacteriovorax stolpii HI3 and Myxococcus sp. MH1, Isolated from a Freshwater Pond: Prey Range, and Predatory Dynamics and Efficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Culture Media and Cultivation Conditions
2.3. Isolation of Predatory Bacteria
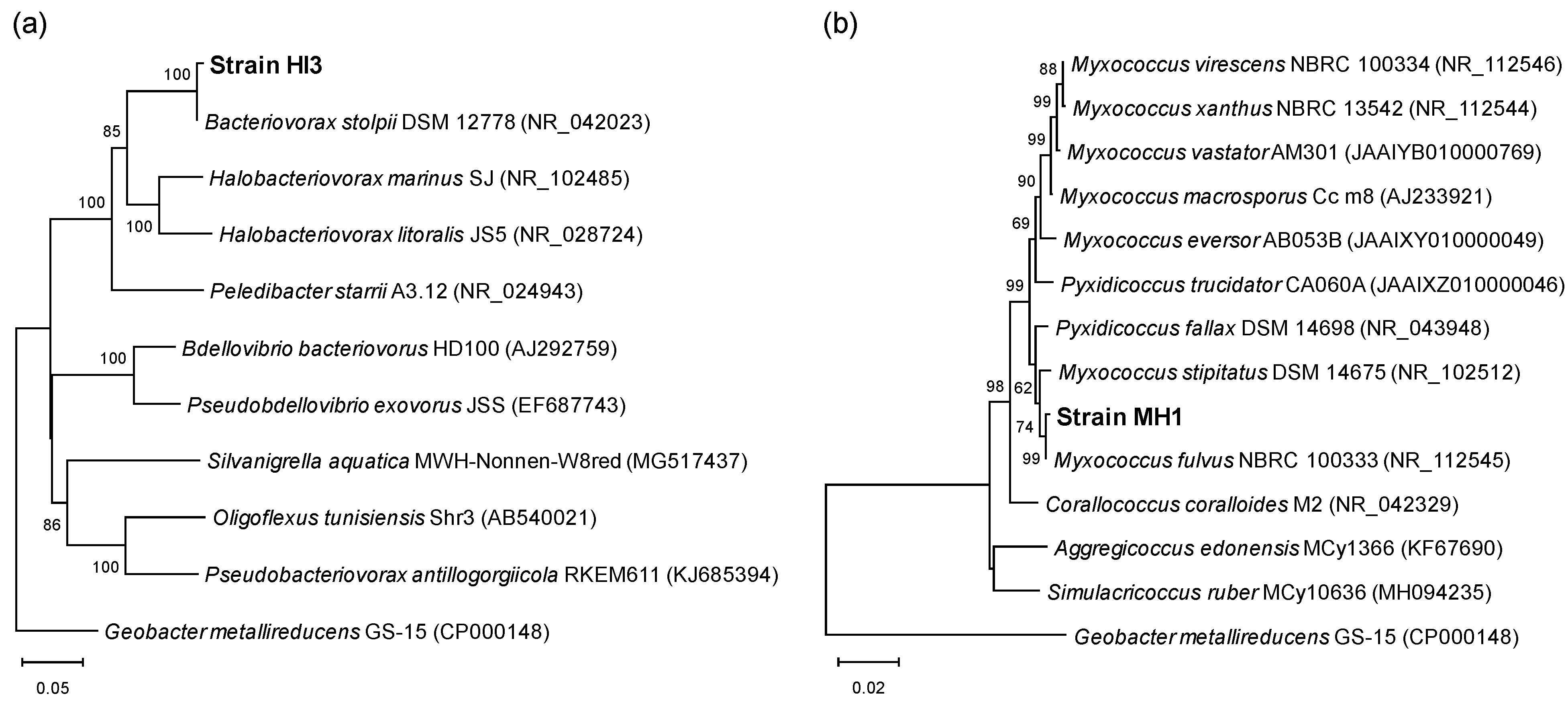
2.4. Phylogenetic Identification
2.5. Prey Range Determination
2.6. Liquid Culture Predation Assay
2.7. Viability qPCR
3. Results and Discussion
3.1. Isolation and Identification of Two Predatory Bacterial Strains
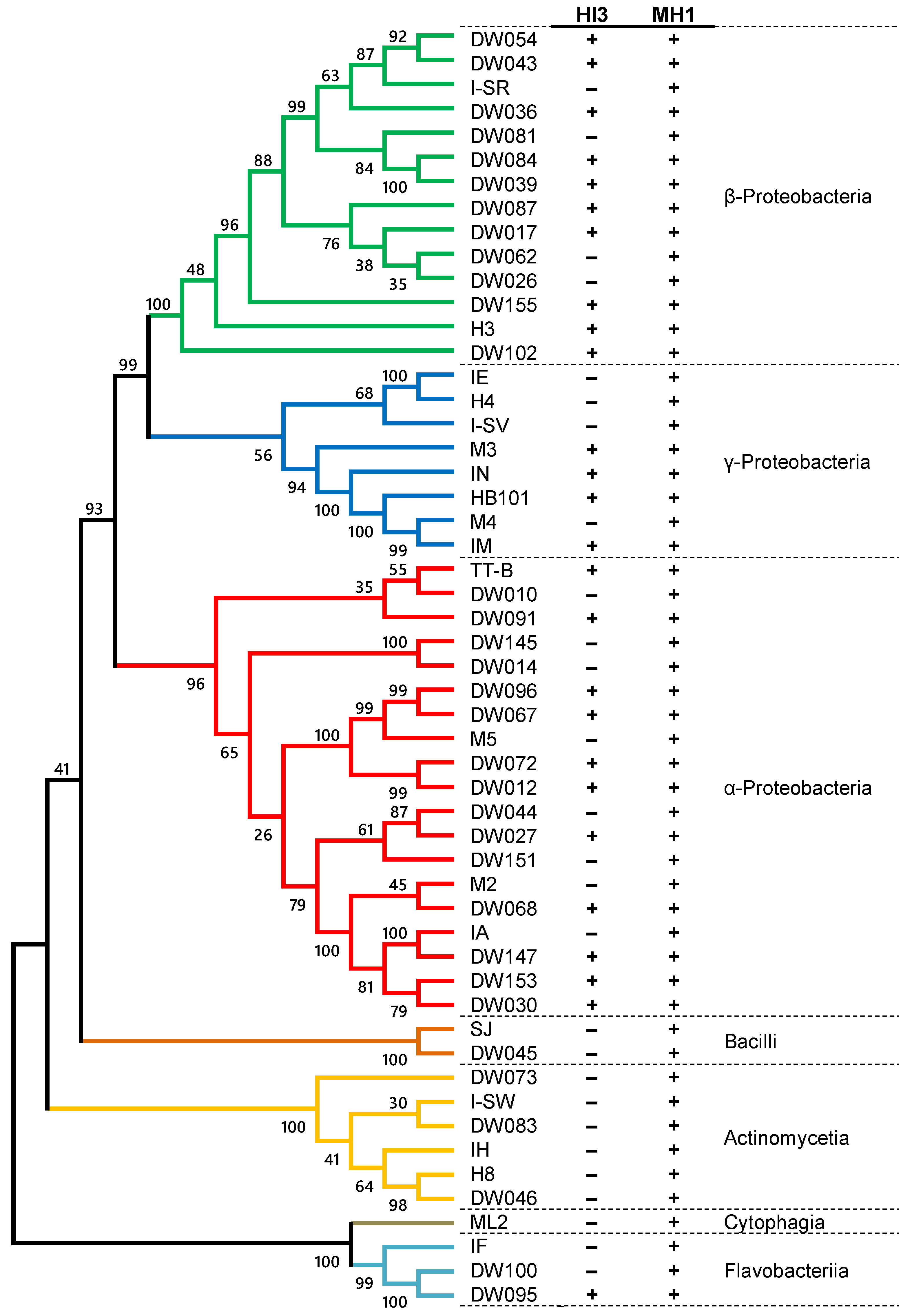
3.2. Prey Range of Isolated Strains
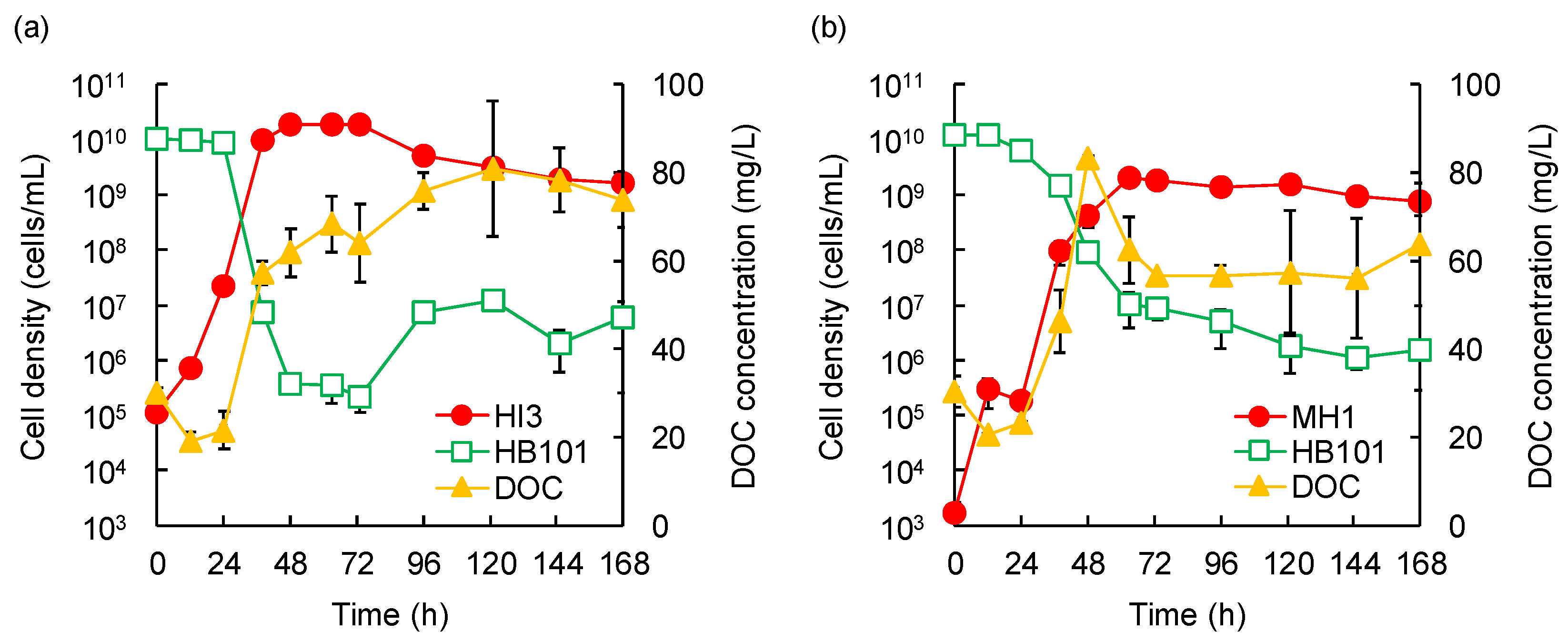
3.3. Temporal Dynamics of Predation by the Isolated Strains
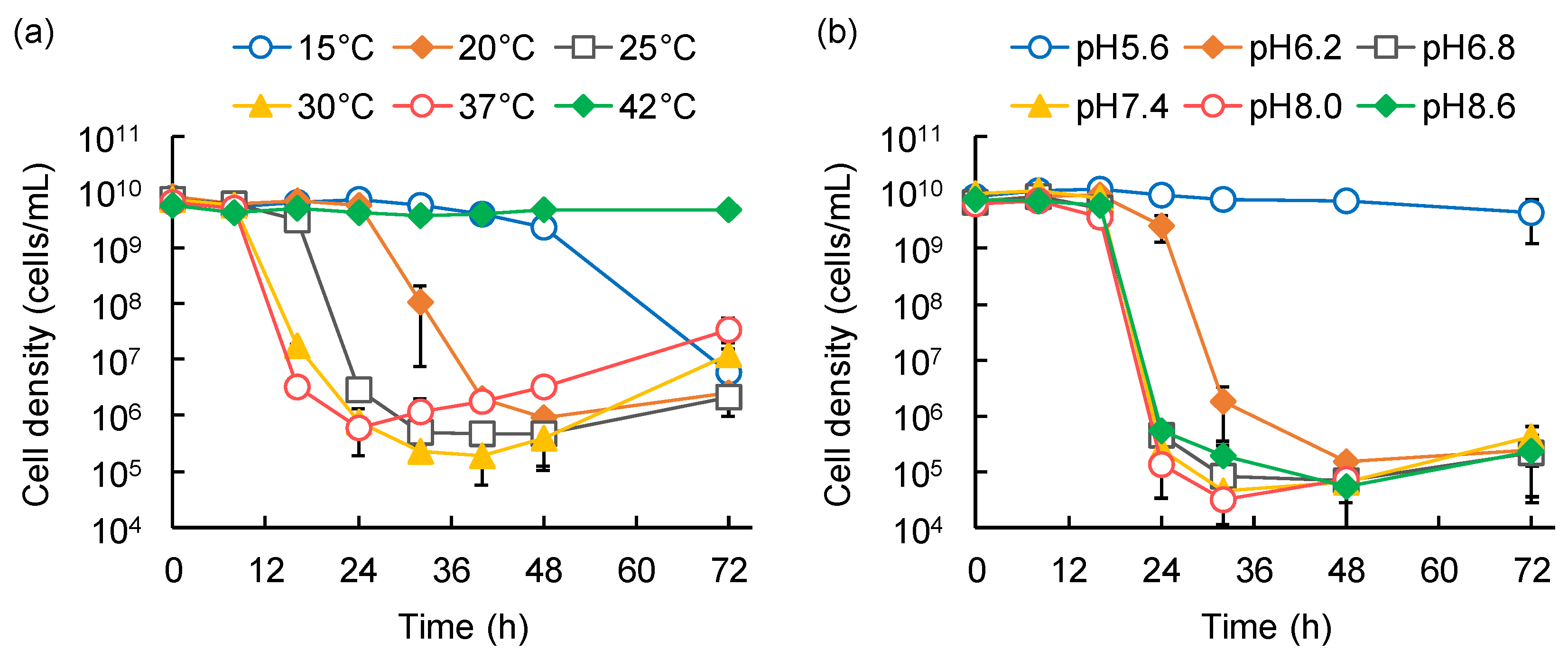
3.4. Effects of Temperature and pH on the Predation Activities of the Isolated Strains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Petters, S.; Groß, V.; Söllinger, A.; Pichler, M.; Reinhard, A.; Bengtsson, M.M.; Urich, T. The soil microbial food web revisited: Predatory myxobacteria as keystone taxa? ISME J. 2021, 15, 2665–2675. [Google Scholar] [CrossRef] [PubMed]
- Ezzedine, J.A.; Desdevises, Y.; Jacquet, S. Bdellovibrio and like organisms: Current understanding and knowledge gaps of the smallest cellular hunters of the microbial world. Crit. Rev. Microbiol. 2022, 48, 428–449. [Google Scholar] [CrossRef] [PubMed]
- Hungate, B.A.; Marks, J.C.; Power, M.E.; Schwartz, E.; van Groenigen, K.J.; Blazewicz, S.J.; Chuckran, P.; Dijkstra, P.; Finley, B.K.; Firestone, M.K.; et al. The functional significance of bacterial predators. mBio 2021, 12, e00466-21. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, A.; Jurkevitch, E. Interactions between Bdellovibrio and like organisms and bacteria in biofilms: Beyond predator-prey dynamics. Environ. Microbiol. 2022, 24, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Pérez, J.; Moraleda-Muñoz, A.; Marcos-Torres, F.J.; Muños-Dorado, J. Bacterial predation: 75 years and counting! Environ. Microbiol. 2016, 18, 766–779. [Google Scholar] [CrossRef]
- Bratanis, E.; Andersson, T.; Lood, R.; Bukowska-Faniband, E. Biotechnological potential of Bdellovibrio and like organisms and their secreted enzymes. Front. Microbiol. 2020, 11, 662. [Google Scholar] [CrossRef]
- Waso, M.; Reyneke, B.; Havenga, B.; Khan, S.; Khan, W. Insights into Bdellovibrio spp. mechanisms of action and potential applications. World J. Microbiol. Biotechnol. 2021, 37, 85. [Google Scholar] [CrossRef]
- Jurkevitch, E.; Davidov, Y. Phylogenetic diversity and evolution of predatory prokaryotes. In Predatory Prokaryotes. Microbiology Monograph; Jurkevitch, E., Ed.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4, pp. 12–56. [Google Scholar] [CrossRef]
- Thiery, S.; Kaimer, C. The predation strategy of Myxococcus xanthus. Front. Microbiol. 2020, 11, 2. [Google Scholar] [CrossRef]
- Reichenbach, H. The ecology of the myxobacteria. Environ. Microbiol. 1999, 1, 15–21. [Google Scholar] [CrossRef]
- Muñoz-Dorado, J.; Marcos-Torres, F.J.; García-Bravo, E.; Moraleda-Muñoz, A.; Pérez, J. Myxobacteria: Moving, killing, feeding, and surviving together. Front. Microbiol. 2016, 7, 781. [Google Scholar] [CrossRef]
- Wang, W.; Luo, X.; Ye, X.; Chen, Y.; Wang, H.; Wang, L.; Wang, Y.; Yang, Y.; Li, Z.; Cao, H.; et al. Predatory Myxococcales are widely distributed in and closely correlated with the bacterial community structure of agricultural land. Appl. Soil Ecol. 2020, 146, 103365. [Google Scholar] [CrossRef]
- Velicer, G.J.; Vos, M. Sociobiology of the Myxobacteria. Annu. Rev. Microbiol. 2009, 63, 599–623. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.D.; MacLean, R.C.; Hillesland, K.L.; Velicer, G.J. Comparative analysis of Myxococcus predation on soil bacteria. Appl. Environ. Microbiol. 2010, 76, 6920–6927. [Google Scholar] [CrossRef] [PubMed]
- Waso-Reyneke, M.; Khan, S.; Khan, W. Interaction of Bdellovibrio bacteriovorus with Gram-negative and Gram-positive bacteria in dual species and polymicrobial communities. Microorganisms 2022, 10, 793. [Google Scholar] [CrossRef] [PubMed]
- Chanyi, R.M.; Ward, C.; Pechey, A.; Koval, S.F. To invade or not to invade: Two approaches to a prokaryotic predatory life cycle. Can. J. Microbiol. 2013, 59, 273–279. [Google Scholar] [CrossRef]
- Feng, S.; Tan, C.H.; Cohen, Y.; Rice, S.A. Isolation of Bdellovibrio bacteriovorus from a tropical wastewater treatment plant and predation of mixed species biofilms assembled by the native community members. Environ. Microbiol. 2016, 18, 3923–3931. [Google Scholar] [CrossRef]
- Yu, R.; Zhang, S.; Chen, Z.; Li, C. Isolation and application of predatory Bdellovibrio-and-like organisms for municipal waste sludge biolysis and dewaterability enhancement. Front. Environ. Sci. Eng. 2017, 11, 10. [Google Scholar] [CrossRef]
- Ezzedine, J.A.; Pavard, G.; Gardillon, M.; Jacquet, S. Bdellovibrio sp: An important bacterial predator in lake Geneva? Open Access J. Microbiol. Biotechnol. 2020, 5, 000157. [Google Scholar] [CrossRef]
- Boyer, H.W.; Roullan-Dussoix, D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 1969, 41, 459–472. [Google Scholar] [CrossRef]
- Cho, G.; Kwon, J.; Soh, S.M.; Jang, H.; Mitchell, R.J. Sensitivity of predatory bacteria to different surfactants and their application to check bacterial predation. Appl. Microbiol. Biotechnol. 2019, 103, 8169–8178. [Google Scholar] [CrossRef]
- Li, Y.; Qiu, F.; Yan, H.; Wan, X.; Wang, M.; Ren, K.; Xu, Q.; Lv, L.; Yin, C.; Liu, X.; et al. Increasing the autotrophic growth of Chlorella USTB-01 via the control of bacterial contamination by Bdellovibrio USTB-06. J. Appl. Microbiol. 2018, 124, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, P.G.; Millard, A.D.; Swain, M.T.; Whitworth, D.E. Transcriptional changes when Myxococcus xanthus preys on Escherichia coli suggest myxobacterial predators are constitutively toxic but regulate their feeding. Microb. Genom. 2018, 4, e000152. [Google Scholar] [CrossRef] [PubMed]
- Jurkevitch, E. Isolation and classification of Bdellovibrio and like organisms. Curr. Protoc. Microbiol. 2012, 26, 7B1.1–7B.1.20. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Fittipaldi, M.; Nocker, A.; Codony, F. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 2012, 91, 276–289. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Inoue, D.; Tsutsui, H.; Yamazaki, Y.; Sei, K.; Soda, S.; Fujita, M.; Ike, M. Application of real-time polymerase chain reaction (PCR) coupled with ethidium monoazide treatment for selective quantification of viable bacteria in aquatic environment. Water Sci. Technol. 2008, 58, 1107–1112. [Google Scholar] [CrossRef]
- Inoue, D.; Hisada, K.; Okumura, T.; Yabuki, Y.; Yoshida, G.; Kuroda, M.; Ike, M. Carbon sources that enable enrichment of 1,4-dioxane-degrading bacteria in landfill leachate. Biodegradation 2020, 31, 23–34. [Google Scholar] [CrossRef]
- Dashiff, A.; Junka, R.A.; Libera, M.; Kadouri, D.E. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J. Appl. Microbiol. 2010, 110, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Oyedara, O.O.; De Luna-Santillana, E.J.; Olguin-Rodriguez, O.; Guo, X.; Mendoza-Villa, M.A.; Menchaca-Arredondo, J.L.; Elufisan, T.O.; Garza-Hernandez, J.A.; Garcia Leon, I.; Rodriguez-Perez, M.A. Isolation of Bdellovibrio sp. from soil samples in Mexico and their potential applications in control of pathogens. MicrobiologyOpen 2016, 5, 992–1002. [Google Scholar] [CrossRef]
- Jurkevitch, E.; Minz, D.; Ramati, B.; Barel, G. Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl. Environ. Microbiol. 2000, 66, 2365–2371. [Google Scholar] [CrossRef]
- Kongrueng, J.; Mitraparp-arthorn, P.; Bandpanwimon, K.; Robins, W.; Vuddhakul, V.; Mekalanos, J. Isolation of Bdellovibrio and like organisms and potential to reduce acute hepatopancreatic necrosis disease caused by Vibrio parahaemolyticus. Dis. Aquat. Org. 2017, 124, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Strack, S.N.; Hoefler, B.C.; Straight, P.D.; Kearns, D.B.; Kirby, J.R. Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus. Appl. Environ. Microbiol. 2014, 80, 5603–5610. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Choi, S.Y.; Son, S.; Mitchell, R.J. Combined application of bacterial predation and violacein to kill polymicrobial pathogenic communities. Sci. Rep. 2017, 7, 14415. [Google Scholar] [CrossRef] [PubMed]
- Rogosky, A.M.; Moak, P.L.; Emmert, E.A. Differential predation by Bdellovibrio bacteriovorus 109J. Curr. Microbiol. 2006, 52, 81–85. [Google Scholar] [CrossRef]
- Waso, M.; Khan, S.; Khan, W. Assessment of predatory bacteria and prey interactions using culture-based methods and EMA-qPCR. Microbiol. Res. 2019, 228, 126305. [Google Scholar] [CrossRef]
- Shemesh, Y.; Jurkevitch, E. Plastic phenotypic resistance to predation by Bdellovibrio and like organisms in bacterial prey. Environ. Microbiol. 2004, 6, 12–18. [Google Scholar] [CrossRef]
- Baer, M.L.; Ravel, J.; Chun, J.; Hill, R.T.; Williams, H.N. A proposal for the reclassification of Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax gen. nov. as Bacteriovorax stolpii comb. nov. and Bacteriovorax starrii comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2000, 50, 219–224. [Google Scholar] [CrossRef][Green Version]
- Williams, H.N.; Baer, M.L. Bacteriovorax. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 1–8. [Google Scholar] [CrossRef]
- Reichenbach, H. Myxococcus. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 1–8. [Google Scholar] [CrossRef]
- Chen, X.J.; Han, K.; Feng, J.; Zhuo, L.; Li, Y.J.; Li, Y.Z. The complete genome sequence and analysis of a plasmid-bearing myxobacterial strain Myxococcus fulvus 124B02 (M 206081). Stand. Genom. Sci. 2016, 11, 1. [Google Scholar] [CrossRef] [PubMed]

| Strain | Sequence (5′–3′) | Products of the Targeted Genes | Annealing and Extension Temp. (°C) | Amplicon Size (bp) | Amplification Efficiency (%) | Correlation Coefficient (r2) | Reference |
|---|---|---|---|---|---|---|---|
| HI3 | TAAACGAGGGAGTGCCCTTC | 16S rRNA | 60 | 160 | 88.2–92.6 | 0.994–0.999 | This study |
| GTTAGCCCAGGCAGTCTTTCTA | |||||||
| MH1 | ACGAAAACCCGTAGCCCAAC | 16S rRNA | 65 | 160 | 82.5–93.4 | 0.992–1.000 | This study |
| TTCACACCCGACTTGTCACG | |||||||
| HB101 | GCATCCATAGCAACAGACCCA | Thiamine-binding periplasmic protein | 66 | 105 | 81.9–92.7 | 0.990–0.999 | [30] |
| CGCCACAAAGCCTGAAAGAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inoue, D.; Hiroshima, N.; Nakamura, S.; Ishizawa, H.; Ike, M. Characterization of Two Novel Predatory Bacteria, Bacteriovorax stolpii HI3 and Myxococcus sp. MH1, Isolated from a Freshwater Pond: Prey Range, and Predatory Dynamics and Efficiency. Microorganisms 2022, 10, 1816. https://doi.org/10.3390/microorganisms10091816
Inoue D, Hiroshima N, Nakamura S, Ishizawa H, Ike M. Characterization of Two Novel Predatory Bacteria, Bacteriovorax stolpii HI3 and Myxococcus sp. MH1, Isolated from a Freshwater Pond: Prey Range, and Predatory Dynamics and Efficiency. Microorganisms. 2022; 10(9):1816. https://doi.org/10.3390/microorganisms10091816
Chicago/Turabian StyleInoue, Daisuke, Naoto Hiroshima, So Nakamura, Hidehiro Ishizawa, and Michihiko Ike. 2022. "Characterization of Two Novel Predatory Bacteria, Bacteriovorax stolpii HI3 and Myxococcus sp. MH1, Isolated from a Freshwater Pond: Prey Range, and Predatory Dynamics and Efficiency" Microorganisms 10, no. 9: 1816. https://doi.org/10.3390/microorganisms10091816
APA StyleInoue, D., Hiroshima, N., Nakamura, S., Ishizawa, H., & Ike, M. (2022). Characterization of Two Novel Predatory Bacteria, Bacteriovorax stolpii HI3 and Myxococcus sp. MH1, Isolated from a Freshwater Pond: Prey Range, and Predatory Dynamics and Efficiency. Microorganisms, 10(9), 1816. https://doi.org/10.3390/microorganisms10091816






