Phylogeny of Regulators of G-Protein Signaling Genes in Leptographium qinlingensis and Expression Levels of Three RGSs in Response to Different Terpenoids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain
2.2. Fungal Media and Growth Condition
2.3. Terpenoid Treatments
2.4. RNA Isolation and cDNA Synthesis
2.5. Gene Amplification and Cloning
2.6. Analysis of Full-Length cDNA Sequences
2.7. Effects of Terpenoids on Expression Levels of three L. qinlingensis RGSs (Real Time-qPCR)
2.8. Statistical Analysis
3. Results
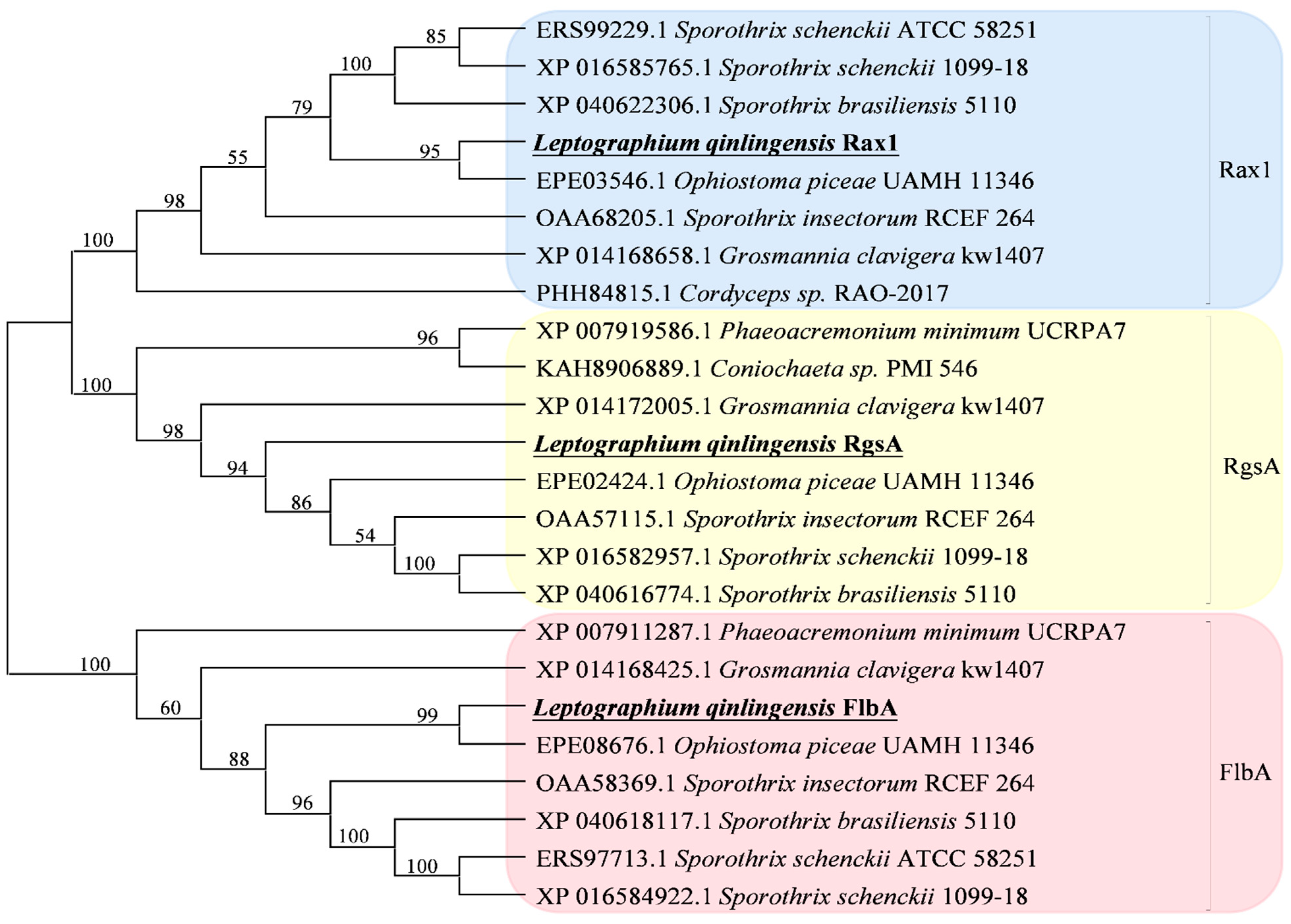
3.1. Identification of L. qinlingensis RGS Genes
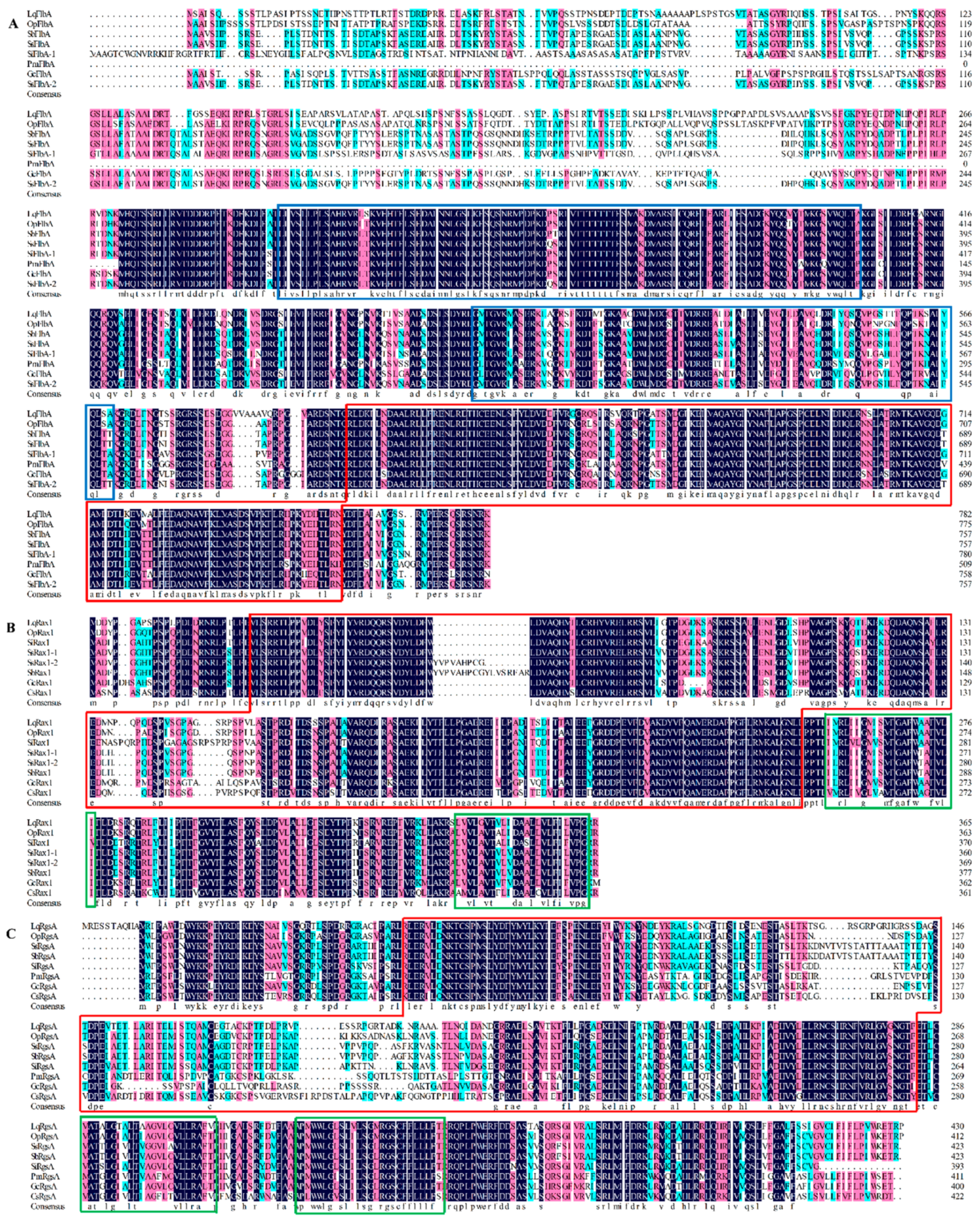
3.2. Physicochemical Properties and Bioinformation Analysis
3.3. Effect of Terpenoids on L. qinlingensis Growth and Reproduction
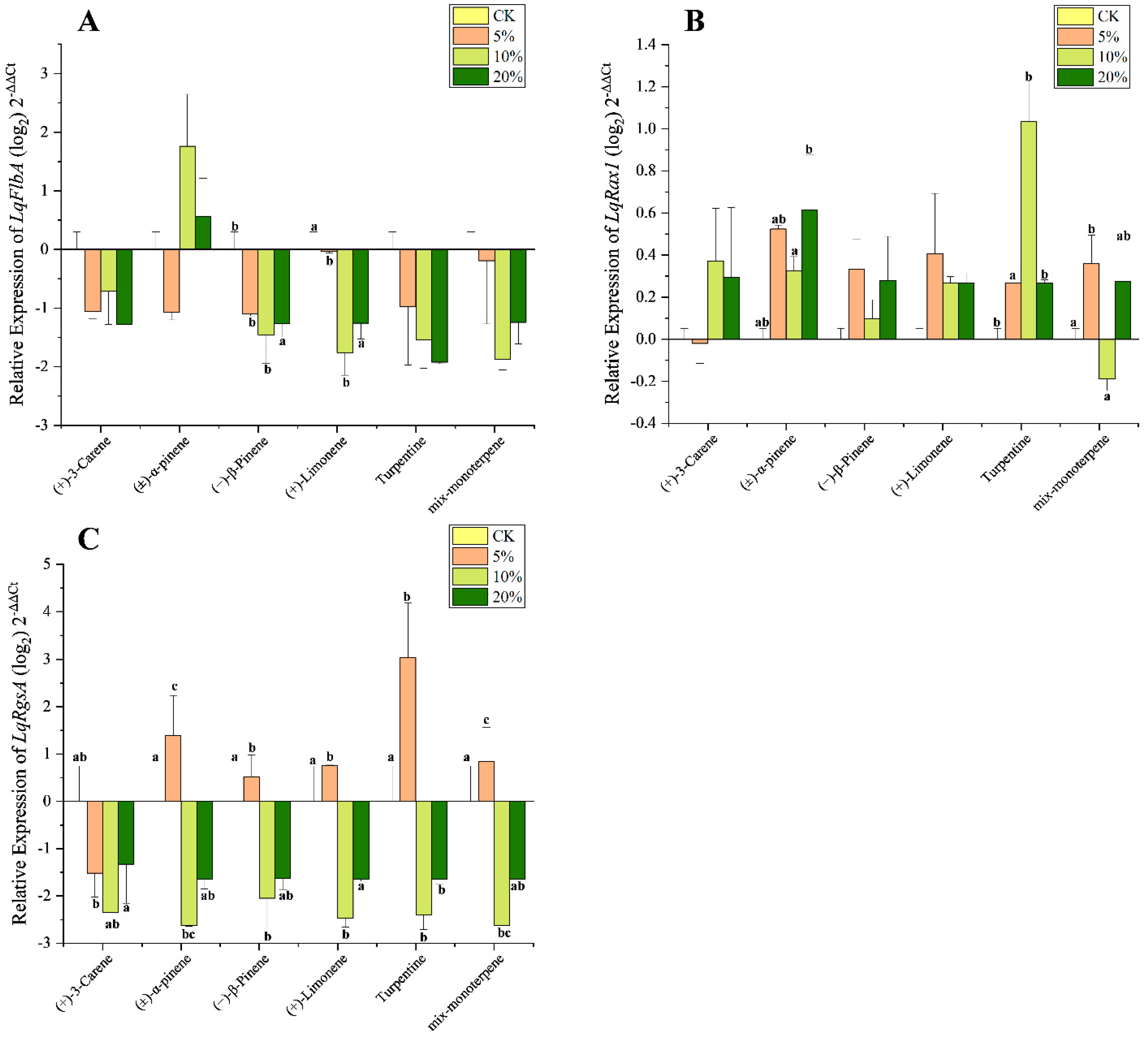
3.4. Effect of Terpenoids on Expression Level of LqRGS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kurz, W.A.; Dymond, C.C.; Stinson, G.; Rampley, G.J.; Neilson, E.T.; Carroll, A.L.; Ebata, T.; Safranyik, L. Mountain pine beetle and forest carbon feedback to climate change. Nature 2008, 452, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Yu, J.; Wang, C.; Chen, H. Cellulolytic bacteria associated with the gut of Dendroctonus armandi larvae (Coleoptera: Curculionidae: Scolytinae). Forests 2014, 5, 455–465. [Google Scholar] [CrossRef]
- Chen, H.; Tang, M. Spatial and temporal dynamics of bark beetles in Chinese white pine in Qinling Mountains of Shaanxi province, China. Environ. Entomol. 2014, 36, 1124–1130. [Google Scholar] [CrossRef]
- Chen, H.; Tang, M.; Zhu, C.; Hu, J. The enzymes in the secretions of Dendroctonus armandi (Scolytidae) and their symbiotic fungus of Leptographium qinlingensis. Sci. Silvae Sin. 2004, 40, 123–126. (In Chinese) [Google Scholar]
- Tang, M.; Chen, H. Effect of symbiotic fungi of Dendroctonus armandi on host trees. Sci. Silvae Sin. 1999, 35, 63–66. (In Chinese) [Google Scholar]
- Hofstetter, R.W.; Dinkins-Bookwalter, J.; Davis, T.S.; Klepzig, K.D. Symbiotic Associations of Bark Beetles. In Bark Beetles; Vega, F.E., Hofstetter, R.W., Eds.; Elsevier: San Diego, CA, USA, 2015; pp. 209–245. [Google Scholar] [CrossRef]
- Lehenberger, M.; Benkert, M.; Biedermann, P.H. Ethanol-enriched substrate facilitates ambrosia beetle fungi, but inhibits their pathogens and fungal symbionts of bark beetles. Front. Microbiol. 2021, 11, 3487. [Google Scholar] [CrossRef]
- Wang, Y.; Lim, L.; DiGuistini, S.; Robertson, G.; Bohlmann, J.; Breuil, C. A specialized ABC efflux transporter GcABC-G1 confers monoterpene resistance to Grosmannia clavigera, a bark beetle-associated fungal pathogen of pine trees. New Phytol. 2013, 197, 886–898. [Google Scholar] [CrossRef]
- Erbilgin, N.; Cale, J.A.; Lusebrink, I.; Najar, A.; Klutsch, J.G.; Sherwood, P.; Bonello, P.; Evenden, M.L. Water-deficit and fungal infection can differentially affect the production of different classes of defense compounds in two host pines of mountain pine beetle. Tree Physiol. 2017, 37, 338–350. [Google Scholar] [CrossRef] [Green Version]
- Raffa, K.F.; Aukema, B.H.; Erbilgin, N.; Klepzig, K.D.; Wallin, K.F. Interactions among conifer terpenoids and bark beetles across multiple levels of scale: An attempt to understand links between population patterns and physiological processes. Recent Adv. Phytochem. 2005, 39, 79–118. [Google Scholar] [CrossRef]
- Dai, L.; Li, Z.; Yu, J.; Ma, M.; Zhang, R.; Chen, H.; Pham, T. The CYP51F1 gene of Leptographium qinlingensis: Sequence characteristic, phylogeny and transcript levels. Int. J. Mol. Sci. 2015, 16, 12014–12034. [Google Scholar] [CrossRef] [Green Version]
- Lah, L.; Haridas, S.; Bohlmann, J.; Breuil, C. Biology The cytochromes P450 of Grosmannia clavigera: Genome organization, phylogeny, and expression in response to pine host chemicals. Fungal Genet. Biol. 2013, 50, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Neves, S.R.; Ram, P.T.; Iyengar, R. G protein pathways. Science 2002, 296, 1636–1639. [Google Scholar] [CrossRef] [PubMed]
- Syrovatkina, V.; Alegre, K.O.; Dey, R.; Huang, X.-Y. Regulation, signaling, and physiological functions of G-proteins. J. Mol. Biol. 2016, 428, 3850–3868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worzfeld, T.; Wettschureck, N.; Offermanns, S. G(12)/G(13)-mediated signalling in mammalian physiology and disease. Trends Pharmacol. Sci. 2008, 29, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Goddard, A.; Davey, J.; Ladds, G. Investigating RGS proteins in yeast. Semin. Cell Dev. Biol. 2006, 17, 352–362. [Google Scholar] [CrossRef]
- Park, H.-S.; Kim, M.-J.; Yu, J.-H.; Shin, K.-S. Heterotrimeric G-protein signalers and RGSs in Aspergillus fumigatus. Pathogens 2020, 9, 902. [Google Scholar] [CrossRef]
- Kimple, A.J.; Bosch, D.E.; Giguère, P.M.; Siderovski, D.P. Regulators of G-protein signaling and their Gα substrates: Promises and challenges in their use as drug discovery targets. Pharmacol. Rev. 2011, 63, 728–749. [Google Scholar] [CrossRef] [Green Version]
- Moretti, M.; Wang, L.; Grognet, P.; Lanver, D.; Link, H.; Kahmann, R. Three regulators of G protein signaling differentially affect mating, morphology and virulence in the smut fungus Ustilago maydis. Mol. Microbiol. 2017, 105, 901–921. [Google Scholar] [CrossRef] [Green Version]
- McCudden, C.; Hains, M.; Kimple, R.; Siderovski, D.; Willard, F. G-protein signaling: Back to the future. Cell. Mol. Life Sci. 2005, 62, 551–577. [Google Scholar] [CrossRef] [Green Version]
- Chasse, S.A.; Flanary, P.; Parnell, S.C.; Hao, N.; Cha, J.Y.; Siderovski, D.P.; Dohlman, H.G. Genome-Scale analysis reveals Sst2 as the principal regulator of mating pheromone signaling in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 2006, 5, 330–346. [Google Scholar] [CrossRef] [Green Version]
- Versele, M.; de Winde, J.H.; Thevelein, J.M. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 1999, 18, 5577–5591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, K.-H.; Seo, J.-A.; Yu, J.-H. Regulators of G-protein signalling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (Gα) signalling. Mol. Microbiol. 2004, 53, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.H. Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J. Microbiol. 2006, 44, 145–154. Available online: https://pubmed.ncbi.nlm.nih.gov/16728950 (accessed on 18 August 2022).
- Yu, J.H.; Wieser, J.; Adams, T. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 1996, 15, 5184–5190. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Yang, K.; Tumukunde, E.; Guo, Z.; Zhang, B.; Liu, Y.; Zhuang, Z.; Yuan, J.; Wang, S.; Buan, N.R. Regulator of G protein signaling contributes to the development and aflatoxin biosynthesis in Aspergillus flavus through the regulation of Gα activity. Appl. Environ. Microbiol. 2022, 88, e00244-22. [Google Scholar] [CrossRef]
- De Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28–72. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.G.; Kim, S.S.; Yu, J.H.; Shin, K.-S. Characterization of gprK encoding a putative hybrid G-protein-coupled receptor in Aspergillus fumigatus. PLoS ONE 2016, 11, e0161312. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, M.-W.; Jun, S.-C.; Choi, Y.-H.; Yu, J.-H.; Shin, K.-S. RgsD negatively controls development, toxigenesis, stress response, and virulence in Aspergillus fumigatus. Sci. Rep. 2019, 9, 811–825. [Google Scholar] [CrossRef]
- Mah, J.H.; Yu, J.H. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell 2006, 5, 1585–1595. [Google Scholar] [CrossRef] [Green Version]
- An, H.; Gan, T.; Tang, M.; Chen, H. Molecular mechanism of overcoming host resistance by the target of rapamycin gene in Leptographium qinlingensis. Microorganisms 2022, 10, 503. [Google Scholar] [CrossRef]
- Dai, L.; Zheng, J.; Ye, J.; Chen, H. Phylogeny of Leptographium qinlingensis cytochrome P450 genes and transcription levels of six CYPs in response to different nutrition media or terpenoids. Arch. Microbiol. 2021, 204, 16. [Google Scholar] [CrossRef] [PubMed]
- Di Guistini, S.; Wang, Y.; Liao, N.Y.; Taylor, G.; Tanguay, P.; Feau, N.; Henrissat, B.; Chan, S.K.; Hesse-Orce, U.; Alamouti, S.M.; et al. Genome and transcriptome analyses of the mountain pine beetle-fungal symbiont Grosmannia clavigera, a lodgepole pine pathogen. Proc. Natl. Acad. Sci. USA 2011, 108, 2504–2509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, L.; Li, H.; Zheng, J.; Chen, H. Transcriptome analyses of the Chinese white pine beetle-fungal symbiont Leptographium qinlingensis under terpene stress or growth on host pine sawdust. Symbiosis 2022, 86, 17–31. [Google Scholar] [CrossRef]
- Ballon, D.R.; Flanary, P.L.; Gladue, D.P.; Konopka, J.B.; Dohlman, H.G.; Thorner, J. DEP-domain-mediated regulation of GPCR signaling responses. Cell 2006, 126, 1079–1093. [Google Scholar] [CrossRef] [Green Version]
- Fujita, A.; Lord, M.; Hiroko, T.; Hiroko, F.; Chen, T.; Oka, C.; Misumi, Y.; Chant, J. Rax1, a protein required for the establishment of the bipolar budding pattern in yeast. Gene 2004, 327, 161–169. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, W.; Liu, K.; Huang, Q.; Zhang, X.; Yan, X.; Chen, Y.; Wang, J.; Qi, Z.; Wang, Z. Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog. 2011, 7, e1002450. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Wright, S.J.; Krystofova, S.; Park, G.; Borkovich, K.A. Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 2007, 61, 423–452. [Google Scholar] [CrossRef]
- Ma, N.; Zhao, Y.; Wang, Y.; Yang, L.; Li, D.; Yang, J.; Jiang, K.; Zhang, K.-Q.; Yang, J. Functional analysis of seven regulators of G protein signaling (RGSs) in the nematode-trapping fungus Arthrobotrys oligospora. Virulence 2021, 12, 1825–1840. [Google Scholar] [CrossRef]
- Dohlman, H.G.; Song, J.; Ma, D.; Courchesne, W.E.; Thorner, J. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: Expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit). Mol. Cell. Biol. 1996, 16, 5194–5209. [Google Scholar] [CrossRef] [Green Version]
- Burchett, S.A. Regulators of G protein signaling: A bestiary of modular protein binding domains. J. Neurochem. 2000, 75, 1335–1351. [Google Scholar] [CrossRef]
- Yu, J.H.; Keller, N. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 2005, 43, 437–458. [Google Scholar] [CrossRef] [PubMed]
- Lafuente, M.J.; Gancedo, C. Disruption and basic functional analysis of six novel ORFs of chromosome XV from Saccharomyces cerevisiae. Yeast 1999, 15, 935–943. Available online: https://pubmed.ncbi.nlm.nih.gov/10407273 (accessed on 18 August 2022). [CrossRef]
- Kang, P.J.; Angerman, E.; Nakashima, K.; Pringle, J.R.; Park, H.O. Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast. Mol. Biol. Cell 2004, 15, 5145–5157. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Snyder, M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 2001, 12, 2147–2170. [Google Scholar] [CrossRef] [Green Version]
- Igbalajobi, O.A.; Yu, J.H.; Shin, K.S. Characterization of the rax1 gene encoding a putative regulator of G protein signaling in Aspergillus fumigatus. Biochem. Biophys. Res. Commun. 2017, 487, 426–432. [Google Scholar] [CrossRef]
- Choi, Y.H.; Lee, M.W.; Igbalajobi, O.A.; Yu, J.H.; Shin, K.S. Transcriptomic and functional studies of the RGS protein Rax1 in Aspergillus fumigatus. Pathogens 2020, 9, 36. [Google Scholar] [CrossRef] [Green Version]
- Haridas, S.; Wang, Y.; Lim, L.; Massoumi Alamouti, S.; Jackman, S.; Docking, R.; Robertson, G.; Birol, I.; Bohlmann, J.; Breuil, C. The genome and transcriptome of the pine saprophyte Ophiostoma piceae, and a comparison with the bark beetle-associated pine pathogen Grosmannia clavigera. BMC Genom. 2013, 14, 373. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.; Xu, L.; Xu, D.; Lou, Q.; Lu, M.; Sun, J. Does cryptic microbiota mitigate pine resistance to an invasive beetle-fungus complex? Implications for invasion potential. Sci. Rep. 2016, 6, 33110. [Google Scholar] [CrossRef] [Green Version]
- Howe, M.; Keefover-Ring, K.; Raffa, K.F. Pine engravers carry bacterial communities whose members reduce concentrations of host monoterpenes with variable degrees of redundancy, specificity, and capability. Environ. Entomol. 2018, 47, 638–645. [Google Scholar] [CrossRef]
- Briones-Roblero, C.I.; Rodríguez-Díaz, R.; Santiago-Cruz, J.A.; Zúñiga, G.; Rivera-Orduña, F.N. Degradation capacities of bacteria and yeasts isolated from the gut of Dendroctonus rhizophagus (Curculionidae: Scolytinae). Folia Microbiol. 2017, 62, 1–9. [Google Scholar] [CrossRef]

| Reagents | Reagent Source | Purity | V1 (DMSO) * |
|---|---|---|---|
| (+)-α-pinene/(−)-α-pinene | Shanghai Aladdin Bio-Technology Co., Ltd. (Shanghai, China) | 98% | 390 mL |
| (+)-3-carene | Shanghai Aladdin Bio-Technology Co., Ltd. (Shanghai, China) | 90% | 350 mL |
| (−)-β-pinene | Shanghai Aladdin Bio-Technology Co., Ltd. (Shanghai, China) | 99% | 395 mL |
| (+)-limonene | Shanghai Aladdin Bio-Technology Co., Ltd. (Shanghai, China) | 98% | 390 mL |
| turpentine | Shanghai Aladdin Bio-Technology Co., Ltd. (Shanghai, China) | AR | 400 mL |
| Gene | Primer Direction 5′→3′ | Purpose | |
|---|---|---|---|
| Forward | Reverse | ||
| FlbA | ACTTGCGCGAGACCCA | AATTTCGGCACCGAGTCGCT | cDNA |
| GCCAGGACGGTGCTATGAT | CGCGGATCCTCCACTAGTGATTTCACTATAGG | 3′RACE | |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | AAAGACAGGTTCTCCTCGCAG | 5′RACE | |
| ATGTCCGCCATCTCTCAGTCCTCTT | TTACTTGCGGTTCGACCGGCTCTGG | Full-length | |
| GCTATGATCGACACCCTGAAG | GTAGTTTCTCAGCGTATGGTCG | qPCR | |
| Rax1 | CTGGCTCGATGTTGCCCAGCACATG | ACTGGAACGAGGCCAGAAAGTACAC | cDNA |
| CTGCCTTTGTGCTCATCTT | CGCGGATCCTCCACTAGTGATTTCACTATAGG | 3′RACE | |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | GCTGAGGTCGCCCAGGTTC | 5′RACE | |
| ATGGACGACTACCCCGGCG | CTACAGCCGGCGGC | Full-length | |
| CACCGGTCGATCTGTACTCG | CGCAGTTCACGCACATAGTG | qPCR | |
| RgsA | ACCCCGCCCACCTCAAGCCC | TGGGCGTCCTTGACGCGCAGCTT | cDNA |
| GCTTCTTCTTACTGCTGTTCA | CGCGGATCCTCCACTAGTGATTTCACTATAGG | 3′RACE | |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | AAAGGTGCCATTGCCGACG | 5′RACE | |
| ATGCGAGAATCATCAACAG | TTAAAGAGGCCGCGTCTC | Full-length | |
| ACAAGAAGCCAGAGTACCGC | TTGTTCTCGAGGACACGCTC | qPCR | |
| EF1 | CCGCTGGTACGGGTGAGTT | CTTGGTGGTGTCCATCTTGTT | qPCR |
| Gene | BLAST Matches in Genbank | Identity in the Full Length * | ||
|---|---|---|---|---|
| Species | Gene | Accession No. | Blastp (%) | |
| LqFlbA | Ophiostoma piceae UAMH 11346 | FlbA | EPE08676.1 | 87.98 |
| Sporothrix brasiliensis 5110 | FlbA | XP_040618117.1 | 83.63 | |
| Sporothrix schenckii ATCC 58251 | FlbA | ERS97713.1 | 83.12 | |
| Sporothrix schenckii 1099-18 | FlbA | XP_016584922.1 | 82.86 | |
| Sporothrix insectorum RCEF 264 | FlbA | OAA58369.1 | 81.89 | |
| Phaeoacremonium minimum UCRPA7 | FlbA | XP_007911287.1 | 79.64 | |
| Grosmannia clavigera kw1407 | FlbA | XP_014168425.1 | 77.18 | |
| LqRax1 | Ophiostoma piceae UAMH 11346 | Rax1 | EPE03546.1 | 92.35 |
| Sporothrix insectorum RCEF 264 | Rax1 | OAA68205.1 | 86.02 | |
| Sporothrix schenckii ATCC 58251 | Rax1 | ERS99229.1 | 88.25 | |
| Sporothrix schenckii 1099-18 | Rax1 | XP_016585765.1 | 86.13 | |
| Sporothrix brasiliensis 5110 | Rax1 | XP_040622306.1 | 84.6 | |
| Grosmannia clavigera kw1407 | Rax1 | XP_014168658.1 | 83.74 | |
| Cordyceps sp. RAO-2017 | Rax1 | PHH84815.1 | 79.78 | |
| LqRgsA | Ophiostoma piceae UAMH 11346 | Rgs | EPE02424.1 | 79.38 |
| Sporothrix brasiliensis 5110 | Rgs | XP_040616774.1 | 75.41 | |
| Sporothrix schenckii 1099-18 | Rgs | XP_016582957.1 | 74.70 | |
| Sporothrix insectorum RCEF 264 | Rgs | OAA57115.1 | 72.84 | |
| Grosmannia clavigera kw1407 | Rgs | XP_014172005.1 | 65.87 | |
| Phaeoacremonium minimum UCRPA7 | Rgs | XP_007919586.1 | 63.25 | |
| Coniochaeta sp. PMI_546 | Rgs | KAH8906889.1 | 61.31 | |
| Gene Name | Full Length (bp) * | ORF Size (aa/bp) * | Mw (kDa) * | I.P. * | Signal Peptide Prediction ** |
|---|---|---|---|---|---|
| LqFlbA | 2699 | 782/2349 | 85.09 | 9.04 | SP 0 mTP 0 other 1 |
| LqRax1 | 1884 | 366/1101 | 41.23 | 5.91 | SP 0 mTP 0 other 1 |
| LqRgsA | 1894 | 431/1296 | 48.46 | 9.60 | SP 0.0001 mTP 0.0001 other 0.9998 |
| Media | K | a | b | R2 | Inflection Day | |
|---|---|---|---|---|---|---|
| DMSO | 91.167 | 45.003 | 4.625 | 0.997 | 12.35 | |
| MEA | 68.201 | 241.959 | 9.157 | 0.983 | 8.99 | |
| 5% | (+)-3-Carene | 91.613 | 65.04 | 5.152 | 0.978 | 12.16 |
| (±)-α-Pinene | 81.624 | 416.282 | 7.54 | 0.978 | 12.00 | |
| (−)-β-Pinene | 79.843 | 419.731 | 7.663 | 0.982 | 11.82 | |
| (+)-Limonene | 112.499 | 111.587 | 4.605 | 0.979 | 15.36 | |
| Turpentine | 93.140 | 124.781 | 5.762 | 0.973 | 12.56 | |
| Mix-monoterpene | 91.486 | 126.549 | 5.84 | 0.974 | 12.43 | |
| 10% | (+)-3-Carene | 89.768 | 125.2 | 5.9 | 0.976 | 12.28 |
| (±)-α-Pinene | 81.116 | 488.797 | 7.672 | 0.986 | 12.11 | |
| (−)-β-Pinene | 81.752 | 192.869 | 6.733 | 0.983 | 11.72 | |
| (+)-Limonene | 88.877 | 179.027 | 6.114 | 0.999 | 12.73 | |
| Turpentine | 89.513 | 203.785 | 6.406 | 0.972 | 12.45 | |
| Mix-monoterpene | 91.040 | 86.253 | 5.46 | 0.977 | 12.25 | |
| 20% | (+)-3-Carene | 91.995 | 87.883 | 5.45 | 0.975 | 12.32 |
| (±)-α-Pinene | 81.688 | 344.178 | 7.291 | 0.986 | 12.02 | |
| (−)-β-Pinene | 82.197 | 353.544 | 7.274 | 0.987 | 12.10 | |
| (+)-Limonene | 80.416 | 691.62 | 7.984 | 0.995 | 12.29 | |
| Turpentine | 93.776 | 115.599 | 5.659 | 0.972 | 12.59 | |
| Mix-monoterpene | 86.014 | 322.104 | 7.035 | 0.973 | 12.31 | |
| Gene | (+)-3-Carene | (±)-α-Pinene | (−)-β-Pinene | (+)-Limonene | Turpentine | Mix-Monoterpene | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | Sig | F | Sig | F | Sig | F | Sig | F | Sig | F | Sig | |
| LqFlbA | 5.817 | 0.061 | 0.455 | 0.728 | 9 | 0.029 | 20.224 | 0.007 | 4.257 | 0.098 | 4.452 | 0.092 |
| LqRax1 | 1.753 | 0.294 | 7.841 | 0.038 | 2569 | 0.192 | 2.642 | 0.186 | 43.679 | 0.002 | 8.591 | 0.032 |
| LqRgsA | 5.045 | 0.045 | 18.965 | 0.008 | 9.841 | 0.026 | 29.271 | 0.004 | 23.106 | 0.005 | 17.967 | 0.009 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, T.; An, H.; Tang, M.; Chen, H. Phylogeny of Regulators of G-Protein Signaling Genes in Leptographium qinlingensis and Expression Levels of Three RGSs in Response to Different Terpenoids. Microorganisms 2022, 10, 1698. https://doi.org/10.3390/microorganisms10091698
Gan T, An H, Tang M, Chen H. Phylogeny of Regulators of G-Protein Signaling Genes in Leptographium qinlingensis and Expression Levels of Three RGSs in Response to Different Terpenoids. Microorganisms. 2022; 10(9):1698. https://doi.org/10.3390/microorganisms10091698
Chicago/Turabian StyleGan, Tian, Huanli An, Ming Tang, and Hui Chen. 2022. "Phylogeny of Regulators of G-Protein Signaling Genes in Leptographium qinlingensis and Expression Levels of Three RGSs in Response to Different Terpenoids" Microorganisms 10, no. 9: 1698. https://doi.org/10.3390/microorganisms10091698
APA StyleGan, T., An, H., Tang, M., & Chen, H. (2022). Phylogeny of Regulators of G-Protein Signaling Genes in Leptographium qinlingensis and Expression Levels of Three RGSs in Response to Different Terpenoids. Microorganisms, 10(9), 1698. https://doi.org/10.3390/microorganisms10091698






