Abstract
Validation studies conducted within a food processing facility using surrogate organisms could better represent the manufacturing process than controlled laboratory studies with pathogenic bacteria on precision equipment in a BSL-2 lab. The objectives of this project were to examine potential surrogate bacteria during biltong processing, conduct biltong surrogate validation lethality studies, and measure critical factors and intrinsic parameters during processing. Beef pieces (1.9 cm × 5.1 cm × 7.6 cm) were inoculated with four-strain mixtures of Carnobacterium divergens/C. gallinarum, Pediococcus acidilactici/P. pentosaceous, and Biotype 1 E. coli ATCC BAA (-1427, -1428, -1429, and -1430), as well as a two-strain mixture of Latilactobacillus sakei and other commercially available individual bacterial cultures (P. acidilactici Saga200/Kerry Foods; Enterococcus faecium 201224-016/Vivolac Cultures). Inoculated beef was vacuum-tumbled in marinade and dried in a humidity-controlled oven for 8–10 days (24.9 °C; 55% relative humidity). Microbial enumeration of surviving surrogate bacteria and evaluation of intrinsic factors (water activity, pH, and salt concentration) were performed post inoculation, post marination, and after 2, 4, 6, 8, and 10 days of drying. Trials were performed in duplicate replication with triplicate samples per sampling time and analyzed by one-way RM-ANOVA. Trials conducted with E. faecium, Pediococcus spp., and L. sakei never demonstrated more than 2 log reduction during the biltong process. However, Carnobacterium achieved a >5 log (5.85 log) reduction over a drying period of 8 days and aligned with the reductions observed in previous trials with pathogenic bacteria (Salmonella, E. coli O157:H7, L. monocytogenes, and S. aureus) in biltong validation studies. Studies comparing resuspended freeze-dried or frozen cells vs. freshly grown cells for beef inoculation showed no significant differences during biltong processing. Carnobacterium spp. would be an effective nonpathogenic in-plant surrogate to monitor microbial safety that mimics the response of pathogenic bacteria to validate biltong processing within a manufacturer’s own facility.
1. Introduction
Biltong is a South African style dried beef product that is growing in popularity in the United States. This dried meat product is traditionally made using lean strips of beef that are marinated in a mixture of traditional spices (coriander and pepper), salt, and vinegar and then dried at low or ambient temperature and humidity. Dried beef processing guidelines, as issued by the United States Department of Agriculture Food Safety and Inspection Service (USDA-FSIS), require dried beef products to be heated to an internal temperature of 160 °F (71.1 °C) in a sealed oven or steam injector with a relative humidity greater than 90% during the cooking/heating process [1]. Since biltong does not have a heat lethality step during processing and deviates from these guidelines, biltong manufacturers must conduct a validation or challenge study to evaluate the ability of their process to sufficiently inactive bacterial pathogens such as Salmonella spp. which have been historically linked to outbreaks and recalls of dried meat and poultry products [2]. USDA-FSIS does give processors two different options to safely produce these alternative dried meat products. The first option requires Salmonella testing of every lot of edible ingredients used during processing and an overall process reduction of a ‘pathogen of concern’ of at least 2 log. Alternatively, processors can forego ingredient testing if they can demonstrate that their process can achieve ≥ 5 log reduction of Salmonella by the end of processing [3].
USDA-FSIS regulatory guidance for manufacture and sale of biltong requires processors to demonstrate product safety by process validation against a ‘pathogen of concern’. In recent BSL-2 in-lab studies, this was performed with Salmonella serovars [4], E. coli O157:H7, L. monocytogenes, and S. aureus [5]. These experiments, while successful in achieving a >5 log reduction of foodborne pathogens where the data are currently used by processors in support of their in-plant food safety (HACCP) processes, are often conducted in highly controlled BSL-2 laboratory environments with research-grade equipment. The food processing environment is extremely variable between small and large processors, and both likely have greater variability of process parameters than that found in BSL-2 lab equipment. USDA-FSIS has recognized this difference and has allowed consideration of ‘in-plant’ validation studies using surrogate organisms if the surrogate can mimic a pathogen’s response to a process [6,7,8]. The intention is that in-plant data would more likely reflect the actual process variability and conditions than scientific equipment from a BSL-2 lab. Conducting a validation study within a processor’s own facility would allow for a more accurate representation of the impact of a commercial process on pathogenic bacteria. Due to food safety concerns, it is unsafe to introduce pathogenic bacteria into a manufacturing facility to test whether the process achieves sufficient microbial reduction. Therefore, nonpathogenic surrogate bacteria would be better suited to mimic the response of pathogens to actual processing conditions [8]. This presents the following question: what surrogate organism should be used for the biltong process?
A surrogate organism for a challenge study is typically a nonpathogenic organism that has similar survival capabilities and susceptibility to injury as the target pathogen and closely mimics how the pathogen would react under similar processing conditions [9,10]. A variety of organisms have been used as surrogates in place of pathogens to mimic pathogenic responses in commercial food processes, predominantly E. faecium, Pediococcus spp., and Biotype 1 E. coli. Enterococcus faecium ATCC 8459 (NRRL B-2354), used as a surrogate for Salmonella Enteritidis PT 30 in the thermal processing of wheat flour [11], as a S. enterica surrogate for storage time and temperature of milk powders [12], in thermal extrusion of low-moisture foods [13], and in plant-level validation of thermal processes for peanuts and pecans [14]. Investigators also found that Pediococcus strains had similar heat tolerances to Salmonella spp. and would be suitable surrogates for validation studies of jerky-style dried meat products [15,16,17]. Pediococcus acidilactici ATCC 8042 was examined as a Salmonella surrogate for thermal processing of toasted oats for cereal and peanuts for peanut butter [18], and for processing of low-moisture pet food [19]. Biotype 1 E. coli ATCC BAA-1427, BAA-1428, BAA-1429, and BAA-1430 have been used as thermal surrogates for E. coli O157:H7 in meat processes [20], as Salmonella surrogates for thermal processing of ground beef [21], and for thermal treatment of almonds and pistachios [22,23]. These strains have been recommended by USDA-FSIS as surrogate indicator organisms for food process validation studies [8].
Despite the prevalence of studies performed with surrogate bacteria for various food processes, no surrogate organisms have been proven to suitably represent the response of pathogens during biltong processing. The objective of this study was to examine potential nonpathogenic lactic acid bacteria and generic E. coli strains that could be used for in-plant studies to mimic pathogen lethality during biltong processing.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
Bacterial cultures used in this study were obtained from various sources including our laboratory culture collection, commercial starter cultures, and bacteria isolated from biltong trials as listed in Table 1.

Table 1.
List of strains used as challenge organisms for biltong processing in this study.
Bacterial isolates obtained from previous biltong beef trials after marination and drying for 8 days at 24.9 °C (75 °F) and 55% relative humidity (RH) were identified by 16S rRNA PCR/sequencing [24] as Carnobacterium gallinarum, Carnobacterium divergens, and Latilactobacillus sakei for examination as biltong process surrogates (Table 1).
Other lactic acid bacteria used in this study included Pedicoccus acidilatcici ATCC 8042, P. acidilactici P02K5, P. pentosaceus FBB61-2, and P. pentosaceus ATCC 43200, which are maintained in our laboratory culture collection. Some of these strains have been evaluated in other surrogate studies [19,25]. Nonpathogenic E. coli ATCC BAA-1427, BAA-1428, BAA-1429, and BAA-1430 have been used as Biotype 1 surrogate strains in various process validation studies and recommended for such use by USDA-FSIS [8,20,26]. P. acidilactici Saga200, used as a protective starter culture, was obtained as a frozen slurry from Kerry Foods (Beloit, WI, USA). Enterococcus faecium 201224-016 was obtained as a freeze-dried powder from Vivolac Cultures (Indianapolis, IN, USA) and is sold as a probiotic.
Carnobacterium spp., E. faecium, and E. coli cultures were inoculated into tryptic soy broth (TSB, BD Bacto, Franklin Laes, NJ, USA) and grown at 30 °C for 24 h. L. sakei and Pedicoccus spp. were inoculated into De Man, Rogosa and Sharpe broth (MRS, BD Bacto) and grown at 30 °C for 24 h. Cultures were prepared for storage by centrifugation (7200× g, 5 °C) of 9 mL of fresh, overnight culture, and the resulting pellet was resuspended with 2–3 mL of fresh, sterile TSB or MRS broth containing 10% glycerol. The cells in freezing media were then placed in 8 mL sterile glass vials and stored in an ultralow-temperature freezer (−80 °C) until use. Prior to use, frozen stocks were revived by transferring 100 µL of partially thawed culture into 9 mL of either TSB or MRS broth and incubated overnight at 30 °C.
Several cultures were used directly after suspension from the freeze-dried or frozen state for comparison of biltong process performance with metabolically active forms grown in liquid media. Prior to use, P. acidilactici Saga200 (frozen) was resuspended by adding 0.5 g of the frozen culture to 9 mL of 0.1% buffered peptone water (BPW, BD Difco) and vortexing until completely incorporated. E. faecium 201224-016 was resuspended by adding 0.1 g of the freeze-dried culture to 9 mL of 0.1% BPW and vortexing until completely mixed.
2.2. Acid Adpation of Cultures
Acid adaptation of active four-strain mixtures of Carnobacterium spp., Pediococcus spp., and E. coli BAA-strains was conducted as first described by Wilde et al. [27] and as used in previous biltong studies [4,28]. In brief, individual cultures were inoculated into TSB or MRS containing 1% glucose, incubated overnight at 30 °C, and harvested by centrifugation; cell pellets were then resuspended with 0.1% BPW. For mixed culture biltong inocula, individual strains were cultured, centrifuged, resuspended, and then combined in equal proportions to create a mixed inoculum cocktail. The commercial starter cultures (P. acidilactici Saga200 and E. faecium 201224-016) were not acid-adapted and used as a single-strain inoculum.
2.3. Beef Sample Preparation and Inoculation
USDA select-grade boneless beef rounds were obtained from a local meat processor (Ralph’s Perkins, OK, USA) who obtains beef from a wholesale beef broker. Beef rounds were trimmed of fat and cut into approximately 5.1 cm wide × 1.9 cm thick × 7.6 cm long beef squares and held overnight at 5 °C on foil-lined trays wrapped in plastic bags. Beef pieces were inoculated the following morning with the respective inoculum depending on the trials being performed that day. Beef pieces were inoculated with the Carnobacterium spp. mixture (C. divergens GO-R2E-B, GO-R1B; C. gallinarum NB-R2A, NB-R2B), the L. sakei mixture (L. sakei GO-R2C, GO-R2D), the Pediococcus spp. mixture (P. acidilactici ATCC 8042, PO2K5; P. pentosaceous ATCC 43200, FBB61-2), P. acidilactici Saga200, or E. faecium 201224-016. The inoculum suspension (150 µL) was applied to each side of the beef pieces and immediately spread with a gloved finger. Inoculated beef pieces were then allowed to incubate for 30 min at 5 °C to allow for bacterial attachment prior to use.
2.4. Biltong Processing, Marination, and Drying
Biltong processing was conducted as described whereby trials were performed in duplicate and triplicate samples were harvested at each timepoint (n = 6) [4,29]. Following inoculation and attachment, the beef pieces were then dipped in sterile water to mimic rinse treatments that processors often apply using antimicrobials or water during processing. The inoculated pieces were placed in a plastic basket, dipped in sterile water in a stainless-steel tub for 30 s, and drained for 60 s to release excess liquid. The beef pieces were then placed into a chilled metal tumbling vessel containing a biltong marinade. The biltong marinade consisted of 2.2% salt, 0.8% black pepper, 1.1% coarse ground coriander, and 4% red wine vinegar (100-grain; 10% acetic acid) in relation to the total meat weight. Beef pieces were vacuum-tumbled (15 inches Hg) in a Biro VTS-43 vacuum-tumbler (Marblehead, OH, USA) for 30 min and then hung to dry in a humidity-controlled oven (Hotpack, Model 435315, Warminster, PA, USA) at 55% relative humidity and 24.9 °C (75 °F) for 8–10 days.
2.5. Selective Recovery of Inoculum Bacteria from Biltong-Inoculated Beef
The bacteria assessed in this study as potential biltong processing surrogates were inoculated onto raw beef, and initial and residual inoculum enumeration had to preclude other natural contaminants also found on raw beef, those contributed during trimming of beef, or from the marinade spice mix. Prior studies indicated that such processing conditions induce stresses, and injured cells may not be recovered on harsh selective media, thereby giving a falsely lower count [28]. To eliminate the possibility of inhibiting injured-but-viable cells, we used generic growth media (TSA, MRS agar) supplemented with antibiotics to which the strains are resistant as a selective medium to enumerate our inoculated organisms from samples taken during biltong processing [4,28]. Antibiotic resistance was determined using antibiotic susceptibility discs (BD BBL Sensi-Discs, BD Labs, Franklin Lakes, NJ, USA) to determine innate antibiotic resistance (Table 1). After identification of antibiotic resistances, cultures were then enumerated on media with and without antibiotics to ensure the absence of inhibition from the use of antibiotics in the media as described previously [4,28,30]. For some strains used as inoculum cocktails that did not have consensus of the same antibiotic resistances, antibiotic resistance was acquired by plating on low level antibiotics known to generate spontaneous antibiotic resistance (i.e., gentamycin and rifamycin).
2.6. Comparison of Commerically Available Starter Cultures as Biltong Inoculants in their Lyophilized and Metabolically Active Forms
2.6.1. Culture Preparation
Lactic acid bacteria obtained as freeze-dried cultures from starter culture companies for use in validation studies may present a facile method of use as validation inocula by simply resuspending the cells in buffer and directly inoculating beef samples [15,17]. Freeze-drying or lyophilization of bacteria exposes them to stressful conditions that can affect subsequent cell viability or activity [20,21]. Therefore, the activity of lyophilized (E. faecium 201224-016) and frozen (P. acidilactici Saga200) starter cultures and their metabolically active forms (i.e., after growth in media) were compared in their response to biltong processing.
For the lyophilized culture (E. faecium 201224-016), 0.1 g of freeze-dried powder was added to 9 mL of sterile 0.1% BPW and vortexed until completed suspended. The resuspended mixture was then used to inoculate each beef piece (300 µL; 150 µL/side) prior to marination.
For the frozen starter culture (P. acidilactici Saga200), a sterile hollow hole puncher was used to core ~0.8 g of frozen Saga200 from the manufacture’s container which was added to 9 mL of sterile 0.1% BPW and vortexed until mixed. The culture suspension was kept chilled on ice and used shortly thereafter to inoculate beef pieces.
Metabolically active versions of these cultures were obtained by growth in 150 mL of the appropriate media (TSB, MRS) for 24 h at 30 °C, centrifugation, and resuspension of the recovered cell pellet with 5 mL of sterile 0.1% BPW. The resuspended culture was then used to inoculate beef pieces prior to use in the validation study. The lyophilized and metabolically active forms of E. faecium 201224-016 and P. acidilactici Saga200 were used in parallel and simultaneous biltong trials to reduce any variables that might influence the observed effect of the marinade and drying process.
2.6.2. Lyophilization of Carnobacterium gallinarum NB-R2A
To the authors’ knowledge, there is no commercially available Carnobacterium strain available in the United States. Therefore, C. gallinarum NB-R2A, isolated from biltong, was lyophilized via freeze-drying to examine a lyophilized version for comparison with the actively grown culture. Carnobacterium gallinarum NB-R2A was inoculated into 9 mL of TSB from frozen stock and incubated for 18 h at 30 °C. Following incubation, the 9 mL culture was transferred to 190 mL of TSB and incubated again for 18 h at 30 °C. The culture was then centrifuged at 7200× g for 20 min. The supernatant was removed, and the cell pellet was resuspended with 5 mL of sterile BPW and repeated. The supernatant was removed following centrifugation, and the final cell pellet was resuspended with 10 mL of autoclaved milk-based freeze-drying medium consisting of 11 g of skim milk powder, 1 g of dextrose, 1 g of trehalose, and 0.2 g of yeast extract per 100 mL. The milk/cell suspension was added to Oak Ridge tubes (5 mL each) and freeze-dried using a Heto vacuum centrifuge (Model VR-maxi) connected to a Heto freezing condensor (Model CT 60E) and a Leybold Trivac vacuum pump (Model D2.5F) setup for 24 h under vacuum. The freeze-dried powder was then stored at −80 °C until use in our biltong study. Just before use, 0.25 g of powder was added to 9 mL of sterile 0.1% BPW, vortexed until mixed, and used to inoculate beef pieces for biltong processing.
2.7. Evaluation of Critical Parameters and Intrinsic Factors in the Biltong Process
2.7.1. Water Activity
Uninoculated beef pieces were sampled for water activity (Aw) measurements at various stages throughout processing (in triplicate) including the initial raw beef, beef after marination, and then beef after drying for 2, 4, 6, 8, and 10 days. To obtain measurements, beef pieces were cut in half and placed in a sampling cup with the interior portion of the sample facing upward (toward the sensor). Samples were then covered with sampling cup cover containing the sensor and allowed to equilibrate to the temperature of the room. Water activity was measured using a HC2-AW-USB probe with a direct PC interface and HW4-P-Quick software (Rotronic Corp., Hauppauge, NY, USA). Measurements were taken in triplicate for each sample at each timepoint.
2.7.2. Moisture Loss
Following marination, each beef piece was individually weighed and labeled prior to being hung in the humidity-controlled oven. Three pieces were selected and weighed prior to processing, and then sampled every 2 days while drying. The weight at the time of sampling was compared to the initial weight of the same piece recorded prior to drying. The determination of percent moisture loss was calculated as per Equation (1).
.
2.7.3. Measurement of Biltong Beef pH
Measurements of beef pH were obtained at various points in the biltong process including raw beef, beef following marination, and beef after 2, 4, 6, 8, and 10 days of drying. At each timepoint, three pieces of uninoculated beef were collected, weighed, and then added to a laboratory blender with steel blades (Waring Commercial, New Harford, CT, USA) with sterile water of equal weight to the weight of the beef pieces. The water and beef mixtures were blended until a finely ground mixture was formed. The pH of the homogenized meat mixture was measured in triplicate using an H-series pH meter and probe (Hach, Loveland, CO, USA).
2.7.4. Salt Concentration
The homogenized meat mixture used to measure pH was also used to obtain salt concentrations of each sample. Horiba LAQUA Twin Pocket Meter (Horiba Instruments, Irvine, CA, USA) was used to quantify sodium ion concentration. Approximately 300 µL of the homogenized sample was placed in the sample chamber and allowed to stabilize before recording. Readings (in ppm) were taken in triplicate for each sample. To determine the salt (NaCl) concentration from the sodium ion concentration, the following equations were used:
2.8. Microbial Sampling and Inoculum Enumeration of Biltong Beef
At each sampling timepoint of biltong beef processing (raw beef, after marinade, and after every 2, 4, 6, 8, and 10 days of drying), three beef pieces were selected at random and placed in a sterile Whirl-pak filter stomaching bag (Nasco, Fort Atkinson, WI, USA) in combination with 100 mL of 1% neutralizing buffered peptone water (nBPW, Criterion, Hardy Diagnostics, Santa Maria, CA, USA). Samples were stomached for 60 s in a paddle-blender masticator (IUL Instruments, Barcelona, Spain). Serial dilutions were made with 1% BPW and plated on TSA containing gentamicin and rifamycin (2.5 µg/mL each) for Carnobacterium, on MRSA containing gentamicin and rifamycin (2.5 µg/mL each) for L. sakei, on MRSA containing gentamicin (10 µg/mL) and rifamycin (5 µg/mL) for Pediococcus spp., on TSA containing naldixic acid and colistin (10 µg/mL each) for E. faecium 201224-016, or on MRS containing nalidixic acid and colistin (10 µg/mL each) for P. acidilactici Saga200; the filter bag dilution was considered the 10° dilution. Plates were incubated at 30 °C for 48 h and enumerated as log CFU/mL. Samples were collected in triplicate replication and plated in duplicate at each sampling timepoint.
2.9. Statistical Analysis
Validation trials were conducted in duplicate with triplicate sampling at each timepoint (n = 6) as per validation criteria established by the National Advisory Committee on Microbial Criteria for Foods (NACMCF) [9] and supported by the USDA-FSIS [31]. Data are presented as the mean of multiple replications with standard deviation of the mean represented by error bars. Statistical analysis of data collected over time was performed using one-way repeated-measures analysis of variance (RM-ANOVA). Pairwise multiple comparisons were performed using the Holm–Sidak test to determine significant differences. Data treatments with the same letter are not significantly different (p > 0.05); treatments with different letters are significantly different (p < 0.05).
3. Results and Discussion
3.1. Critical Parameters and Intrinsic Factors
3.1.1. Water Activity, Moisture Loss, and Salt Concentrations
To complement the surrogate validation trials, we measured and recorded critical operational parameters and intrinsic factors at each key stage of processing (raw beef, inoculation, marination, and every 2 days of drying) as recommended by USDA-FSIS [1]. Water activity (Aw) is a measure of free, unbound water available for bacterial growth. USDA-FSIS considers vacuum-tumbled beef as ‘nonintact beef’, whereby Aw is a primary safety factor as there is no heat lethality step in biltong processing and biltong is processed as thick beef samples [32,33]. Therefore, Aw is a critical safety factor for control of bacteria that might be internalized due to vacuum tumbling. S. aureus that can tolerate low Aw and high salt levels would be a concern for possible production of staphylococcal enterotoxin. The targeted Aw for shelf-stable beef jerky is <0.85 which was achieved after 7 days of drying (Figure 1) [1,2]. Water activity after 8 and 10 days of drying ranged from 0.82 to 0.79 respectively. Similarly, beef samples showed incremental moisture loss with 59% and 62.5% loss at 8 and 10 days, respectively (Figure 1).
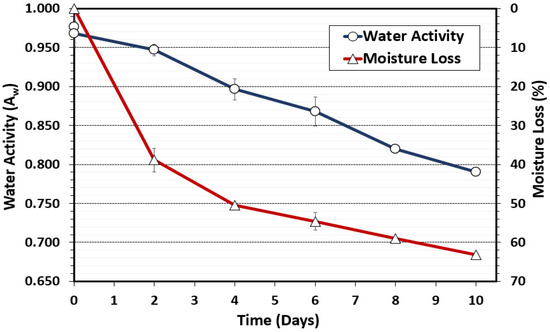
Figure 1.
Water activity (Aw) and moisture loss during biltong processing at 24.9 °C (75 °F) and 55% RH. The data represent the average of measurements taken during duplicate trials with triplicate samples taken at each time interval (n = 6).
Salt concentration was also determined during the biltong process. Salt concentration was calculated from sodium readings obtained with the LAQUAtwin NA-11 sodium ion meter (Horiba Inc, Irvine, CA, USA). The initial calculated salt concentration determined on raw beef was 0.12% NaCl; then, following the marination step, the beef salt concentration shot up to 2.17% (2.17 g NaCl/100 g beef). The initial salt level falls in line with expectations given that the biltong marinade was formulated at 2.2% salt (w/w). The salt concentration increased over time and was indirectly proportional to moisture loss during the drying process (Figure 2). As expected, as moisture loss occurred, Aw was also reduced to below 0.85 Aw (Figure 1) and the salt concentration increased to above 4% (Figure 2); both conditions are inhibitory to most bacteria, helping to ensure a safe product for consumers [34]. Biltong safety involves an interplay among moisture, salt concentration, and Aw since moisture loss increases salt concentration, while salt binds water and helps to draw it out of the interior of the beef, thereby reducing Aw. For consumer issues regarding high sodium levels, the use of alternative salts (CaCl2, KCl) instead of NaCl can help lower sodium levels in finished biltong while still maintaining a 5 log reduction of pathogen (Salmonella) [29].
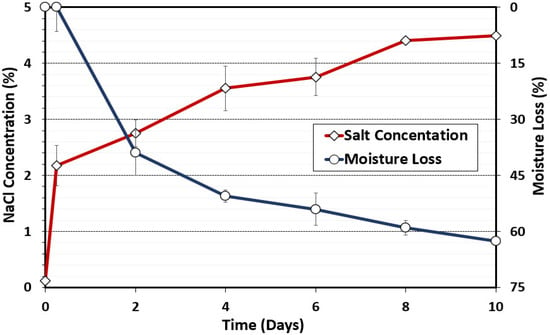
Figure 2.
Moisture loss (%) and salt concentration (%) during biltong processing. Measurements were taken with initial beef samples, after marination, and after 2, 4, 6, 8, and 10 days of drying at 24.9 °C (75 °F) and 55% RH. Data points represent the mean of duplicate trials with triplicate samples taken at each time interval (n = 6).
3.1.2. The pH of Beef during Biltong Processing
The initial pH of the raw meat pieces was approximately pH 5.43 (Figure 3), which was determined by blending beef samples in sterile water in a laboratory blender. The pH of the samples then decreased following the marination step to 5.02, which can be attributed to the presence of residual 100-grain red wine vinegar in the marinade. After removal from the marinade, the pH of biltong beef samples then equilibrated slightly higher to ~5.18–5.20 for the remainder of the drying process in the humidity-controlled oven (Figure 3). The pH of the marination solution was much lower (pH 2.5–2.7); during 30 min vacuum tumbling, the surface bacteria were immersed in the low-pH marinade solution, which could lead to cell death and inactivation of pathogenic bacteria [35,36], as observed in the current study and prior biltong trials where levels of inoculated pathogens were reduced after marination [4]. After removal from the vacuum tumbler, the residual marinade on the surface was absorbed, and the pH of biltong beef samples equilibrated to ~5.18–5.20 for the remainder of the drying process in the humidity oven (Figure 3).
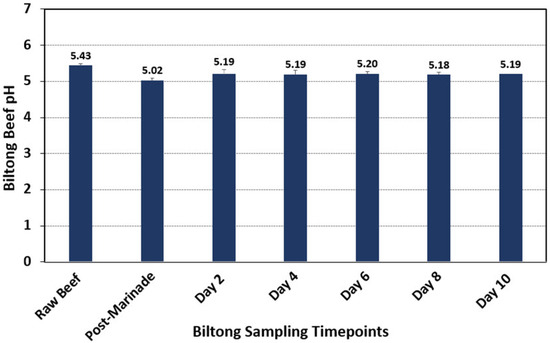
Figure 3.
The pH of meat at each sampling timepoint during biltong processing. Samples were taken in triplicate at each timepoint following blending with sterile water in a laboratory blender (n = 6).
3.1.3. Temperature and Relative Humidity during Biltong Processing
Temperature and RH measurements were recorded by computer software connected to the handheld temperature and humidity recorders to which the probes in the oven chamber were connected (Figure 4). Two temperature probes were inserted separately into two beef pieces to measure the internal beef temperature during processing, while the humidity probe was placed midway within the chamber. Air temperature and humidity were set to 23.9 °C (75 °F) and 55% throughout the duration of each trial but cycled above and below the set points. The internal temperature of the beef was more consistent and steadily increased from their initial temperature to match the temperature of the chamber. Long-term storage at low RH helps to evaporate moisture from the beef.
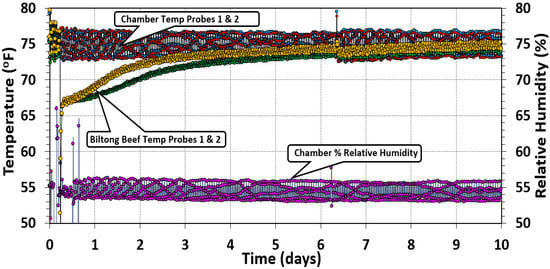
Figure 4.
Oven temperature and relative humidity measurements. The temperature was set to 24.9 °C (75 °F), and the relative humidity setpoint was 55% RH during the drying process over a period of 10 days. Graphical data show the typical cycling of oven control above/below setpoint. Two temperature probes were placed in various places in the chamber and two additional probes were inserted into separate pieces of beef to track the internal temperature of the biltong product over the same drying period.
3.2. Surrogate Log Reductions during Biltong Processing
Various bacteria were considered for examination as possible nonpathogenic surrogates, including strains recovered from biltong after processing. These included a two-strain mixture of L. sakei GO-R2C and GO-R2D and a four-strain mixture of C. divergens GO-R2E-B and GO-R1B and C. gallinarum NB-R2A and NB-R2B (Figure 5). We also examined a four-strain mixture of P. acidilactici and P. pentosaceous strains (P. acidilactici ATCC 8042 and PO2K5; P. pentosaceous ATCC 43200 and FBB61-2) vs. starter cultures that were available through culture companies (E. faecium 201224-016 and P. acidilactici Saga200) as surrogate organisms (Figure 5).
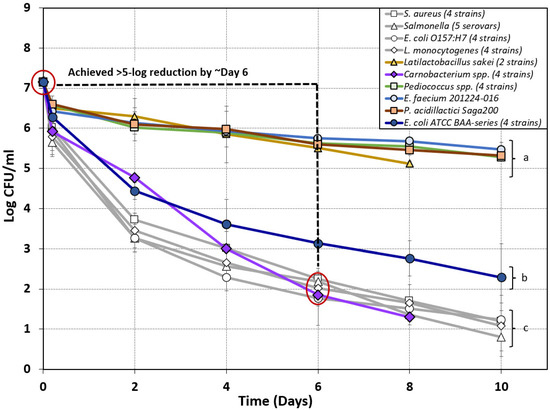
Figure 5.
Composite graph of biltong processing data of nonpathogenic bacteria attempting to mimic the biltong process log reduction of pathogenic bacteria (light-gray lines) to be considered a possible ‘biltong processing surrogate’ organism for in-plant validation. Log reduction curves of various lactic acid bacteria (Carnobacterium spp., Pediococcus spp., L. sakei, and E. faecium) and Biotype I E. coli strains tested as potential surrogate organisms for biltong processing over a period of 8–10 days. Strains were compared to the log reduction curves observed during previous biltong validation studies using pathogenic bacteria including Salmonella serovars [4], S. aureus, E. coli O157:H7, and L. monocytogenes [5]. Data points are the mean of duplicate trials sampled in triplicate (n = 6). Statistical analysis was performed using one-way repeated-measures analysis of variance (RM-ANOVA) of the entire time course of data; curves with the same letter are not significantly different (p > 0.05); isolates with different letters are significantly different (p < 0.05).
Only a slight reduction from inoculated levels was observed following vinegar/spice/salt marination (0.65, 0.58, 0.75, and 0.61 log reduction) with all cultures used, except for the four-strain mixtures of Carnobacterium spp. and E. coli ATCC BAA series (Figure 5). A larger log reduction was observed after marination of the four-strain mixtures of Carnobacterium spp. (1.23 log) and E. coli ATCC BAA-strains (0.86 log) (Figure 5). Trials using E. coli ATCC BAA (four-strain mix), L. sakei (two-strain mix), Pediococcus spp. (four-strain mix), E. faecium 201224-016, and P. acidilactici Saga200 failed to achieve a 5 log reduction during biltong processing with overall reductions of 4.86 log, 2.03 log, 1.87 log, 1.68 log, and 1.83 log respectively. Of all the nonpathogenic strains examined, only the four-strain mixture of Carnobacterium spp. achieved an overall reduction of greater than 5 log (5.85 log) during the 8 day drying period (Figure 5). On the basis of these results, Carnobacterium spp. were the only organisms that achieved a 5 log reduction (within 6–8 days) comparable to that observed for the pathogenic strains, and they presented the best case for use as a Salmonella, L. monocytogenes, E. coli O157:H7, or S. aureus surrogate for biltong processing (Figure 5).
3.3. Comparison of Lyophilized/Frozen Starter Cultures with Metabolically Active (Grown) Versions in Biltong Processing Trials
Several reports in the literature have used freeze-dried or frozen cultures, resuspended directly in buffer, to inoculate food samples in process trials for direct comparison to pathogens grown in microbiological media (which we describe as ‘active cultures’) [15,17]. The ease of availability of freeze-dried/frozen cultures from culture companies would facilitate the use of such cultures for in-plant validation studies; however, we were interested to see if they could provide the same response in a biltong process as the actively grown cultures (Figure 6). The comparisons were between two commercially available starter cultures, E. faecium 201224-016 (Vivolac Cultures; freeze-dried) and P. acidilactici Saga200 (Kerry Foods; frozen), and a lyophilized C. divergens NB R2A, which was chosen from among the Carnobacterium mixed strains demonstrating >5 log reduction in Figure 5.
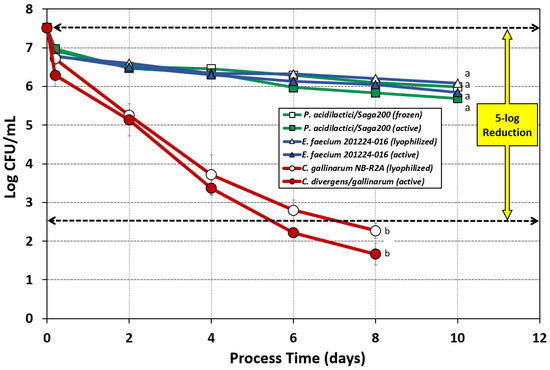
Figure 6.
Biltong processing of beef inoculated with lyophilized/frozen cells vs. metabolically active cells (freshly grown) of E. faecium 201224-016, P. acidilactici, and C. gallinarum NB-R2A. Lyophilized C. gallinarum NB-R2A was compared to a four-strain cocktail of metabolically active C. divergens/gallinarum. Graph curves of frozen or lyophilized cultures have hollow symbols. Statistical analysis was performed using one-way repeated-measures analysis of variance (RM-ANOVA) over the entire time course of the datasets; graphs with the same letter are not significantly different (p > 0.05); isolates with different letters are significantly different (p < 0.05).
Neither the lyophilized version of E. faecium 201224-016 (1.43 log reduction) nor the frozen version of P. acidilactici Saga200 (1.54 log reduction) achieved the 5 log reduction target; survival curves of the lyophilized/frozen forms were also not significantly different when compared to their metabolically active forms, i.e., 1.68 and 1.83 log reduction, respectively (Figure 6). The lyophilized single strain C. divergens NB R2A also showed no significant difference from the metabolically active culture and again achieved 5 log reduction during the biltong process (Figure 6). The data show that lyophilized or frozen versions of E. faecium, P. acidilactici, or C. gallinarum do not respond differently than actively grown cultures to biltong processing conditions and, when possible, their use might facilitate inoculated studies.
4. Conclusions
The lethality observed in the biltong process with Carnobacterium spp. aligned with that observed with four major pathogenic organisms indicating that Carnobacterium spp. could be an effective in-plant surrogate organism to monitor the effectiveness of biltong processing within a manufacturer’s facility. Enterococcus faecium, L. sakei, and Pediococcus spp. were not reduced much (<2 log) and were resilient toward the acid, salt, and low Aw experienced during 10 days of biltong processing. The use of lyophilized/frozen cells as inoculum for validation of biltong processing was not significantly different than using actively grown cells. This work helps to fill USDA-FSIS knowledge gaps in air-dried shelf-stable dried beef (biltong) processing with regard to potential surrogate organisms and critical factors involved in the biltong process. Future studies on biltong processing may include whether pathogens such as Salmonella, known to survive long periods of low water activity, can survive the extended shelf-life of biltong products to ensure that this does not become a possible (overlooked) problem.
Author Contributions
Conceptualization, P.M.M.; methodology, P.M.M. and C.E.K.; software, P.M.M.; validation, C.E.K. and P.M.M.; formal analysis, C.E.K. and J.W.; investigation, C.E.K. and J.W.; resources, P.M.M.; data curation, P.M.M.; writing—original draft preparation, C.E.K.; writing—review and editing, P.M.M.; visualization, P.M.M.; supervision, P.M.M.; project administration, P.M.M.; funding acquisition, P.M.M. All authors have read and agreed to the published version of the manuscript.
Funding
Funding for the study was contributed in part by the Robert M. Kerr Food & Ag Products Center, the Advance Foods—SE Gilliland Professorship in Microbial Food Safety (#21-57200), the USDA National Institute of Food and Agriculture (Hatch Project #OKL03090), the Oklahoma Agricultural Experiment Station, the Division of Agricultural Sciences and Natural Resources at Oklahoma State University, and various industry sources.
Institutional Review Board Statement
The work performed in this study was performed under Protocol #17-8 approved by the OSU Institutional Biosafety Committee within the Office of University Research Compliance.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to acknowledge the ‘mentoring team’ supporting Caitlin Karolenko’s USDA-FSIS 2021–2022 Graduate Fellowship Award, including Emilio Esteban (USDA-FSIS), Isabel Walls (USDA-FSIS; Washington, DC, USA), Meryl Silverman (USDA-FSIS; OPPD, Washington, DC, USA), Kristina Barlow (USDA-FSIS; Risk Management and Innovations Staff, OPPD, Washington, DC, USA), Terry Campbell (USDA-FSIS; OPHS, Eastern Laboratory, Athens, GA), and Steve W. Mamber (USDA-FSIS; OPARM, Washington, DC, USA), for their technical assistance in meat processing and USDA-FSIS regulatory policy with respect to dried beef.
Conflicts of Interest
The authors declare no conflict of interest.
References
- USDA-FSIS. FSIS Compliance Guideline for Meat and Poultry Jerky Produced by Small and Very Small Establishments; U.S. Food Safety and Inspection Service: Washington, DC, USA, 2014; pp. 1–54.
- USDA-FSIS. Salmonella Compliance Guidelines for Small and Very Small Meat and Poultry Establishments That Produce Ready-to-Eat (RTE) Products and Revised Appendix A, June 2017; U.S. Food Safety and Inspection Service: Washington, DC, USA, 2017; pp. 1–37.
- Nickelson, R.; Luchansky, J.B.; Kaspar, C.; Johnson, E. An executive summary prepared by The Blue Ribbon Task Force of the National Cattlemen’s Beef Association. In Dry Fermented Sausage and Escherichia coli O157:H7 Validation Research; Research Report No. 11-316; National Cattlemen’s Beef Association: Chicago, IL, USA, 1996. [Google Scholar]
- Karolenko, C.E.; Bhusal, A.; Nelson, J.L.; Muriana, P.M. Processing of biltong (dried beef) to achieve USDA-FSIS 5-log reduction of Salmonella without a heat lethality step. Microorganisms 2020, 8, 791. [Google Scholar] [CrossRef]
- Gavai, K.; Karolenko, C.; Muriana, P.M. Effect of biltong dried beef processing on the reduction of Listeria monocytogenes, E. coli O157:H7, and Staphylococcus aureus, and the contribution of the major marinade components. Microorganisms 2022, 10, 1308. [Google Scholar] [CrossRef] [PubMed]
- USDA-FSIS. Guidelines for preparing and submitting experimental protocols for in-plant trials of new technologies and procedures. Fed. Reg. 1995, 60, 2. [Google Scholar]
- USDA-FSIS. FSIS procedures for notification of new technology. Fed. Reg. 2003, 68, 3. [Google Scholar]
- USDA-FSIS. Use of Non-Pathogenic Escherichia coli (E. coli) Cultures as Surrogate Indicator Organisms in Validation Studies. Available online: https://ask.usda.gov/s/article/Use-of-Non-pathogenic-Escherichia-coli-E-coli-Cultures-as-Surrogate-Indicator-Organisms-in-Validation-Studies#:~:text=Can%20establishments%20use%20non-pathogenic%20E.%20coli%20cultures%20as,establishment%20to%20measure%20changes%20in%20microbial%20counts%3F%20Yes (accessed on 13 June 2022).
- National Advisory Committee on Microbiological Criteria for Foods. Parameters for determining inoculated pack/challenge study protocols. J. Food Prot. 2010, 73, 140–202. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Gurtler, J.B. Selection of surrogate bacteria for use in food safety challenge studies: A review. J. Food Prot. 2017, 80, 1506–1536. [Google Scholar] [CrossRef]
- Liu, S.; Rojas, R.V.; Gray, P.; Zhu, M.-J.; Tang, J. Enterococcus faecium as a Salmonella surrogate in the thermal processing of wheat flour: Influence of water activity at high temperatures. Food Microbiol. 2018, 74, 92–99. [Google Scholar] [CrossRef]
- Wei, X.; Agarwal, S.; Subbiah, J. Evaluation of Enterococcus faecium NRRL B-2354 as a surrogate for Salmonella enterica in milk powders at different storage times and temperatures. J. Dairy Sci. 2021, 104, 198–210. [Google Scholar] [CrossRef]
- Bianchini, A.; Stratton, J.; Weier, S.; Hartter, T.; Plattner, B.; Rokey, G.; Hertzel, G.; Gompa, L.; Martinez, B.; Eskridge, K.M. Use of Enterococcus faecium as a surrogate for Salmonella enterica during extrusion of a balanced carbohydrate-protein meal. J. Food Prot. 2014, 77, 75–82. [Google Scholar] [CrossRef]
- Brar, P.K.; Danyluk, M.D. Validation of Enterococcus faecium as a surrogate for Salmonella under different processing conditions for peanuts and pecans. Food Microbiol. 2019, 80, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Borowski, A.G.; Ingham, S.C.; Ingham, B.H. Validation of ground-and-formed beef jerky processes using commercial lactic acid bacteria starter cultures as pathogen surrogates. J. Food Prot. 2009, 72, 1234–1247. [Google Scholar] [CrossRef] [PubMed]
- Dierschke, S.; Ingham, S.C.; Ingham, B.H. Destruction of Escherichia coli O157:H7, Salmonella, Listeria monocytogenes, and Staphylococcus aureus achieved during manufacture of whole-muscle beef jerky in home-style dehydrators. J. Food Prot. 2010, 73, 2034–2042. [Google Scholar] [CrossRef]
- Borowski, A.G.; Ingham, S.C.; Ingham, B.H. Lethality of home-style dehydrator processes against Escherichia coli O157:H7 and Salmonella serovars in the manufacture of ground-and-formed beef jerky and the potential for using a pathogen surrogate in process validation. J. Food Prot. 2009, 72, 2056–2064. [Google Scholar] [CrossRef] [PubMed]
- Deen, B.; Diez-Gonzalez, F. Assessment of Pediococcus acidilactici ATCC 8042 as potential Salmonella surrogate for thermal treatments of toasted oats cereal and peanut butter. Food Microbiol. 2019, 83, 187–192. [Google Scholar] [CrossRef]
- Ceylan, E.; Bautista, D.A. Evaluating Pediococcus acidilactici and Enterococcus faecium NRRL B-2354 as thermal surrogate microorganisms for Salmonella for in-plant validation studies of low-moisture pet food products. J. Food Prot. 2015, 78, 934–939. [Google Scholar] [CrossRef]
- Keeling, C.; Niebuhr, S.E.; Acuff, G.R.; Dickson, J.S. Evaluation of Escherichia coli Biotype I as a Surrogate for Escherichia coli O157:H7 for Cooking, Fermentation, Freezing, and Refrigerated Storage in Meat Processes. J. Food Prot. 2009, 72, 728–732. [Google Scholar] [CrossRef]
- Redemann, M.A.; Brar, J.; Niebuhr, S.E.; Lucia, L.M.; Acuff, G.R.; Dickson, J.S.; Singh, M. Evaluation of thermal process lethality for non-pathogenic Escherichia coli as a surrogate for Salmonella in ground beef. LWT 2018, 90, 290–296. [Google Scholar] [CrossRef]
- Ma, L.; Kornacki, J.L.; Zhang, G.; Lin, C.M.; Doyle, M.P. Development of thermal surrogate microorganisms in ground beef for in-plant critical control point validation studies. J. Food Prot. 2007, 70, 952–957. [Google Scholar] [CrossRef]
- Almond Board of California. Guidelines for Process Validation Using Enterococcus faecium NRRL B-2354; Almond Board of California: Modesto, CA, USA, 2007. [Google Scholar]
- Shah, K.; Muriana, P.M. Efficacy of a next generation quaternary ammonium chloride sanitizer on Staphylococcus and Pseudomonas biofilms and practical application in a food processing environment. Appl. Microbiol. 2021, 1, 89–103. [Google Scholar] [CrossRef]
- de Souza de Azevedo, P.O.; de Azevedo, H.F.; Figueroa, E.; Converti, A.; Domínguez, J.M.; de Souza Oliveira, R.P. Effects of pH and sugar supplements on bacteriocin-like inhibitory substance production by Pediococcus pentosaceus. Molec. Biol. Rep. 2019, 46, 4883–4891. [Google Scholar] [CrossRef]
- Niebuhr, S.E.; Laury, A.; Acuff, G.R.; Dickson, J.S. Evaluation of Nonpathogenic Surrogate Bacteria as Process Validation Indicators for Salmonella enterica for Selected Antimicrobial Treatments, Cold Storage, and Fermentation in Meat. J. Food Prot. 2008, 71, 714–718. [Google Scholar] [CrossRef]
- Wilde, S.; Jørgensen, F.; Campbell, A.; Rowbury, R.; Humphrey, T. Growth of Salmonella enterica Serovar Enteritidis PT4 in media containing glucose results in enhanced RpoS-independent heat and acid tolerance but does not affect the ability to survive air-drying on surfaces. Food Microbiol. 2000, 17, 679–686. [Google Scholar] [CrossRef]
- Karolenko, C.E.; Bhusal, A.; Gautam, D.; Muriana, P.M. Selenite cystine agar for enumeration of inoculated Salmonella serovars recovered from stressful conditions during antimicrobial validation studies. Microorganisms 2020, 8, 338. [Google Scholar] [CrossRef] [PubMed]
- Karolenko, C.; Muriana, P. Quantification of process lethality (5-Log reduction) of Salmonella and salt concentration during sodium replacement in biltong marinade. Foods 2020, 9, 1570. [Google Scholar] [CrossRef]
- Bhusal, A.; Nelson, J.; Pletcher, D.; Muriana, P.M. Comparison of sodium nitrite and ‘natural’ nitrite on the inhibition of spore germination and outgrowth of Clostridium sporogenes in low- and high-fat frankfurters. Appl. Microbiol. 2021, 1, 104–122. [Google Scholar] [CrossRef]
- USDA-FSIS (Ed.) FSIS Compliance Guideline HACCP Systems Validation (April 2015); USDA-FSIS: Washington, DC, USA, 2015; pp. 1–68.
- Pokharel, S.; Brooks, J.C.; Martin, J.N.; Brashears, M.M. Antimicrobial susceptibility and internalization of Salmonella Typhimurium in vacuum-tumbled marinated beef products. Lett. Appl. Microbiol. 2016, 63, 412–418. [Google Scholar] [CrossRef]
- USDA-FSIS. Non-Intact Beef Products. Available online: https://ask.usda.gov/s/article/Non-intact-beef-products (accessed on 16 June 2022).
- Gurtler, J.B.; Keller, S.E.; Kornacki, J.L.; Annous, B.A.; Jin, T.; Fan, X. Challenges in recovering foodborne pathogens from low-water-activity foods. J. Food Prot. 2019, 82, 988–996. [Google Scholar] [CrossRef]
- Lund, P.A.; De Biase, D.; Liran, O.; Scheler, O.; Mira, N.P.; Cetecioglu, Z.; Fernández, E.N.; Bover-Cid, S.; Hall, R.; Sauer, M.; et al. Understanding how microorganisms respond to acid pH Is central to their control and successful exploitation. Front. Microbiol. 2020, 11, 556140. [Google Scholar] [CrossRef]
- Jin, Q.; Kirk, M.F. pH as a primary control in environmental microbiology: 1. thermodynamic perspective. Front. Environ. Sci. 2018, 6, 21. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).