Abstract
Antibiosis has been proposed to contribute to the beneficial bacteria-mediated biocontrol against pea Aphanomyces root rot caused by the oomycete pathogen Aphanomyces euteiches. However, the antibiotics required for disease suppression remain unknown. In this study, we found that the wild type strains of Pseudomonas protegens Pf-5 and Pseudomonas fluorescens 2P24, but not their mutants that lack 2,4-diacetylphloroglucinol, strongly inhibited A. euteiches on culture plates. Purified 2,4-diacetylphloroglucinol compound caused extensive hyphal branching and stunted hyphal growth of A. euteiches. Using a GFP-based transcriptional reporter assay, we found that expression of the 2,4-diacetylphloroglucinol biosynthesis gene phlAPf-5 is activated by germinating pea seeds. The 2,4-diacetylphloroglucinol producing Pf-5 derivative, but not its 2,4-diacetylphloroglucinol non-producing mutant, reduced disease severity caused by A. euteiches on pea plants in greenhouse conditions. This is the first report that 2,4-diacetylphloroglucinol produced by strains of Pseudomonas species plays an important role in the biocontrol of pea Aphanomyces root rot.
1. Introduction
Biological control of soil-borne plant diseases by beneficial bacteria is a promising ecofriendly disease management strategy. Commercial application of beneficial bacteria-mediated biological control can be improved by increasing the disease control efficacy and reducing the control variations [1]. Understanding how beneficial bacteria suppress plant diseases will help us increase the efficacy and consistency of biological control [2].
Aphanomyces root rot of pea (Pisum sativum), caused by the oomycete pathogen Aphanomyces euteiches, is one of the most damaging soil-borne diseases that constrain pea production worldwide [3]. Interest in using beneficial bacteria to control A. euteiches is growing because of the unavailability of resistant pea cultivars, limited chemical management options, concerns of potential fungicide resistance by the pathogens, and the increasing requests for ecofriendly management alternatives [4,5]. Several beneficial bacteria, including strains of Pseudomonas spp., Bacillus mycoides, Rhizobium spp., Pantoea agglomerans, Lysobacter capsici, and Burkholderia cepacia, were found to inhibit the A. euteiches growth in culture media and/or reduce the disease incidence in greenhouse [6,7,8]. Results of these studies suggest that antibiosis likely contributes to the pathogen suppression and disease control. For example, the beneficial bacteria that reduced the disease severity of pea Aphanomyces root rot in growth camber biocontrol assays were also found to inhibit the A. euteiches mycelial growth on culture plates [8]. However, the antibiotic(s) required by the beneficial bacteria to inhibit the A. euteiches growth and control the disease remain(s) unknown. Considering the well-recognized and important roles of antibiotics in biocontrol [9], filling this knowledge gap will advance our understanding of the biocontrol mechanism of pea Aphanomyces root rot and improve the disease control efficacy of the antibiotic-producing beneficial bacteria.
The goals of this study were to identify beneficial bacteria-derived antibiotics that inhibit the growth of A. euteiches and characterize the role of those antibiotics in the tri-trophic interactions of A. euteiches, pea plant and the beneficial bacteria. Antagonistic bacterial isolates that belong to different genera, including Pseudomonas, Paenibacillus, Bacillus, Pseudarthrobacter, and Chryseobacterium, were identified from pea rhizosphere soils in this work. We focus on species of the Pseudomonas genus because strains of this group have been constantly identified to suppress pea Aphanomyces root rot [8,10]. Pseudomonas protegens strain Pf-5, a soil bacterium which is well-known for its wide-spectrum antimicrobial activity due to the production of many antibiotics including pyoluteorin, pyrrolnitrin, 2,4-diacetylphloroglucinol (2,4-DAPG), orfamide A, rhizoxin, hydrogen cyanide [11,12,13,14], and Pseudomonas fluorescens strain 2P24, a wheat rhizosphere bacterium with antibacterial and antifungal activities [15,16], were used in this study. The inhibitory effects of P. protegens Pf-5 and P. fluorescens 2P24 on the growth of A. euteiches were measured. The antibiotic required by P. protegens Pf-5 and P. fluorescens 2P24 to inhibit A. euteiches was identified and the roles of the antibiotic in biological control of pea Aphanomyces root rot were investigated.
2. Materials and Methods
2.1. Strains and Cultural Conditions
The oomycete and bacterial strains, plasmids, and sequences of oligonucleotides used in this study are listed in Table 1. Strains of P. protegens Pf-5 and P. fluorescens 2P24 were cultured at 28 °C on King’s medium B (KB) [17] or Nutrient Broth (Becton, Dickenson and Company, Franklin Lakes, NJ, USA) with or without agar (15%). Liquid cultures were grown with shaking at 250 rpm. A. euteiches was cultured at room temperature on half strength potato dextrose agar (½ PDA) (Becton, Dickenson and Company) plates that are commonly used to culture plant-associated oomycetes [18,19].
Phloroglucinol was purchased from Sigma-Aldrich (St. Louis, MO, USA), monoacetylphloroglucinol was purchased from Sigma-Aldrich Chemie GmbH (Schnelldorf, Germany), and 2,4-diacetylphloroglucinol was purchased from Toronto Research Chemicals (North York, ON, Canada). The compounds were dissolved in methanol.
2.2. Isolation and Identification of Pea-Associated Bacterial Antagonists
Rhizosphere soils were collected in June 2020 from lentil-growing fields at Valley County (GPS: 48°43.352′ N; 106°05.213′ W), Montana. The rhizosphere soils were suspended in 10 mL sterilized water and the suspensions were diluted onto ½ and 1/10 TSA (tryptone soy agar) plates that were incubated at 28 °C for 2–5 days. Bacterial isolates with diverse colony morphologies were collected from the plates and their inhibition against the growth of A. euteiches was tested as described below. The isolates that showed clear inhibition against A. euteiches were confirmed in a repeat experiment and stored at −80 °C in a freezer and their taxonomies were identified via 16s DNA analysis using oligonucleotide pairs 27f/1492r [20].
The DNA sequences of the PCR amplified 16s rRNA genes were determined by standard Sanger sequencing at GeneWiz (Azenta Life Sciences, Chelmsford, MA, USA), deposited into GenBank (accession numbers were shown in the phylogenetic tree in Figure 1C), and analyzed using BLAST with the NCBI nr/nt database to identify the closely related bacterial strains. The 16s rRNA sequences of the bacterial isolates that were identified in this study and the closely related strains that were retrieved from the NCBI database were aligned using a web-based sequence aligner program MAFFT v 7.490 [21]. The aligned sequences in the Phylip (.phy) format were then used for the phylogenetic analysis using CIPRES Science Gateway [22]. The phylogenetic tree was generated using the IQ-tree on XSEDE v 2.1.2 [23] based on the maximum likelihood approach. The GTR nucleotide substitution model was used, and the number of bootstrap iterations was set as 1000. The numbers at the tree nodes represent bootstrap support. The Figtree v 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree) was used for the visualization of the phylogenetic tree. The scale at the bottom represents the number of nucleotide substitutions per site.

Table 1.
Bacterial strains, plasmid, and primers used in this study.
Table 1.
Bacterial strains, plasmid, and primers used in this study.
| Strains, Plasmids, and Primers | Genotypes, Relevant Characteristics, and DNA Sequences # | Reference or Source |
|---|---|---|
| A. euteiches | Oomycete pathogen isolated from pea Aphanomyces root rot disease sample in Montana. | [24] |
| P. protegens strains: | ||
| LK099 | Wild type Pf-5, soil isolate, DAPG+, Ofa+, Prn+, Plt+, HCN+, Rzx+. | [25] |
| JL4975 | ΔgacAPf-5, altered in the many phenotypes regulated by GacA, DAPG−, Ofa−, Prn−, Plt−, HCN−, Rzx−. | [11] |
| LK147 | 6-fold mutant of Pf-5, ΔpltAΔpcnCΔofaAΔrzxBΔhcnBΔphlA, DAPG−, Ofa−, Prn−, Plt−, HCN−, Rzx−. | [26] |
| LK107 | 5-fold mutant of Pf-5, ΔpltAΔpcnCΔofaAΔrzxBΔhcnB, produces DAPG+, Ofa−, Prn−, Plt−, HCN−, Rzx−. | This study |
| LK023 | ΔphlAPf-5, DAPG−. | [27] |
| LK557 | phlA repaired, DAPG+. | This study |
| LK410 | ΔphlFPf-5, overproduces DAPG, DAPG++ | This study |
| JL4776 | ΔofaAPf-5, Ofa− | [28] |
| JL4793 | ΔpcnCPf-5, Prn− | [11] |
| JL4805 | ΔpltAPf-5, Plt− | [11] |
| JL4808 | ΔrzxBPf-5, Rzx− | [11] |
| JL4809 | ΔhcnBPf-5, HCN− | [29] |
| P. fluorescens strains: | ||
| LK500 | Wild type strain 2P24, wheat rhizosphere isolate, wild type, produce DAPG and HCN. | [30] |
| LK501 | ΔgacA2P24, altered in the many phenotypes regulated by GacA, DAPG−, HCN− | [31] |
| LK506 | ΔphlACBD2P24, DAPG−. This mutant also has a deleted phlG gene and lacks PhlG that degrades DAPG to MAPG. | [32] |
| Plasmids | ||
| pEX18Tx | Gene replacement vector with MCS from pUC18, sacB+ Tcr | [33] |
| pEX18Tc-phlA | pEX18Tc containing wild-type phlAPf-5 gene for repair of phlA mutation in the chromosome. | This study |
| pEX18Tc-phlD | pEX18Tc containing wild-type phlDPf-5 gene for repair of phlD mutation in the chromosome. | This study |
| pEX18Tc-ΔphlF | pEX18Tc containing phlFPf-5 with a 546-bp in-frame deletion. | This study |
| pPROBE-NT | pBBR1 containing a promoter-less gfp, Kmr | [34] |
| pphlApromoter-gfp | pPROBE-NT containing the promoter of phlAPf-5 fused with a promoter-less gfp. Kmr | This study |
| pphlAtranslation-gfp | pPROBE-NT containing the promoter and the first seven codons of phlAPf-5 fused with a promoter-less gfp. Kmr | This study |
| Primers | oligonucleotide sequences (5′ to 3′) * | |
| phlA-F3 | ATAGGATCCTTAAGGATTTCGATGGTGG | |
| phlA-R3 | ATAGGTACCTGTTGCGGTTGATGGTGTCGGCG | |
| phlA_HindIII-F | GGACACAAGCTTCCCTATTTGGAGTCTGCTGT | |
| phlA_HindIII-R | CACACCAAGCTTTTCACATTCAGTGCTGGAGC | |
| gfp-phlA-f1 | CTGCAGGTCGACTCTAGAGTCGATGGTGGAAGTGAGAATG | |
| gfp-phlA-r1 | AGTGAAAAGTTCTTCTCCTTTACTCATGACAATACCTATCTTTTTCAC | |
| phlA-gfp-f1 | GTGAAAAAGATAGGTATTGTCATGAGTAAAGGAGAAGAA CTTTTCAC | |
| phlA-gfp-r1 | CATTCTCACTTCCACCATCGACTCTAGAGTCGACCTGCAG | |
| phlF-UpF-Hind3 | TATAAGCTTGAGGTCGGTGTTTTTCC | |
| phlF-UpR-ovlp | TCAACGTTGCGTACCAGGACAAGAGCCGATGGAGCTGCG | |
| phlF-DnF-ovlp | CGCAGCTCCATCGGCTCTTGTCCTGGTACGCAACGTTGA | |
| phlF-DnR-EcoRI | ATAGAATTCAAGTGGTGGTTCATCTGG C | |
| Pf5-phlD-UpF-Hind | CGACACAAGCTTCAGTGCGAAGAATGCAACGA | |
| Pf5-phlD-DnR-Hind | CCTCTCAAGCTTTGGTGACAATGATGCTGGTG | |
| 27f | AGAGTTTGATCCTGGCTCAG | |
| 1492r | CGGTTACCTTGTTACGACTT | |
#: Phenotype abbreviations: DAPG, 2,4-diacetylphloroglucinol; HCN, hydrogen cyanide; Ofa, orfamide A; Plt, pyoluteorin; Prn, pyrrolnitrin; Rzx, rhizoxin derivatives; abbreviations of antibiotics and their concentrations used in this work are: Tc, tetracycline (10 μg/mL for E. coli, 200 μg/mL for Pf-5); Km, kanamycin (50 μg/mL). *: underlines show DNA restriction enzyme sites that were used for cloning.
2.3. Inhibition Assays against A. euteiches
Inhibition assays were conducted on ½ PDA plates. A 5 mm agar plug from a 4-day old culture of A. euteiches was placed onto the center of the plates. Bacterial cells were washed by sterilized water to make a cell density at OD600 = 0.1. A drop of 3 μL of the bacterial cell suspension was placed on the agar surface about 2.5 cm away from the center of an A. euteiches plug. Inoculated plates were air-dried and incubated at 28 °C without light. Data were collected 2–3 days after inoculation by measuring the distance from the growing center of A. euteiches to the bacterial inoculation site. At least three replicates were used for each of the tested strains in one experiment. The experiment was repeated three times independently.
For the inhibition assay using purified compounds, the design was the same as above except the compounds were dissolved in methanol to make a 100 mM stock. A serial dilution was done using methanol to make appropriate concentrations. One μL of each concentration was added to a 6 mm-diameter filter paper disc and air-dried before placing it onto ½ PDA plates.
To observe the impact of 2,4-DAPG on the hyphal morphology of A. euteiches, the oomycete hyphae from the growing edge closest to the filter paper disc were collected immediately after the growth inhibition started and examined under a microscope. The experiment was repeated two times.
2.4. Construction of Pf-5 Mutant and Derivatives
The ΔphlFPf-5 deletion mutant of Pf-5 was made by following our previous method [35]. Briefly, two DNA fragments flanking the phlFPf-5 gene were PCR amplified from the genome of Pf-5 using two oligonucleotide pairs, phlF-UpF-Hind3/phlF-UpR-ovlp and phlF-DnF-ovlp/phlF-DnR-EcoRI (Table 1). These two DNA fragments were fused together by PCR and resulted in a 1204-bp DNA fragment that contains a 546-bp in-frame deletion of phlFPf-5 gene. The PCR product was digested by HindIII and EcoRI and ligated into pEX18Tc to make a deletion construct pEX18Tc-ΔphlF. This construct was transferred into wild type Pf-5 to delete phlFPf-5 gene in the chromosome. The deletion of phlFPf-5 was confirmed by PCR and subsequent DNA sequencing.
The 5-fold mutant of Pf-5 (ΔpltAΔpcnCΔofaAΔrzxBΔhcnB, strain ID LK107) which produces 2,4-DAPG but lacks pyrrolnitrin, orfamide A, rhizoxin, and hydrogen cyanide was made by introducing a wild type phlDPf-5 gene to replace the deleted phlDPf-5 in a previous made 6-fold mutant (ΔpltAΔphlDΔpcnCΔofaAΔrzxBΔhcnB, strain ID JL4909) [36]. Briefly, a 2103-bp DNA fragment containing the wild type phlDPf-5 gene and its flanking sequences was PCR amplified from the genome of Pf-5 using oligonucleotide pairs Pf5-phlD-UpF-Hind/Pf5-phlD-DnR-Hind. The PCR product was digested by HindIII and ligated into pEX18Tc to make a complementation construct pEX18Tc-phlD. This construct was transferred into the 6-fold mutant (JL4909) to restore the phlDPf-5 gene in the chromosome.
A similar strategy was used to replace the deleted phlAPf-5 gene in the ΔphlAPf-5 mutant’s chromosome with a wild type copy of the phlAPf-5 gene. Briefly, a 1700-bp DNA fragment which contained the wild type phlAPf-5 gene and its flanking sequences was PCR amplified using oligonucleotide pairs phlA_HindIII-F/phlA_HindIII-R, digested by HindIII, and ligated into pEX18Tc to make a complementation construct pEX18Tc-phlA. This construct was transferred into the ΔphlAPf-5 mutant to restore the phlAPf-5 gene in the chromosome.
2.5. Construction of Reporter Strains and GFP Activity Assays
The construction of GFP reporter constructs and the GFP activity assays were modified from our previous report [37]. Briefly, to measure the transcriptional expression of phlAPf-5, a 1001-bp DNA fragment which contained the promoter of phlAPf-5 gene was PCR amplified from the Pf-5 genome using oligonucleotide pair phlA-F3/phlA-R3 (Table 1). The PCR product was digested with BamHI and KpnI and ligated to pPROBE-NT to create the reporter construct pphlApromoter-gfp. To measure the translational expression of phlAPf-5, a 570-bp DNA fragment which contained the promoter and the first seven codons of phlAPf-5 was PCR amplified from the Pf-5 genome using oligonucleotide pair gfp-phlA-f1/gfp-phlA-r1 (Table 1). The vector pPROBE-NT was PCR amplified using primers phlA-gfp-f1/phlA-gfp-r1 and the PCR product was digested with KpnI. The purified two DNA fragments were assembled using NEBuilder HiFi DNA Assembly Master Mix (NEB, catalog no. E2621L) to make pphlAtranslation-gfp. The reporter constructs were confirmed by sequencing analysis and transferred into the strain Pf-5.
To measure the GFP activity of Pf-5 reporter strains, wild type Pf-5 containing the above made construct pphlApromoter-gfp, pphlAtranslation-gfp or the empty vector were cultured overnight on KB plates plus kanamycin at 28 °C. Then the cultures were inoculated into NBGly plus kanamycin, a non-conducive medium for 2,4-DAPG production, at 28 °C with shaking from early morning until late afternoon. The bacterial cells were washed three times with sterilized distilled water and diluted to an optical density of 600 nm (OD600) equal to 0.02 in phosphate buffered saline (PBS). Field pea seeds (P. sativum L., cv. Carousel yellow) were surface sterilized twice with 3% sodium hypochlorite, 3 min per time, and rinsed three times with sterilized distilled water. The seeds were pregerminated for 3 days before use. Germinated pea seeds, five-day old A. euteiches plug (5 mm in diameter) and 5 mL of a bacterial suspension in PBS were added to test tubes (20 mm diameter opening). The test tubes were incubated at 28 °C without shaking. The OD600 was measured using a 96-well plate reader (SpectraMax M2) to monitor growth of bacteria in the PBS. The green fluorescence was monitored at 24 h by measuring emission at 535 nm with an excitation at 485 nm. Background GFP levels of the PBS solutions containing A. euteiches plugs or the germinated seeds were subtracted from the observed GFP activity of the bacterial reporter strains. The GFP levels of wild type Pf-5 with pPROBE-NT empty vector were further subtracted as another background correction. For each measurement, the GFP value was divided by the corresponding OD600 to determine the relative GFP level. The experiment included three replicates for each treatment and was repeated two times independently.
2.6. Biocontrol Assays and Disease Evaluations
Oospores of A. euteiches were prepared in homogenized oatmeal broth (5 g rolled oats per liter of water) using a previous method [38]. Mycelial mats in water were blended for 5 min and the oospore concentration was determined by a hemocytometer. The oospores were diluted in water and mixed with greenhouse soils (pH: 7.3; NO3-N: 68 ppm; PO4-P: 3 ppm; potassium: 47 ppm) to make a final concentration of 50 oospores per gram soil. Soils mixed with the same volume of water served as non-inoculated control. The mixed soils were evenly filled in 10-cm square pots. Field pea seeds (P. sativum L., cv., Carousel yellow) were surface sterilized twice with 3% sodium hypochlorite, 3 min per time, and rinsed three times with sterilized distilled water. The seeds were then air-dried in a biosafety cabinet. Pf-5 derivative strains used in greenhouse assays included the 5-fold mutant (LK107), which produces 2,4-DAPG but lacks pyoluteorin, pyrrolnitrin, orfamide A, rhizoxin, and hydrogen cyanide, and the 6-fold mutant (LK147), which lacks DAPG and the above five antibiotics. The bacterial strains were cultured on KB plates at 28 °C overnight. The cultures were transferred to KB liquid medium and incubated at 28 °C overnight with shaking at 250 rpm. Then the cells were washed with sterilized, distilled water three times and diluted to OD600 = 0.1. The seeds were treated with the bacterial suspension for three hours at room temperature with gentle shaking (around 60 rpm). Then, the seeds were removed from the suspension and planted in pots in the greenhouse. The planted pots were placed in a shallow tray and were watered the next day after planting by filling the tray with water. Greenhouse conditions were set to 22 °C day/18 °C night with a 16-h photoperiod. Each treatment contained six replicates and each replicate contained six seeds. The experiment was repeated two times.
The disease severity was evaluated by following a previous method [39]. Briefly, pea plants were harvested three weeks after planting. The roots were washed with tap water and examined for root rot symptoms. The disease severity scales were: 0 = healthy roots with no visible symptoms of root rot; 1 = slight water-soaking on the primary or secondary roots; 2 = moderate water-soaking on the primary or secondary roots or epicotyls with light-brown areas and more extensive discoloration; 3 = infected areas extensive and soft, but the entire root was not collapsed and the epicotyl was not markedly shriveled; 4 = extensive discoloration of the roots with tissue collapse and disintegration, or plant dead.
2.7. Statistical Analysis
The statistical analysis of the data collected in this study was performed by Student t-test or one-factor ANOVA analysis.
3. Results
3.1. Isolation and Identification of Bacterial Antagonists
Ten bacterial isolates that show clear antagonistic activities against A. euteiches on ½ PDA plates were identified from lentil field soil samples in this study (Figure 1). Among these ten isolates, four were identified as Pseudomonas spp., three were identified as Paenibacillus spp., and the other three were identified as Bacillus sp., Pseudarthrobacter sp. and Chryseobacterium sp., respectively, via 16s DNA sequencing and phylogenetic analysis (Figure 1C, Figure S1 and Table S1). Overall, the isolates of the Pseudomonas group showed stronger inhibition than strains of the other groups (Figure 1A,B).
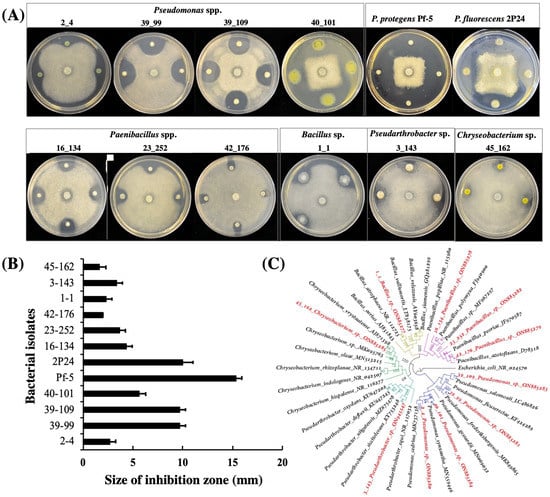
Figure 1.
Bacterial antagonists inhibit the growth of A. euteiches.A. euteiches was cultured in the center of the plates and the bacterial cells were inoculated around the pathogen. All strains were cultured on ½ PDA plates at 28 °C for three days before the results of the inhibition were recorded (A) and the size of the inhibition zone was measured (B). The experiment was repeated three times with similar results. (C); the taxonomy of the bacterial antagonists that were isolated in this work was identified by 16S rRNA analysis. The phylogenetic tree was constructed using IQ-tree and visualized using Figtree v 1.4.4. Five different bacterial genera forming distinct clades are represented by different colors at the branches. The ten bacterial isolates highlighted with red color were isolated in this study. The GenBank accession number of each isolate was shown. The values at the node of the tree represent bootstrap support.
We also included two well-studied Pseudomonas strains, P. protegens Pf-5 and P. fluorescens 2P24, in this work. Our results showed that both strains strongly inhibited the growth of A. euteiches on ½ PDA plates (Figure 1A,B).
3.2. 2,4-DAPG Biosynthesis Pathway Is Required for the Inhibition of A. euteiches by Strains Pf-5 and 2P24
We used P. protegens Pf-5 as a model to identify the antibiotic(s) that inhibit the growth of A. euteiches, because Pf-5 is known to produce many antibiotics including pyoluteorin, pyrrolnitrin, 2,4-diacetylphloroglucinol (2,4-DAPG), orfamide A, rhizoxin, and hydrogen cyanide [40]. We first tested a Pf-5 derivative, the ΔgacAPf-5 mutant, which lacks the regulator protein GacA that globally controls the production of many secondary metabolites including the above-mentioned antibiotics [40]. Compared to the wild-type strain, the ΔgacAPf-5 mutant has no clear inhibition of A. euteiches on the culture plates (0 mm of inhibition zone was recorded, Figure 2A), suggesting that GacA-controlled antibiotic production is required by Pf-5 to inhibit the pathogen.
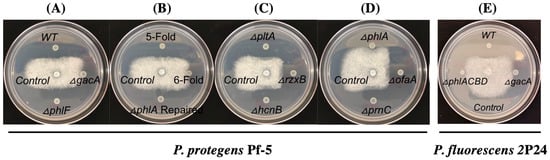
Figure 2.
Inhibition of Pf-5 (A–D) and 2P24 (E) and their derivatives against A. euteiches on plates. A. euteiches was cultured in the center of the plates and the bacterial cells were inoculated around the pathogen. WT: wild type; 5-fold: Pf-5 mutant, ΔpltAΔpcnCΔofaAΔrzxBΔhcnB; 6-fold: Pf-5 mutant, ΔpltAΔpcnCΔofaAΔrzxBΔhcnBΔphlA; control has no bacteria inoculation. All strains were cultured on ½ PDA plates at 28 °C for two days before the results were recorded. Photos are representatives of three replicates and the experiment was repeated three times with similar results.
In strain Pf-5, GacA positively controls the production of at least six different antibiotics including pyoluteorin, 2,4-DAPG, pyrrolnitrin, orfamide A, rhizoxin, and hydrogen cyanide. To test if these antibiotics are involved in the growth inhibition of A. euteiches by Pf-5, a 6-fold mutant which lacks all these six antibiotics was tested in the inhibition assays. The 6-fold mutant did not inhibit the pathogen’s growth (0 mm of inhibition zone was recorded, Figure 2B), indicating that at least one of these six antibiotics contributes to the growth inhibition against A. euteiches by Pf-5. We then tested six single mutants, each lacking the production of one of the six antibiotics due to a mutation of their biosynthesis genes (Figure 2C,D). Among these six mutants, five mutants that lack pyoluteorin, pyrrolnitrin, orfamide A, rhizoxin, or hydrogen cyanide inhibited the pathogen to a similar level (sizes of the inhibition zone were 9.1–10.4 mm) with the wild type strain (10.8 mm of the inhibition zones). The ΔphlAPf-5 mutant, which has an abolished biosynthesis pathway of 2,4-DAPG due to a mutation of its biosynthesis gene phlAPf-5 (Figure 3A), shows almost no inhibition against A. euteiches (0 mm of inhibition zone was recorded, Figure 2D). 2,4-DAPG is a polyketide antibiotic that is toxic to many plant pathogens including bacteria, fungi, oomycetes, and nematodes [41,42,43]. To confirm the role of the 2,4-DAPG pathway in the growth inhibition, we restored the ΔphlAPf-5 mutant by replacing the deleted phlAPf-5 gene with its wild-type copy in the chromosome. The repaired strain phlA+ inhibited A. euteiches to a similar level as the wild type Pf-5 (Figure 2B). Similarly, a 5-fold mutant, which has a restored 2,4-DAPG biosynthesis pathway in the 6-fold mutant background, strongly inhibited A. euteiches. Further, we tested a ΔphlFPf-5 mutant, which overexpresses 2,4-DAPG biosynthesis genes due to a mutation of the transcriptional repressor PhlF (Figure 3A) that negatively regulates the expression of 2,4-DAPG biosynthesis genes [44]. Compared to the wild type, the ΔphlFPf-5 mutant has a slightly stronger inhibition (12.0 mm of inhibition zone was recorded) against the growth of A. euteiches (Figure 2A). Taken together, these results indicate that the 2,4-DAPG biosynthesis pathway of Pf-5 contributes to the inhibition against A. euteiches on culture plates.
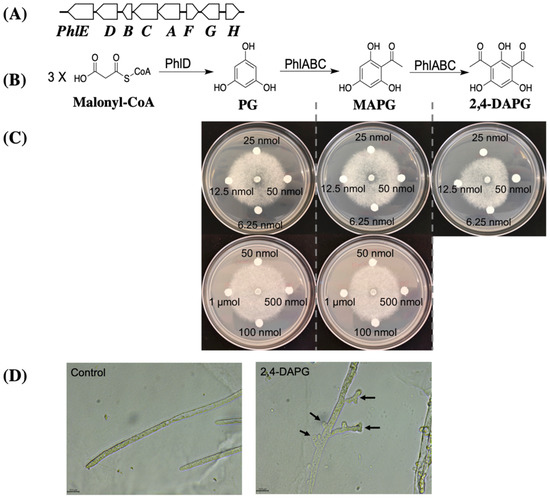
Figure 3.
Impact of metabolites generated in the 2,4-DAPG biosynthesis pathway on hyphal growth of A. euteiches. (A), 2,4-DAPG gene cluster of Pf-5. Genes phlABCD encode four enzymes (PhlABCD) in the 2,4-DAPG biosynthesis pathway (B), and their expression is negatively regulated by the transcriptional repressor PhlF; PhlG degrades 2,4-DAPG to MAPG and its expression is controlled by PhlH; phlE encodes a putative permease of 2,4-DAPG. (C), Filter paper disks containing the indicated amount of the tested compounds were dried and placed around A. euteiches on ½ PDA plates. Representative results from three replicates were recorded two days after inoculation. (D), the oomycete hyphae were sampled at the A. euteiches growth margin on plates of (C) and examined under a microscope immediately after the inhibition was observed. Control indicates no compound treatment. Arrows show the excessive hyphal branching and stunted branches. Size bars indicate 15.9 μm. The experiments were repeated at least two times.
Strain P. fluorescens 2P24 is known to produce 2,4-DAPG and hydrogen cyanide [30]. Based on the results that the 2,4-DAPG biosynthesis pathway is required for Pf-5 to inhibit A. euteiches, we hypothesized that the same antibiotic pathway also contributes to the inhibition by strain 2P24. We tested this hypothesis by comparing the antagonistic activity of the wild type 2P24 and its two derivatives: a ΔphlACBD2P24, mutant, which lacks the four biosynthesis genes phlACBD for 2,4-DAPG production, and a ΔgacA2P24 mutant, which produces neither 2,4-DAPG nor hydrogen cyanide. Our results show that the ΔphlACBD2P24 and the ΔgacA2P24 mutants have similar levels of inhibition (0 mm of inhibition zones were recorded for both mutants), which were much decreased compared to the wild type strain (10.4 mm of inhibition zone was recorded) (Figure 2E). These results show that the 2,4-DAPG biosynthesis pathway contributes to the inhibition of A. euteiches by strain 2P24.
3.3. Purified 2,4-DAPG Suppresses A. euteiches Growth and Alters Its Mycelia Morphology
In Pseudomonas spp., the biosynthesis pathway of 2,4-DAPG generates three metabolites including two intermediates, phloroglucinol (PG) and monoacetylphloroglucinol (MAPG), and the final product 2,4-DAPG (Figure 3B). A previous study showed that the intermediates of the antibiotic pathway may play a role in the inhibition against the target pathogens [45]. The ΔphlACBD2P24 mutant lacks all these three metabolites due to the deletions of their respective biosynthesis genes (Figure 3A,B). The ΔphlAPf-5 mutant lacks MAPG and 2,4-DAPG but produces the first-step intermediate PG because phlD remains functional. The result that ΔphlAPf-5 mutant has an abolished inhibition to A. euteiches (Figure 2D) suggests that PG did not inhibit the pathogen.
We directly tested the toxicity of the three metabolites produced by the 2,4-DAPG biosynthesis pathway against A. euteiches. Growth of A. euteiches was inhibited by 25 nmol of 2,4-DAPG or 500 nmol of MAPG but was not inhibited by PG even at 1 μmol (Figure 3C). The sizes of inhibition zone caused by 25 nmol of 2,4-DAPG, 500 nmol of MAPG, and 1 μmol of PG were 2.9 mm, 0.8 mm, and 0 mm respectively. These data show that, among the three metabolites, 2,4-DAPG is the most potent antibiotic against A. euteiches and likely plays a major role in the pathogen inhibition by strains Pf-5 and 2P24.
We then investigated the impact of 2,4-DAPG on mycelial growth of A. euteiches under a microscope. Compared to the normal hyphal branching and growth of A. euteiches in the control treatment, the 2,4-DAPG-treated pathogen showed excessive branching of the hyphae and stunted growth of the branches (Figure 3D), which is consistent with the reduced mycelial growth caused by the purified antibiotic metabolite and the producing bacteria.
3.4. Expression of the 2,4-DAPG Biosynthesis Genes Is Induced by Germinating Pea Seeds
Antibiotic production of beneficial microorganisms can be strongly regulated by environmental factors, host plants, and target pathogens, which then influences plant disease control suppression. To investigate if the bacterial 2,4-DAPG biosynthesis is regulated by pea seeds and A. euteiches, we measured the expression of 2,4-DAPG biosynthesis genes in the pea-Pseudomonas-A. euteiches tri-trophic system. Strain P. protegens Pf-5 was used as a model of the 2,4-DAPG producing bacteria in this experiment. A reporter construct, pphlApromoter-gfp, which contains the promoter of phlAPf-5 fused with a promoter-less gfp, was made to measure the transcription of the 2,4-DAPG biosynthesis genes. This reporter construct was transferred into the wild type Pf-5. The resultant reporter strain, Pf-5/pphlApromoter-gfp, was incubated with germinating pea seeds with or without A. eut eiches. As shown in Figure 4A, Pf-5/pphlApromoter-gfp had a significant higher GFP level when it was incubated with germinating pea seeds than with the buffer control.
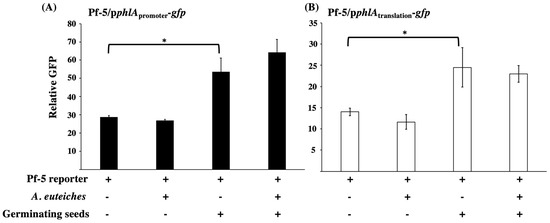
Figure 4.
Expression of phlA by Pf-5 in the interactions with pea germinating seeds and A. euteiches. Wild type Pf-5 containing the GFP-based transcriptional reporter constructs pphlApromoter-gfp (A), or the translational reporter construct pphlAtranslation-gfp (B) was cultured with germinating pea seeds with or without the presence of A. euteiches hyphae. The expression of phlA was measured as relative GFP [fluorescence of GFP divided by (OD600 × 1000)] recorded at 24 h post inoculation. * indicates treatments are significantly different (p < 0.05), as determined by Student t-test. Data are means and standard deviations of three replicates from a representative experiment repeated two times with similar results.
The presence of A. euteiches did not significantly influence phlA transcription by Pf-5, as assessed by the GFP level of the reporter strain. Similarly, the translational expression of phlAPf-5, which was measured using the reporter strain Pf-5/pphlAtranslation-gfp, was also induced by the germinating seeds but not the pathogen (Figure 4B). These results indicate that expression of the 2,4-DAPG biosynthesis genes of Pf-5 is activated at both transcriptional and translational levels by germinating pea seeds which likely results in the production of 2,4-DAPG by the bacterium on pea seed surfaces.
3.5. 2,4-DAPG Plays a Role in the Biocontrol of Pea Aphanomyce Root Rot
Based on the above results that purified 2,4-DAPG and its producing bacteria strongly inhibited A. euteiches on culture plates, and that expression of 2,4-DAPG biosynthesis genes is activated by germinating pea seeds, we hypothesized that this antibiotic plays an important role in the biological control of A. euteiches root rot on pea. In the first step of testing this hypothesis, we set up an A. euteiches-pea root rot system and a biocontrol assay in greenhouse conditions. We found that inoculation of A. euteiches at a concentration of 50 oospores per gram soil caused a significant disease severity on the pea plants (Figure 5).
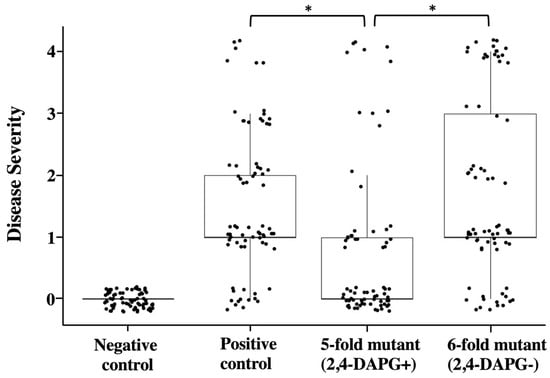
Figure 5.
Biocontrol assays of pea Aphanomyces root rot by P. protegens Pf-5 in greenhouse. Pea seeds without inoculation served as negative control; pea seeds in positive control were planted in soil containing A. euteiches (50 oospores per gram soil) but not the Pf-5 strains. 5-fold mutant: ΔpltAΔpcnCΔofaAΔrzxBΔhcnB; 6-fold mutant: ΔpltAΔpcnCΔofaAΔrzxBΔhcnBΔphlA. Disease severity was evaluated on a 0–4 disease level scale. Scatter plot shows combined data from two independent experiments each had six replicates (i.e., six growth pots) per treatment and each replicate included five to six pea plants. * indicates treatments are significant different (p < 0.05), as determined by one-factor ANOVA analysis.
Next, we tested the disease control efficacy of the 5-fold mutant of Pf-5 which has a functional 2,4-DAPG biosynthesis pathway but lacks the other five antibiotics to avoid their potential impacts on plant development such as seed germination. Results show that pea plants that were treated by the 5-fold mutant have a significantly lower level of disease severity than the control plants treated by only the pathogen. Moreover, pea plants treated by the 6-fold mutant, which has a mutation in the 2,4-DAPG biosynthesis gene phlAPf-5 in the background of the 5-fold mutant, developed a disease level that is significantly higher than the 5-fold mutant. Taken together, these results indicate that 2,4-DAPG plays an important role in the biocontrol of A. euteiches root rot by P. protegens on pea plants.
4. Discussion
In this study, we found that 2,4-DAPG produced by beneficial bacteria of the Pseudomonas genus plays an important role in the growth inhibition of the oomycete pathogen A. euteiches on culture plates and suppression of the pea Aphanomyces root rot disease in greenhouse. Our results are consistent with the previous reports that 2,4-DAPG produced by strains of Pseudomonas spp. is a major determinant in the biocontrol of many soilborne plant diseases due to its broad-spectrum toxicity against bacteria, fungi, oomycetes, and nematodes [41,42,43]. This is the first report that 2,4-DAPG inhibits A. euteiches and contributes to the beneficial bacteria-mediated biocontrol of pea Aphanomyces root rot disease.
Many beneficial bacteria produce multiple antibiotics that inhibit plant pathogens. Two beneficial bacterial strains, P. protegens Pf-5 and P. fluorescens 2P24, were used in this study to determine the role of antibiotics in the growth inhibition of A. euteiches. In addition to the 2,4-DAPG, strain 2P24 is known to produce another antibiotic hydrogen cyanide [30], and strain Pf-5 can produce at least another five antibiotics including pyoluteorin, pyrrolnitrin, orfamide A, rhizoxin, and hydrogen cyanide [40]. The important role of 2,4-DAPG in the inhibition of A. euteiches is clearly supported by the results that the Pf-5 and 2P24 mutants that lack 2,4-DAPG exhibited greatly reduced inhibition against A. euteiches compared to their wild type strains (Figure 2). The anti-oomycete activities of hydrogen cyanide, pyoluteorin, pyrrolnitrin, orfamide A, and rhizoxin have been reported previously in various phytopathogenic systems [46,47,48,49,50]. However, our results show that Pf-5 mutants that lack these five antibiotics retained strong inhibition against A. euteiches, suggesting that these five antibiotics of Pf-5 do not play a major role in the growth inhibition against A. euteiches. The important role of 2,4-DAPG in the inhibition of A. euteiches is also supported by the morphological changes in mycelia (excessive and stunted hyphal branches) of A. euteiches caused by purified 2,4-DAPG (Figure 3) and the reduced disease control efficacy of the 2,4-DAPG nonproducing mutant compared to the 2,4-DAPG producing derivative of Pf-5 (Figure 5). We recognized that the culture conditions used in the inhibition assays may not be conducive for the tested bacterial strains to produce all their antibiotics. For example, the ½ PDA medium used in this study contains a high level of glucose (2%, w/v) which likely represses the biosynthesis of pyoluteorin that is known to inhibit the oomycete pathogen Pythium ultimum [51]. Antibiotics other than 2,4-DAPG may contribute to the inhibition of A. eutriches by Pf-5 in different culture media.
Antibiotic production is critical for many beneficial bacteria to inhibit plant pathogens and suppress plant diseases, but expression of antibiotic biosynthesis genes is often influenced by the tri-trophic interactions between the beneficial bacterium, the pathogen, and the host plant [52,53,54]. Using a GFP-based transcriptional reporter system, we found that expression of the 2,4-DAPG biosynthesis gene phlAPf-5 is strongly induced at both transcriptional and translational levels by germinating pea seeds (Figure 4). These results are consistent with our previous report that the 2,4-DAPG biosynthesis genes such as phlDPf-5 are highly expressed in Pf-5 on the germinating pea seeds (Pisum sativum cv. Sugar Snap) in an RNAseq analysis and that 2,4-DAPG was detected on the Pf-5-treated pea seed surfaces [40]. These data support that production of 2,4-DAPG can be activated by the exudates of germinating pea seeds. Diverse metabolites including amino acids, carbohydrates, and organic acids were detected from pea seed exudates [40]. Our current understanding of how these exudates influence the antibiotic production of beneficial bacteria remains limited. Glucose is a known carbohydrate conducive to 2,4-DAPG production of Pf-5 [27,51] and has been detected in pea seed exudate at a moderate level (81.2 g/seed) [40]. However, the roles of the other more abundant carbohydrates including sucrose (1740.3 g/seed) and galactose (845.8 g/seed) and the most abundant amino acids including glutamate (206.4 g/seed) and arginine (86.1 g/seed) in the regulation of antibiotic production by Pf-5 remain unknown. Metabolites secreted by plant pathogens can also influence the antibiotic production of the associated beneficial bacteria. For example, fusaric acid, which is produced by Fusarium spp., inhibits 2,4-DAPG production of Pf-5 [36]. In this study, we found that the expression of phlAPf-5 was not altered by the presence of A. euteiches hyphae (Figure 4). Infection of plant roots by pathogens can cause a metabolic leak that enhances antibiotic production of rhizosphere-associated beneficial microorganisms. For example, expression of phlA by the beneficial bacterium P. protegens CHA0 was increased on barley roots after infection by Pythium ultimum [55]. No or minimal damage of pea seeds were observed by the inoculated A. euteiches in our transcriptional reporter assays, probably due to the short incubation time (24 h) that was used to collect the GFP data of the bacterial reporter. It will be interesting to investigate the dynamic changes of the pea seed/root exudates, especially the metabolites that are known to regulate 2,4-DAPG production, at the different stages of the disease development of pea Aphanomyces root rot in the future.
In addition to 2,4-DAPG, many other antimicrobial compounds are known to be produced by strains of the Pseudomonas group and contribute to pathogen inhibition and disease biocontrol by the strains [56]. Characterizing different bacterial isolates that inhibit the growth of the pathogens can help us identify novel antibiotics to control the diseases. In this study, four Pseudomonas isolates were isolated from lentil filed soils and showed clear inhibition against A. euteiches (Figure 1). Among these four Pseudomonas isolates, 40_101 is interesting because of its strong inhibition which is comparable to the model strains Pf-5 and 2P24. The antibiotics produced by these strains were not characterized in this study. Future investigations, including whole genome sequencing analysis and bacterial mutagenesis, of these Pseudomonas isolates may identify new antibiotics for the disease manage of pea Aphanomyces root rot. Bacterial antagonists that belong to different genera including Bacillus, Paenibacillus, Pseudarthrobacter, and Chreyseobacterium were also identified in this work, although their inhibition effects are overall less than the Pseudomonas group. The antibiotic(s) that is(are) required for the growth inhibition of A. euteiches by these antagonists was (were) not investigated in this work. However, it is known that strains of Bacillus spp. and Paenibacillus spp. can produce diverse antibiotics such as lipopeptides that are toxic against plant pathogens including oomycetes [57,58]. The antagonistic bacteria were isolated from fields in Montana and have adapted to the local climates and soil/plant environments. Thus, they can serve as useful microbial resources for the development of biocontrol agents to manage pea Aphanomyces root rot disease in Montana.
Molecular identification using 16s rRNA is useful to identify bacteria although its limitation in accurate classification at species level has been recognized especially for strains of the Pseudomonas group [59,60]. The taxonomic classification of the ten bacterial strains isolated in this study was characterized to genus levels via 16s rRNA analysis (Figure 1) and can be further improved by multilocus sequence analysis or whole genome sequencing. Nevertheless, the result that A. euteiches was inhibited by bacterial isolates belonging to distinct genera suggests different antibiotics may be produced by these isolates to suppress the pathogen.
In conclusion, diverse groups of soil bacteria including Pseudomonas spp., Bacillus spp., Paenibacillus sp., Pseudarthrobacter sp., and Chreyseobacterium sp. were found to inhibit the growth of A. euteiches. Evidence obtained via genetic and biochemical analysis and GFP-based reporter assays supports that 2,4-DAPG is the major antibiotic required by model strains of the Pseudomonas, including P. protegens Pf-5 and P. fluorescens 2P24, to inhibit the growth of A. euteiches in culture medium and control the Aphanomyces root rot disease on pea plants. Similar approaches can be used in the future to identify and characterize antibiotics required by the isolated antagonistic bacteria to inhibit the growth of A. euteiches.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms10081596/s1, Table S1: Bacterial strains isolated in this study and their identities to the most closely related strains found in the NCBI database, Figure S1: Phylogenetic analysis of the bacterial antagonists isolated in this study.
Author Contributions
Conceptualization, Q.Y. and X.W.; investigation, X.L., D.N. and Q.Y.; writing—original draft preparation, X.L. and Q.Y.; writing—review and editing, Q.Y., X.L., D.N., X.W. and M.B.; supervision, Q.Y.; project administration, Q.Y.; funding acquisition, Q.Y. and M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by AGR Specialty Crop Block Grant (SCBG) 21SC07013 from the Montana Department of Agriculture.
Acknowledgments
We are grateful to Chiseche Mwanza, Monica Brelsford, Bernard Nyamesorto, Erin Gunnick-Troth, Carmen Murphy, Teresa A. Kidarsa, Brenda Shaffer and Virginia Stockwell for their assistance in this work. We are grateful to Joyce Loper for a helpful review of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- O’Brien, P.A. Biological control of plant diseases. Australas. Plant Pathol. 2017, 46, 293–304. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef]
- Gaulin, E.; Jacquet, C.; Bottin, A.; Dumas, B. Root rot disease of legumes caused by Aphanomyces euteiches. Mol. Plant Pathol. 2007, 8, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Chang, K.-F.; Hwang, S.-F.; Conner, R.; Fredua-Agyeman, R.; Feindel, D.; Strelkov, S.E. Evaluation of host resistance and fungicide application as tools for the management of root rot of field pea caused by Aphanomyces euteiches. Crop J. 2019, 7, 38–48. [Google Scholar] [CrossRef]
- Quillévéré-Hamard, A.; Le Roy, G.; Lesné, A.; Le May, C.; Pilet-Nayel, M.-L. Aggressiveness of Diverse French Aphanomyces euteiches Isolates on Pea Near Isogenic Lines Differing in Resistance Quantitative Trait Loci. Phytopathology® 2021, 111, 695–702. [Google Scholar] [CrossRef]
- Heungens, K.; Parke, J.L. Postinfection biological control of oomycete pathogens of pea by Burkholderia cepacia AMMDR1. Phytopathology 2001, 91, 383–391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wakelin, S.A.; Walter, M.; Jaspers, M.; Stewart, A. Biological control of Aphanomyces euteiches root-rot of pea with spore-forming bacteria. Australas. Plant Pathol. 2002, 31, 401–407. [Google Scholar] [CrossRef]
- Godebo, A.T.; Germida, J.J.; Walley, F.L. Isolation, identification, and assessment of soil bacteria as biocontrol agents of pea root rot caused by Aphanomyces euteiches. Can. J. Soil Sci. 2020, 100, 206–216. [Google Scholar] [CrossRef]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef]
- Parke, J.; Rand, R.; Joy, A.; King, E. Biological control of Pythium damping-off and Aphanomyces root rot of peas by application of Pseudomonas cepacia or P. fluorescens to seed. Plant Dis. 1991, 75, 987–992. [Google Scholar] [CrossRef]
- Henkels, M.D.; Kidarsa, T.A.; Shaffer, B.T.; Goebel, N.C.; Burlinson, P.; Mavrodi, D.V.; Bentley, M.A.; Rangel, L.I.; Davis, E.W.; Thomashow, L.S.; et al. Pseudomonas protegens Pf-5 causes discoloration and pitting of mushroom caps due to the production of antifungal metabolites. Mol. Plant-Microbe Interact. 2014, 27, 733–746. [Google Scholar] [CrossRef]
- Philmus, B.; Shaffer, B.T.; Kidarsa, T.A.; Yan, Q.; Raaijmakers, J.M.; Begley, T.P.; Loper, J.E. Investigations into the biosynthesis, regulation, and self-resistance of toxoflavin in Pseudomonas protegens Pf-5. ChemBioChem 2015, 16, 1782–1790. [Google Scholar] [CrossRef]
- Yan, Q.; Philmus, B.; Chang, J.H.; Loper, J.E. Novel mechanism of metabolic co-regulation coordinates the biosynthesis of secondary metabolites in Pseudomonas protegens. eLife 2017, 6, e22835. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Lopes, L.D.; Shaffer, B.T.; Kidarsa, T.A.; Vining, O.; Philmus, B.; Song, C.; Stockwell, V.O.; Raaijmakers, J.M.; McPhail, K.L.; et al. Secondary Metabolism and Interspecific Competition Affect Accumulation of Spontaneous Mutants in the GacS-GacA Regulatory System in Pseudomonas protegens. mBio 2018, 9, e01845-17. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, Z.; Zhang, B.; Wu, X.-G.; Ren, Z.-G.; Zhang, L.-Q. Posttranscriptional regulation of 2, 4-diacetylphloroglucinol production by GidA and TrmE in Pseudomonas fluorescens 2P24. Appl. Environ. Microbiol. 2014, 80, 3972–3981. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Zhang, B.; Yang, Q.; Zhang, Y.; Zheng, D.; Zhang, L.; Yan, Q.; Wu, X. Cyclic-di-GMP regulates the quorum-sensing system and the biocontrol activity of Pseudomonas fluorescens 2P24 through RsmA and RsmE proteins. Appl. Environ. Microbiol. 2020, 86, e02016-20. [Google Scholar] [CrossRef] [PubMed]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Beckerman, J.; Stone, J.; Ruhl, G.; Creswell, T. First report of Pythium ultimum crown and root rot of industrial hemp in the United States. Plant Dis. 2018, 102, 2045. [Google Scholar] [CrossRef]
- Linaldeddu, B.T.; Bregant, C.; Ruzzon, B.; Montecchio, L. Coniella granati and Phytophthora palmivora the main pathogens involved in pomegranate dieback and mortality in north-eastern Italy. Ital. J. Mycol. 2020, 49, 92–100. [Google Scholar]
- Eden, P.A.; Schmidt, T.M.; Blakemore, R.P.; Pace, N.R. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Evol. Microbiol. 1991, 41, 324–325. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE, 2, New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Parikh, L.; Agindotan, B.O.; Burrows, M.E. Antifungal activity of plant-derived essential oils on pathogens of pulse crops. Plant Dis. 2021, 105, 1692–1701. [Google Scholar] [CrossRef]
- Howell, C.R.; Stipanovic, R.D. Control of Rhizoctonia solani in cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 1979, 69, 480–482. [Google Scholar] [CrossRef]
- Loper, J.E.; Henkels, M.D.; Rangel, L.I.; Olcott, M.H.; Walker, F.L.; Bond, K.L.; Kidarsa, T.A.; Hesse, C.N.; Sneh, B.; Stockwell, V.O.; et al. Rhizoxin analogs, orfamide A and chitinase production contribute to the toxicity of Pseudomonas protegens strain Pf-5 to Drosophila melanogaster. Environ. Microbiol. 2016, 18, 3509–3521. [Google Scholar] [CrossRef]
- Kidarsa, T.A.; Goebel, N.C.; Zabriskie, T.M.; Loper, J.E. Phloroglucinol mediates cross-talk between the pyoluteorin and 2,4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol. Microbiol. 2011, 81, 395–414. [Google Scholar] [CrossRef]
- Gross, H.; Stockwell, V.O.; Henkels, M.D.; Nowak-Thompson, B.; Loper, J.E.; Gerwick, W.H. The genomisotopic approach: A systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 2007, 14, 53–63. [Google Scholar] [CrossRef]
- Loper, J.E.; Hassan, K.A.; Mavrodi, D.V.; Davis II, E.W.; Lim, C.K.; Shaffer, B.T.; Elbourne, L.D.; Stockwell, V.O.; Hartney, S.L.; Breakwell, K.; et al. Comparative genomics of plant-associated Pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012, 8, e1002784. [Google Scholar] [CrossRef]
- Wei, H.-L.; Zhang, L.-Q. Quorum-sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Antonie Leeuwenhoek 2006, 89, 267–280. [Google Scholar] [CrossRef]
- Yan, Q.; Wu, X.-G.; Wei, H.-L.; Wang, H.-M.; Zhang, L.-Q. Differential control of the PcoI/PcoR quorum-sensing system in Pseudomonas fluorescens 2P24 by sigma factor RpoS and the GacS/GacA two-component regulatory system. Microbiol. Res. 2009, 164, 18–26. [Google Scholar] [CrossRef]
- Yan, X.; Yang, R.; Zhao, R.-X.; Han, J.-T.; Jia, W.-J.; Li, D.-Y.; Wang, Y.; Zhang, N.; Wu, Y.; Zhang, L.-Q.; et al. Transcriptional Regulator PhlH Modulates 2, 4-Diacetylphloroglucinol Biosynthesis in Response to the Biosynthetic Intermediate and End Product. Appl. Environ. Microbiol. 2017, 83, e01419-17. [Google Scholar] [CrossRef]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef]
- Miller, W.G.; Leveau, J.H.; Lindow, S.E. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant-Microbe Interact. 2000, 13, 1243–1250. [Google Scholar] [CrossRef]
- Yan, Q.; Liu, M.; Kidarsa, T.; Johnson, C.P.; Loper, J.E. Two Pathway-Specific Transcriptional Regulators, PltR and PltZ, Coordinate Autoinduction of Pyoluteorin in Pseudomonas protegens Pf-5. Microorganisms 2021, 9, 1489. [Google Scholar] [CrossRef]
- Quecine, M.C.; Kidarsa, T.A.; Goebel, N.C.; Shaffer, B.T.; Henkels, M.D.; Zabriskie, T.M.; Loper, J.E. An Interspecies signaling system mediated by fusaric acid has parallel effects on antifungal metabolite production by Pseudomonas protegens Pf-5 and antibiosis of Fusarium spp. Appl. Environ. Microbiol. 2016, 82, 1372–1382. [Google Scholar] [CrossRef]
- Yan, Q.; Philmus, B.; Hesse, C.; Kohen, M.; Chang, J.H.; Loper, J. The rare codon AGA is involved in regulation of pyoluteorin biosynthesis in Pseudomonas protegens Pf-5. Front. Microbiol. 2016, 7, 497. [Google Scholar] [CrossRef]
- Windels, C.E. Aphanomyces Root Rot on Sugar Beet. Plant Health Prog. 2000, 1, 8. [Google Scholar] [CrossRef]
- Papavizas, G.C.; Ayers, W.A. Aphanomyces Species and Their Root Diseases in Pea and Sugar Beet: A Review; Agricultural Research Service, U.S. Department of Agriculture: Washington, DC, USA, 1974.
- Kidarsa, T.A.; Shaffer, B.T.; Goebel, N.C.; Roberts, D.P.; Buyer, J.S.; Johnson, A.; Kobayashi, D.Y.; Zabriskie, T.M.; Paulsen, I.; Loper, J.E. Genes expressed by the biological control bacterium Pseudomonas protegens Pf-5 on seed surfaces under the control of the global regulators GacA and RpoS. Environ. Microbiol. 2013, 15, 716–735. [Google Scholar] [CrossRef]
- Gleeson, O.; O’Gara, F.; Morrissey, J. The Pseudomonas fluorescens secondary metabolite 2,4 diacetylphloroglucinol impairs mitochondrial function in Saccharomyces cerevisiae. Antonie Leeuwenhoek 2010, 97, 261–273. [Google Scholar] [CrossRef]
- Biessy, A.; Filion, M. Phloroglucinol Derivatives in Plant-Beneficial Pseudomonas spp.: Biosynthesis, Regulation, and Functions. Metabolites 2021, 11, 182. [Google Scholar] [CrossRef]
- Ali, M.A.; Luo, J.; Ahmed, T.; Zhang, J.; Xie, T.; Dai, D.; Jiang, J.; Zhu, J.; Hassan, S.; Alorabi, J.A.; et al. Pseudomonas bijieensis Strain XL17 within the P. corrugata Subgroup Producing 2,4-Diacetylphloroglucinol and Lipopeptides Controls Bacterial Canker and Gray Mold Pathogens of Kiwifruit. Microorganisms 2022, 10, 425. [Google Scholar] [CrossRef]
- Delany, I.; Sheehan, M.M.; Fenton, A.; Bardin, S.; Aarons, S.; O’Gara, F. Regulation of production of the antifungal metabolite 2,4-diacetylphloroglucinol in Pseudomonas fluorescens F113: Genetic analysis of phlF as a transcriptional repressor. Microbiology 2000, 146, 537–543. [Google Scholar] [CrossRef]
- Islam, M.T.; Von Tiedemann, A. 2,4-Diacetylphloroglucinol suppresses zoosporogenesis and impairs motility of Peronosporomycete zoospores. World J. Microbiol. Biotechnol. 2011, 27, 2071–2079. [Google Scholar] [CrossRef]
- Howell, C.R.; Stipanovic, R.D. Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology 1980, 70, 712–715. [Google Scholar] [CrossRef]
- Chernin, L.; Brandis, A.; Ismailov, Z.; Chet, I. Pyrrolnitrin production by an Enterobacter agglomerans strain with a broad spectrum of antagonistic activity towards fungal and bacterial phytopathogens. Curr. Microbiol. 1996, 32, 208–212. [Google Scholar] [CrossRef]
- Loper, J.E.; Kobayashi, D.Y.; Paulsen, I.T. The genomic sequence of Pseudomonas fluorescens Pf-5: Insights into biological control. Phytopathology 2007, 97, 233–238. [Google Scholar] [CrossRef]
- Loper, J.E.; Henkels, M.D.; Shaffer, B.T.; Valeriote, F.A.; Gross, H. Isolation and identification of rhizoxin analogs from Pseudomonas fluorescens Pf-5 by using a genomic mining strategy. Appl. Environ. Microbiol. 2008, 74, 3085–3093. [Google Scholar] [CrossRef]
- Anand, A.; Chinchilla, D.; Tan, C.; Mène-Saffrané, L.; L’Haridon, F.; Weisskopf, L. Contribution of hydrogen cyanide to the antagonistic activity of Pseudomonas strains against Phytophthora infestans. Microorganisms 2020, 8, 1144. [Google Scholar] [CrossRef]
- Kraus, J.; Loper, J.E. Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 1995, 61, 849–854. [Google Scholar] [CrossRef]
- Zhao, Y.; Qian, G.; Chen, Y.; Du, L.; Liu, F. Transcriptional and antagonistic responses of biocontrol strain Lysobacter enzymogenes OH11 to the plant pathogenic oomycete Pythium aphanidermatum. Front. Microbiol. 2017, 8, 1025. [Google Scholar] [CrossRef]
- Christiansen, L.; Alanin, K.S.; Phippen, C.B.; Olsson, S.; Stougaard, P.; Hennessy, R.C. Fungal-associated molecules induce key genes involved in the biosynthesis of the antifungal secondary metabolites nunamycin and nunapeptin in the biocontrol strain Pseudomonas fluorescens In5. Appl. Environ. Microbiol. 2020, 86, e01284-20. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-Q.; Yan, X.; Zhang, M.-Y.; Zhang, L.-Q.; He, Y.-X. Flavonoids repress production of antifungal 2,4-DAPG but potentially facilitate root colonization of the rhizobacterium Pseudomonas fluorescens. Environ. Microbiol. 2020, 22, 5073–5089. [Google Scholar] [CrossRef] [PubMed]
- Jousset, A.; Rochat, L.; Lanoue, A.; Bonkowski, M.; Keel, C.; Scheu, S. Plants Respond to Pathogen Infection by Enhancing the Antifungal Gene Expression of Root-Associated Bacteria. Mol. Plant-Microbe Interact. 2011, 24, 352–358. [Google Scholar] [CrossRef]
- Höfte, M. The Use of Pseudomonas spp. as Bacterial Biocontrol Agents to Control Plant Disease; Burleigh Dodds Science Publishing Limited: Cambridge, UK, 2021. [Google Scholar]
- Raaijmakers, J.M.; De Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef]
- Jeong, H.; Choi, S.-K.; Ryu, C.-M.; Park, S.-H. Chronicle of a soil bacterium: Paenibacillus polymyxa E681 as a tiny guardian of plant and human health. Front. Microbiol. 2019, 10, 467. [Google Scholar] [CrossRef]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.-J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef]
- Nikolaidis, M.; Mossialos, D.; Oliver, S.G.; Amoutzias, G.D. Comparative analysis of the core proteomes among the Pseudomonas major evolutionary groups reveals species-specific adaptations for Pseudomonas aeruginosa and Pseudomonas chlororaphis. Diversity 2020, 12, 289. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).