Examination of Genetic Control Elements in the Phototrophic Firmicute Heliomicrobium modesticaldum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Plasmid Construction
2.3. Induction Tests
2.4. Cell Free Extracts
2.5. Reporter Activity Assays
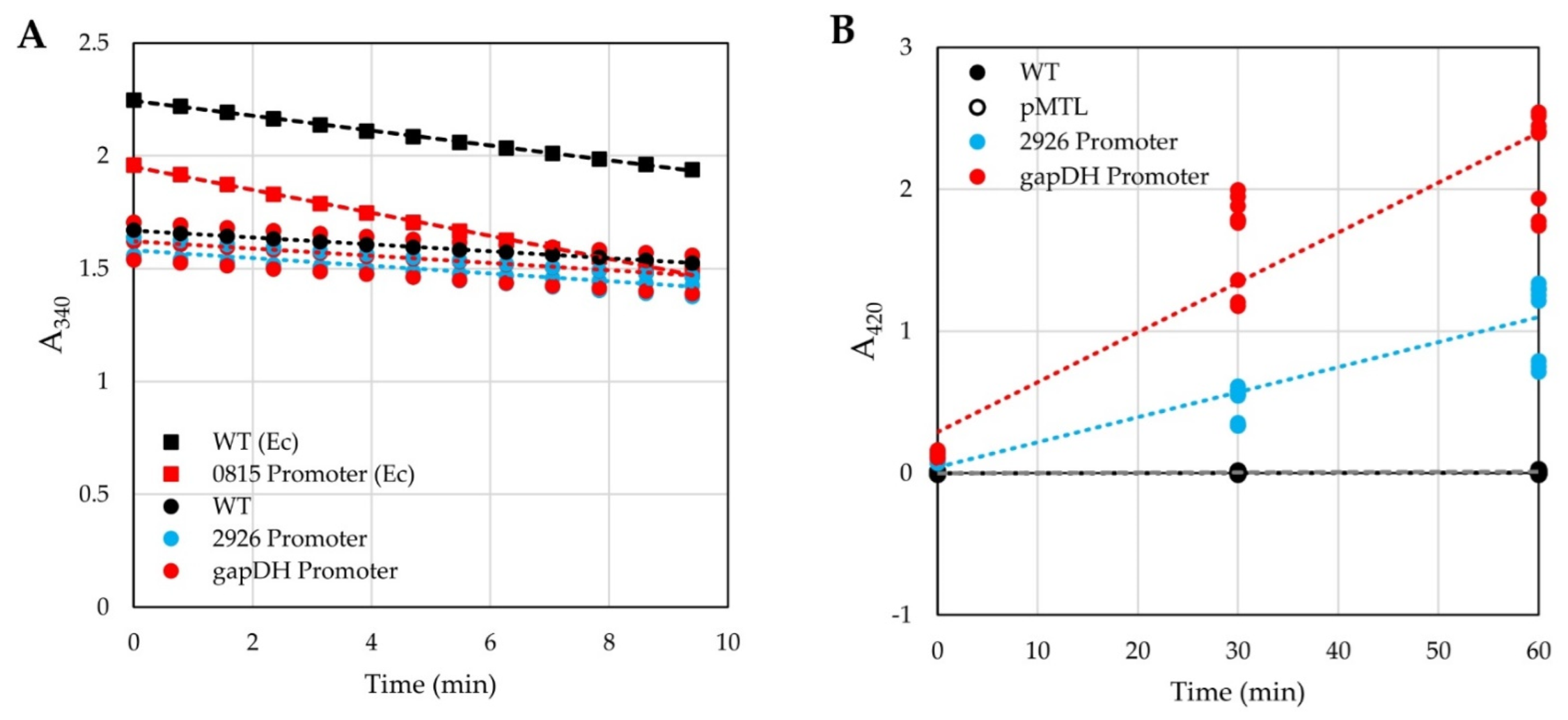
2.5.1. Alcohol Dehydrogenase Assays
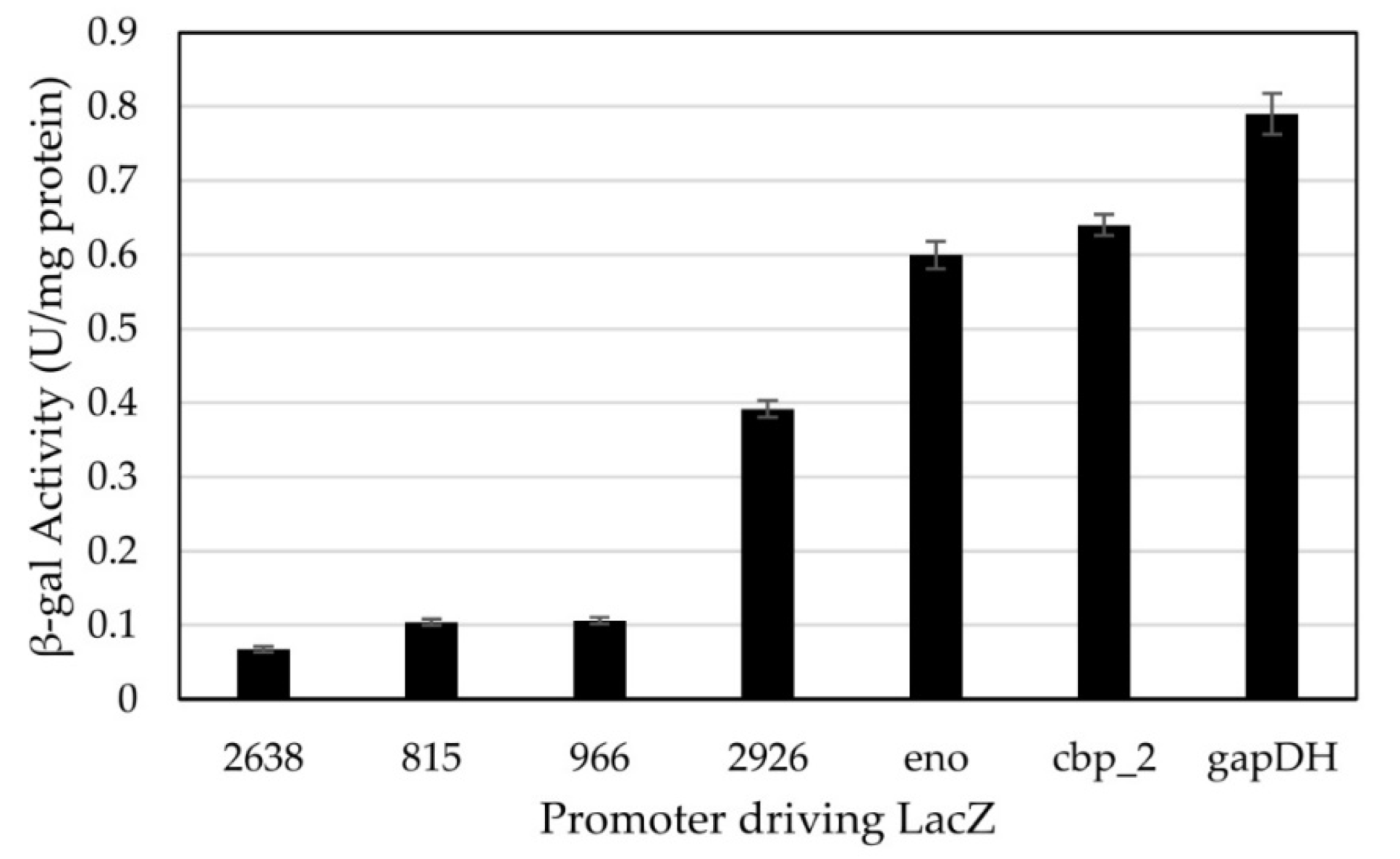
2.5.2. β-Galactosidase Assays
2.6. Calculations—Curve Fittings and Error Calculations
3. Results
3.1. Testing of Reporters in H. modesticaldum
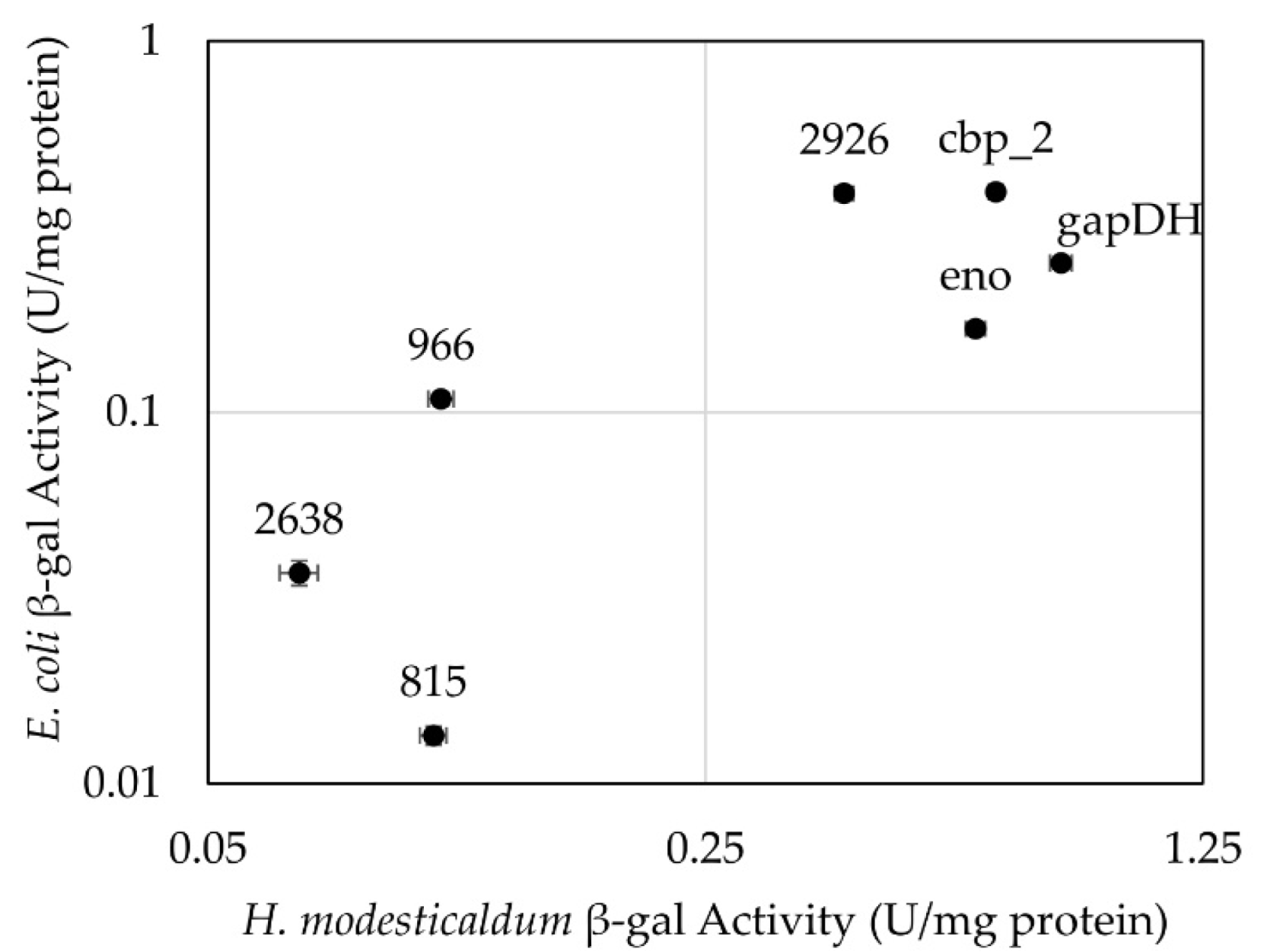
3.2. Testing of Constitutive Promoters from C. thermocellum in H. modesticaldum
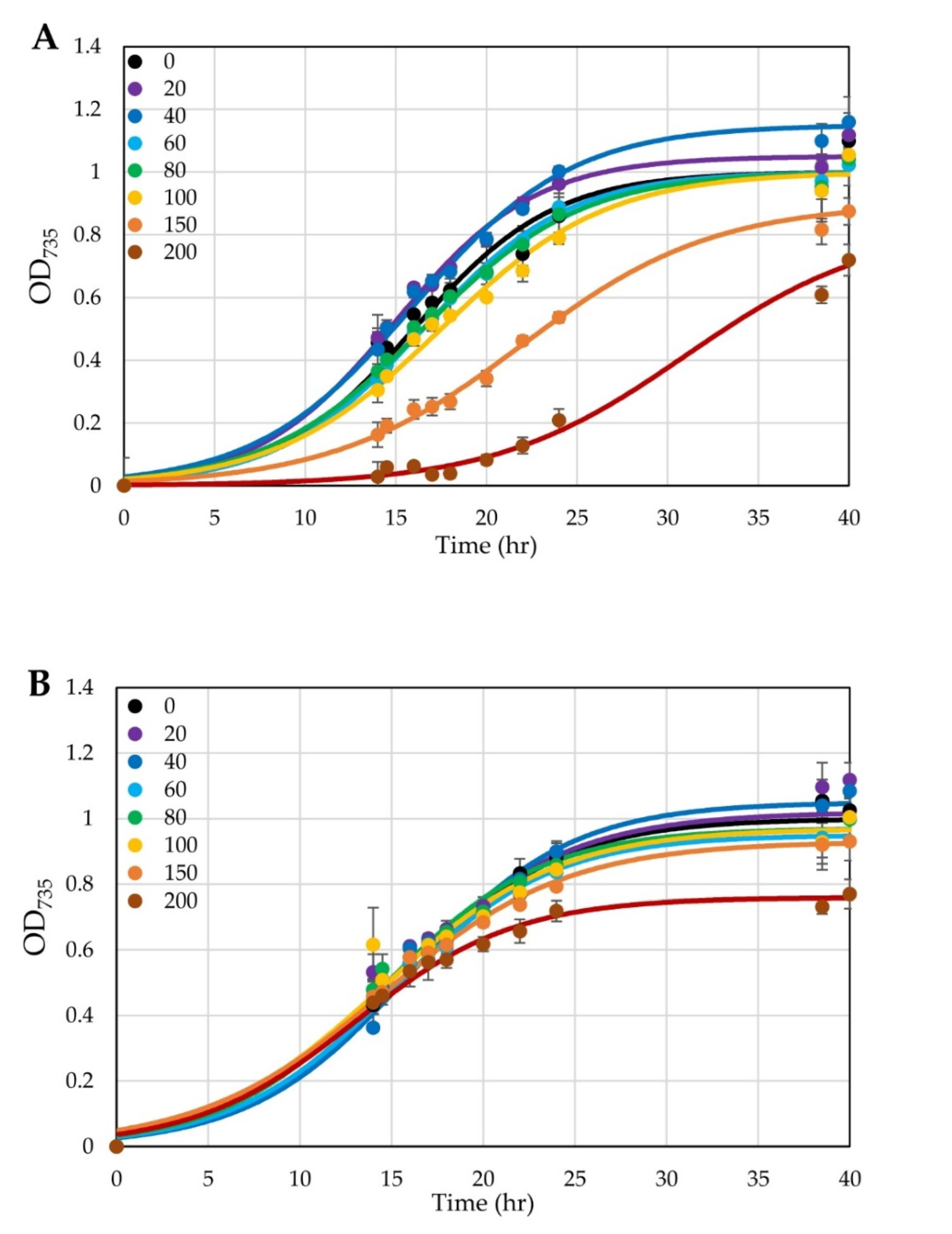
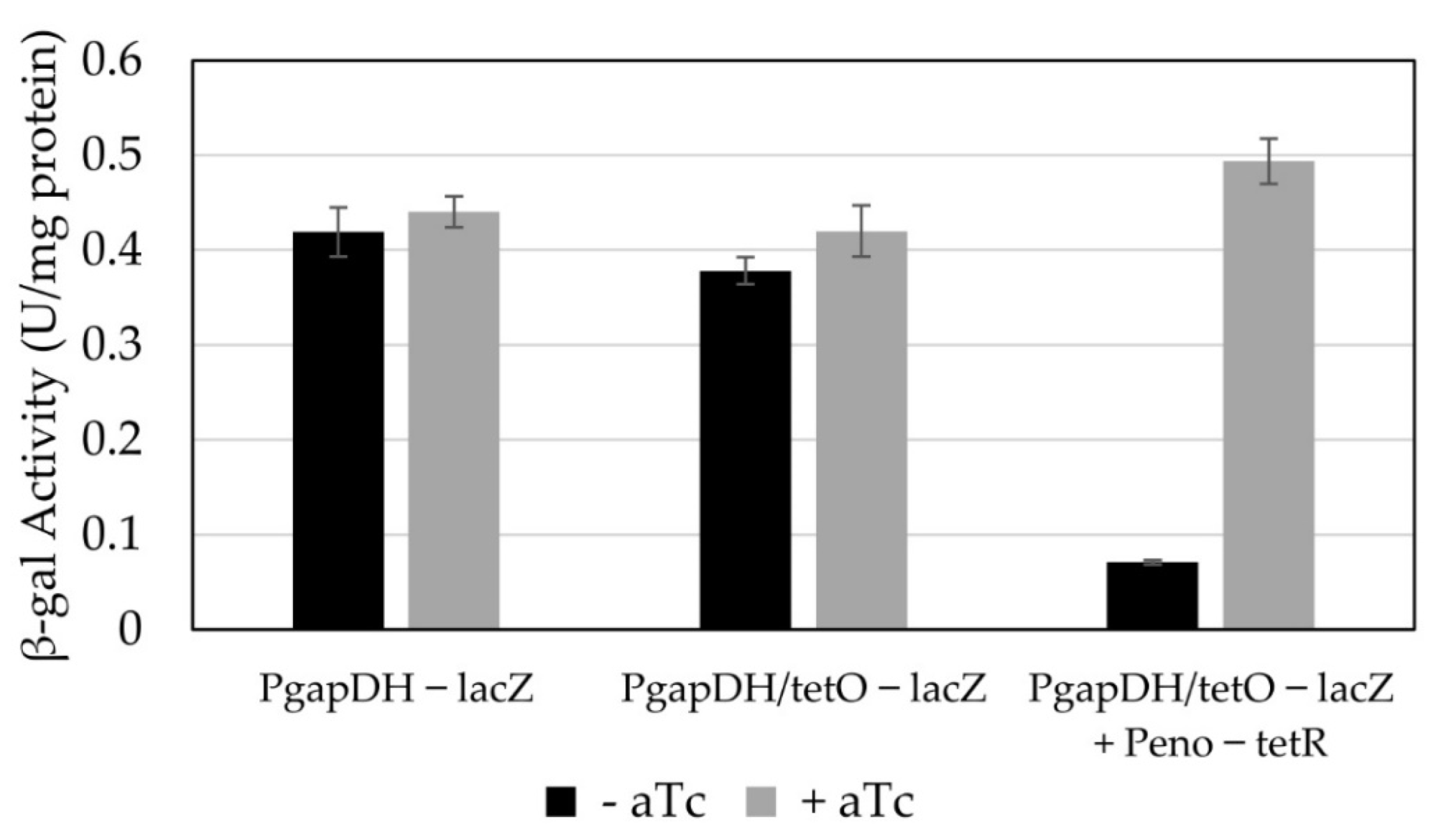
3.3. Conversion of a Constitutive Promoter into an Anhydrotetracycline-Inducible Promoter
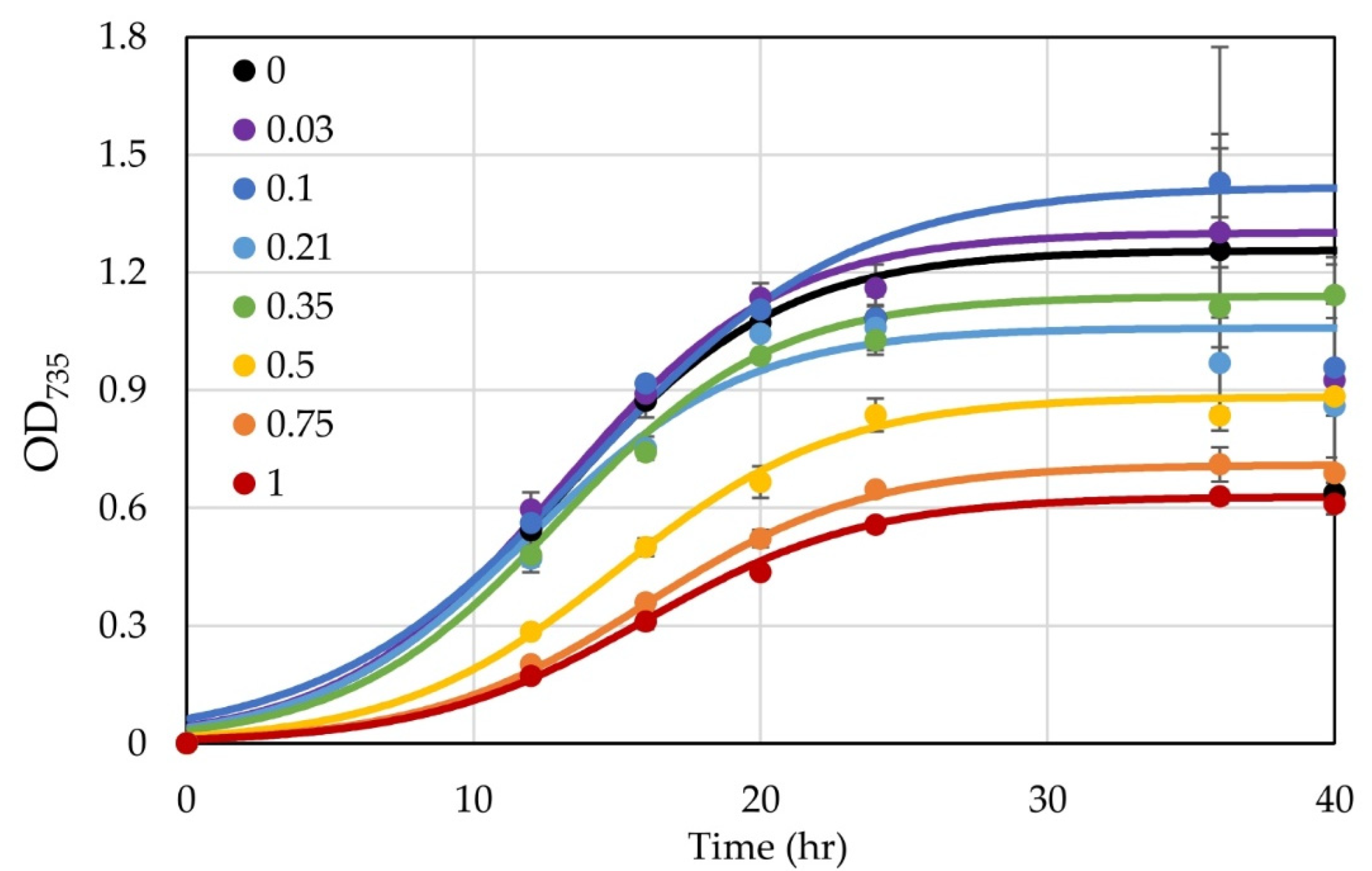
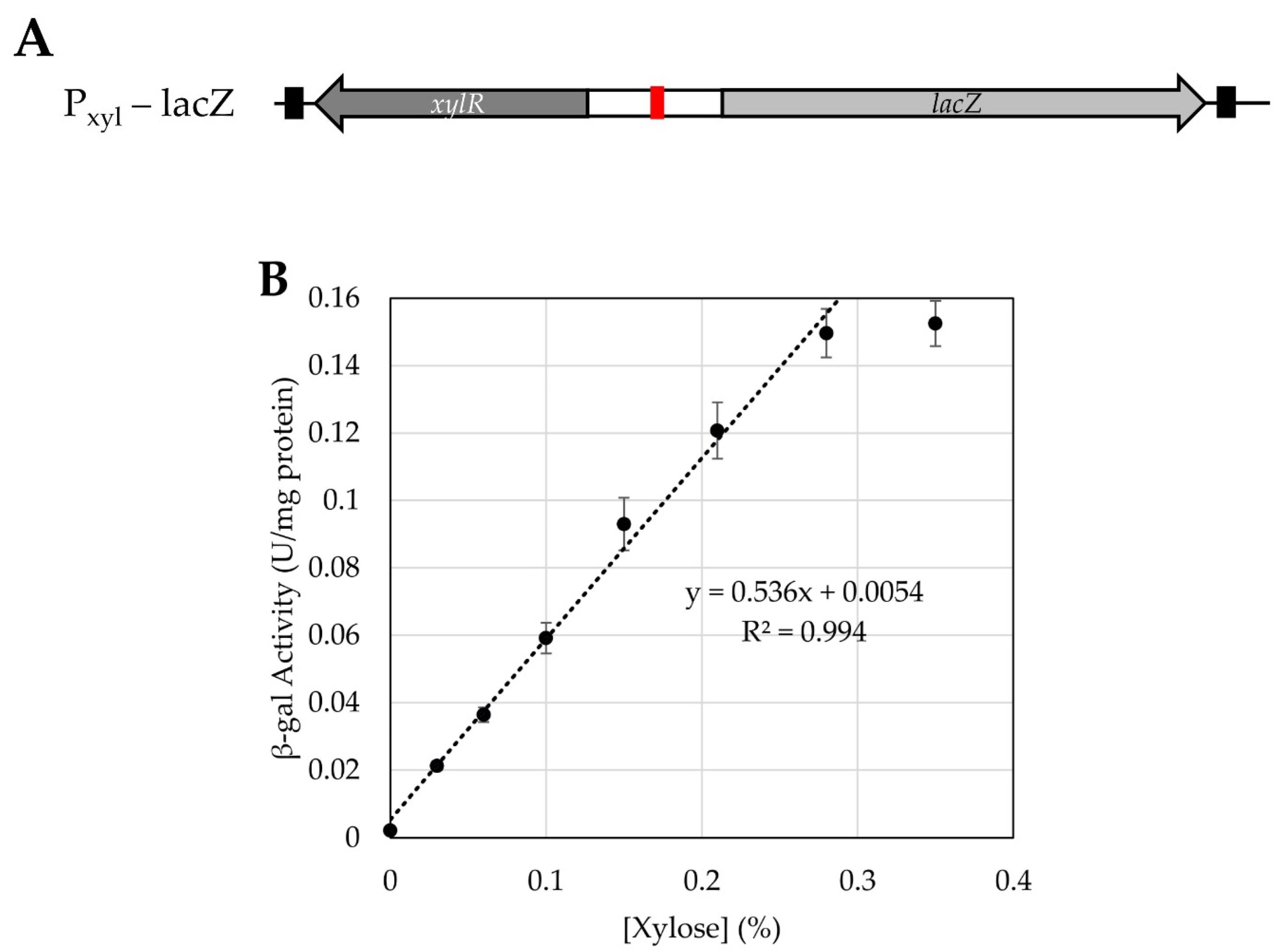
3.4. Testing Use of a Xylose-Inducible Promoter System in H. modesticaldum
4. Discussion
4.1. Reporters
4.2. Constitutive Promoters
4.3. Inducible Promoters
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kimble, L.K.; Stevenson, A.K.; Madigan, M.T. Chemotrophic Growth of Heliobacteria in Darkness. FEMS Microbiol. Lett. 1994, 115, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Kimble, L.K.; Mandelco, L.; Woese, C.R.; Madigan, M.T. Heliobacterium Modesticaldum, Sp. Nov., a Thermophilic Heliobacterium of Hot Springs and Volcanic Soils. Arch. Microbiol. 1995, 163, 259–267. [Google Scholar] [CrossRef]
- Orf, G.S.; Redding, K.E. The Heliobacteria. Encycl. Biochem. 2021, 1–13. [Google Scholar]
- Tang, K.-H.; Yue, H.; Blankenship, R.E. Energy Metabolism of Heliobacterium Modesticaldum during Phototrophic and Chemotrophic Growth. BMC Microbiol. 2010, 10, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, K.H.; Feng, X.; Zhuang, W.Q.; Alvarez-Cohen, L.; Blankenship, R.E.; Tang, Y.J. Carbon Flow of Heliobacteria Is Related More to Clostridia than to the Green Sulfur Bacteria. J. Biol. Chem. 2010, 285, 35104–35112. [Google Scholar] [CrossRef] [Green Version]
- Grégoire, D.S.; Lavoie, N.C.; Poulain, A.J. Heliobacteria Reveal Fermentation as a Key Pathway for Mercury Reduction in Anoxic Environments. Environ. Sci. Technol. 2018, 52, 4145–4153. [Google Scholar] [CrossRef]
- Sattley, W.M.; Madigan, M.T.; Swingley, W.D.; Cheung, P.C.; Clocksin, K.M.; Conrad, A.L.; Dejesa, L.C.; Honchak, B.M.; Jung, D.O.; Karbach, L.E.; et al. The Genome of Heliobacterium Modesticaldum, a Phototrophic Representative of the Firmicutes Containing the Simplest Photosynthetic Apparatus. J. Bacteriol. 2008, 190, 4687–4696. [Google Scholar] [CrossRef] [Green Version]
- Sheehy, D.; Kuang Lu, Y.; Osman, F.; Alattar, Z.; Flores, C.; Sussman, H.; Zaare, S.; Dooling, M.; Meraban, A.; Baker, P.; et al. Genome-Wide Transcriptional Response during the Shift to N2-Fixing Conditions in Heliobacterium modesticaldum. J. Proteom. Bioinform. 2018, 11, 143–160. [Google Scholar] [CrossRef]
- Baker, P.L.; Orf, G.S.; Khan, Z.; Espinoza, L.; Leung, S.; Kevershan, K.; Redding, K.E. A Molecular Biology Tool Kit for the Phototrophic Firmicute, Heliobacterium modesticaldum. Appl. Environ. Microbiol. 2019, 85, e01287-19. [Google Scholar] [CrossRef] [Green Version]
- Baker, P.L.; Orf, G.S.; Kevershan, K.; Pyne, M.E.; Bicer, T.; Redding, K.E. Using the Endogenous CRISPR-Cas System of Heliobacterium Modesticaldum To Delete the Photochemical Reaction Center Core Subunit Gene. Appl. Environ. Microbiol. 2019, 85, e01644-19. [Google Scholar] [CrossRef]
- Gisriel, C.; Sarrou, I.; Ferlez, B.; Golbeck, J.H.; Redding, K.E.; Fromme, R. Structure of a Symmetric Photosynthetic Reaction Center–Photosystem. Science 2017, 357, 1021–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinnickel, M.; Golbeck, J.H. Heliobacterial Photosynthesis. Photosynth. Res. 2007, 92, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Leung, S.W.; Baker, P.L.; Redding, K.E. Deletion of the Cytochrome Bc Complex from Heliobacterium Modesticaldum Results in Viable but Non-Phototrophic Cells. Photosynth. Res. 2021, 148, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Orf, G.S.; Redding, K.E. Expression and Purification of Affinity-Tagged Variants of the Photosynthetic Reaction Center from Heliobacterium modesticaldum. Photosynth. Res. 2019, 142, 335–348. [Google Scholar] [CrossRef]
- Pyne, M.E.; Bruder, M.R.; Moo-Young, M.; Chung, D.A.; Chou, C.P. Harnessing Heterologous and Endogenous CRISPR-Cas Machineries for Efficient Markerless Genome Editing in Clostridium. Sci. Rep. 2016, 6, 25666. [Google Scholar] [CrossRef] [Green Version]
- Olson, D.G.; Maloney, M.; Lanahan, A.A.; Hon, S.; Hauser, L.J.; Lynd, L.R. Identifying Promoters for Gene Expression in Clostridium thermocellum. Metab. Eng. Commun. 2015, 2, 23–29. [Google Scholar] [CrossRef]
- Heap, J.T.; Pennington, O.J.; Cartman, S.T.; Minton, N.P. A Modular System for Clostridium Shuttle Plasmids. J. Microbiol. Methods 2009, 78, 79–85. [Google Scholar] [CrossRef]
- Nariya, H.; Miyata, S.; Kuwahara, T.; Okabe, A. Development and Characterization of a Xylose-Inducible Gene Expression System for Clostridium Perfringens. Appl. Environ. Microbiol. 2011, 77, 8439–8441. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Tao, W.; Zhang, Y.; Li, Y. Development of an Anhydrotetracycline-Inducible Gene Expression System for Solvent-Producing Clostridium acetobutylicum: A Useful Tool for Strain Engineering. Metab. Eng. 2012, 14, 59–67. [Google Scholar] [CrossRef]
- Lin, P.P.; Rabe, K.S.; Takasumi, J.L.; Kadisch, M.; Arnold, F.H.; Liao, J.C. Isobutanol Production at Elevated Temperatures in Thermophilic Geobacillus thermoglucosidasius. Metab. Eng. 2014, 24, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Ramos, J.; Martinez-Bueno, M.; Molina-Henares, A.; Teran, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The TetR Family of Transcriptional Repressors. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertram, R.; Hillen, W. The Application of Tet Repressor in Prokaryotic Gene Regulation and Expression. Microb. Biotechnol. 2008, 1, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Sprengel, R.; Hasan, M.T. Tetracycline-Controlled Genetic Switches. Handb. Exp. Pharmacol. 2007, 178, 49–72. [Google Scholar] [CrossRef]
- Mordaka, P.M.; Heap, J.T. Stringency of Synthetic Promoter Sequences in Clostridium Revealed and Circumvented by Tuning Promoter Library Mutation Rates. ACS Synth. Biol. 2018, 7, 672–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Layton, A.M.; Redding, K.E. Examination of Genetic Control Elements in the Phototrophic Firmicute Heliomicrobium modesticaldum. Microorganisms 2022, 10, 876. https://doi.org/10.3390/microorganisms10050876
Layton AM, Redding KE. Examination of Genetic Control Elements in the Phototrophic Firmicute Heliomicrobium modesticaldum. Microorganisms. 2022; 10(5):876. https://doi.org/10.3390/microorganisms10050876
Chicago/Turabian StyleLayton, Alexandria M., and Kevin E. Redding. 2022. "Examination of Genetic Control Elements in the Phototrophic Firmicute Heliomicrobium modesticaldum" Microorganisms 10, no. 5: 876. https://doi.org/10.3390/microorganisms10050876
APA StyleLayton, A. M., & Redding, K. E. (2022). Examination of Genetic Control Elements in the Phototrophic Firmicute Heliomicrobium modesticaldum. Microorganisms, 10(5), 876. https://doi.org/10.3390/microorganisms10050876





