Genetic Diversity and Relationships of Listeria monocytogenes Serogroup IIa Isolated in Poland
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Isolation and Sequencing
2.3. WGS Analysis
2.4. Comparison of L. monocytogenes Sequences
3. Results and Discussion
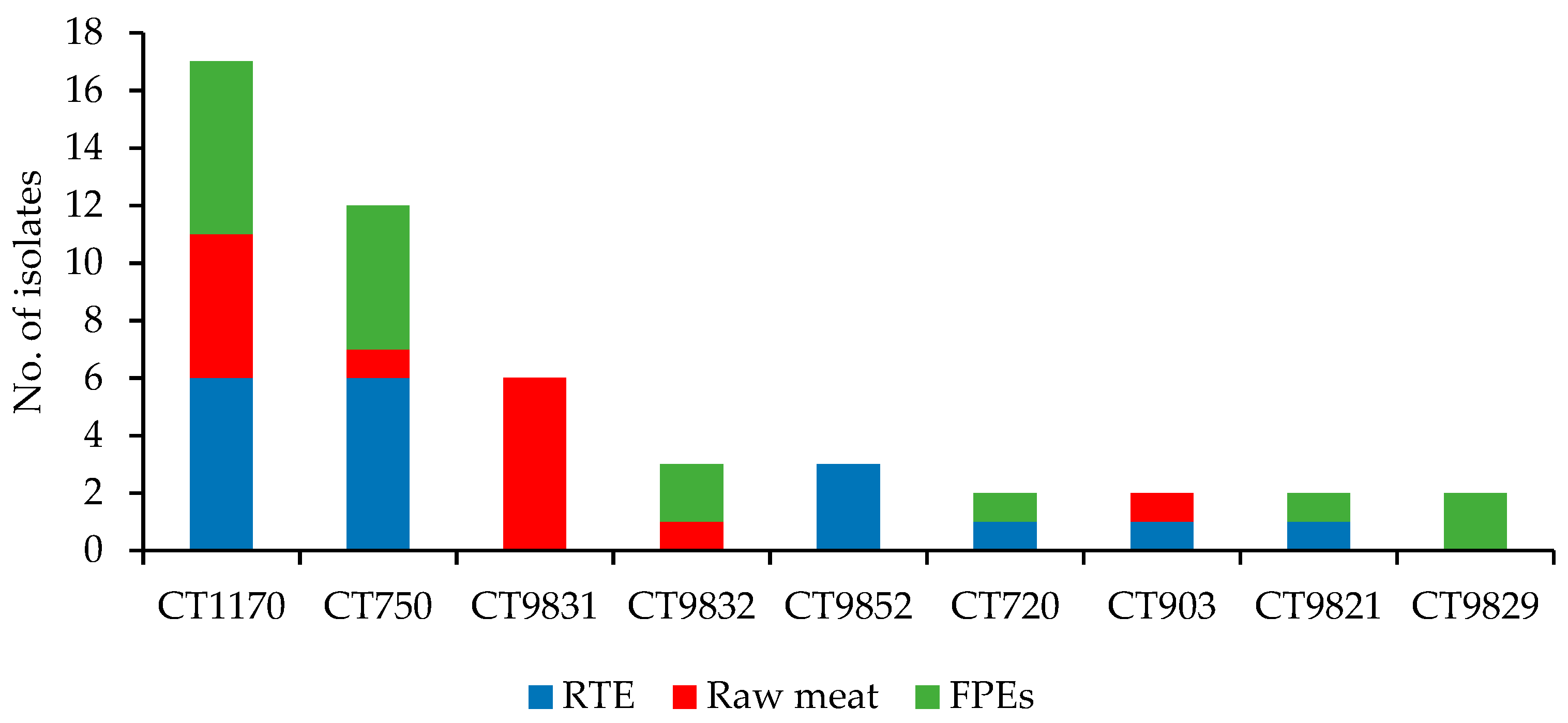
3.1. WGS-Based Typing of L. monocytogenes of IIa Serogroup
3.2. General Molecular Characteristics of L. monocytogenes
3.3. Molecular Characteristics of L. monocytogenes of Different CCs
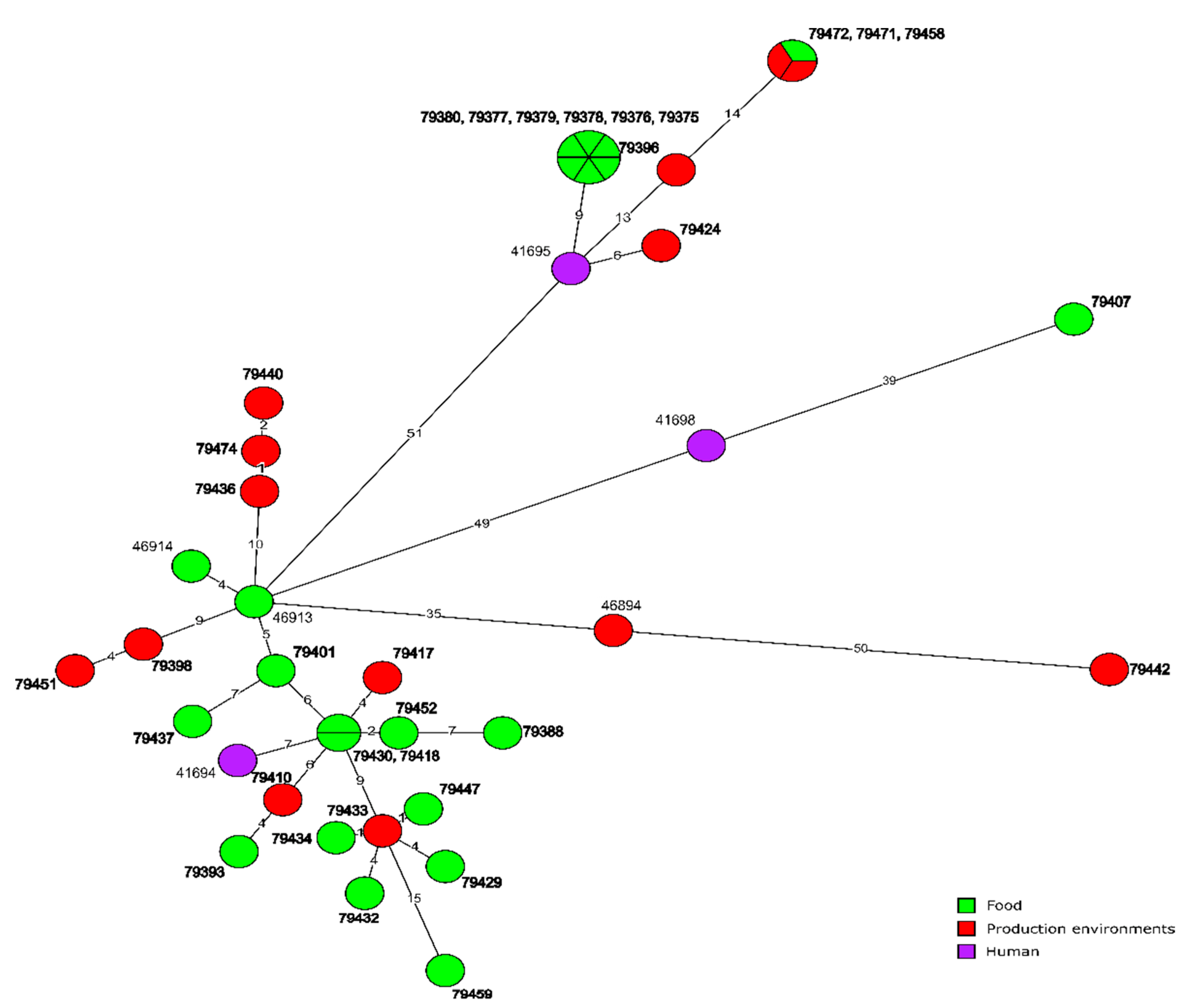
3.3.1. CC155 Isolates
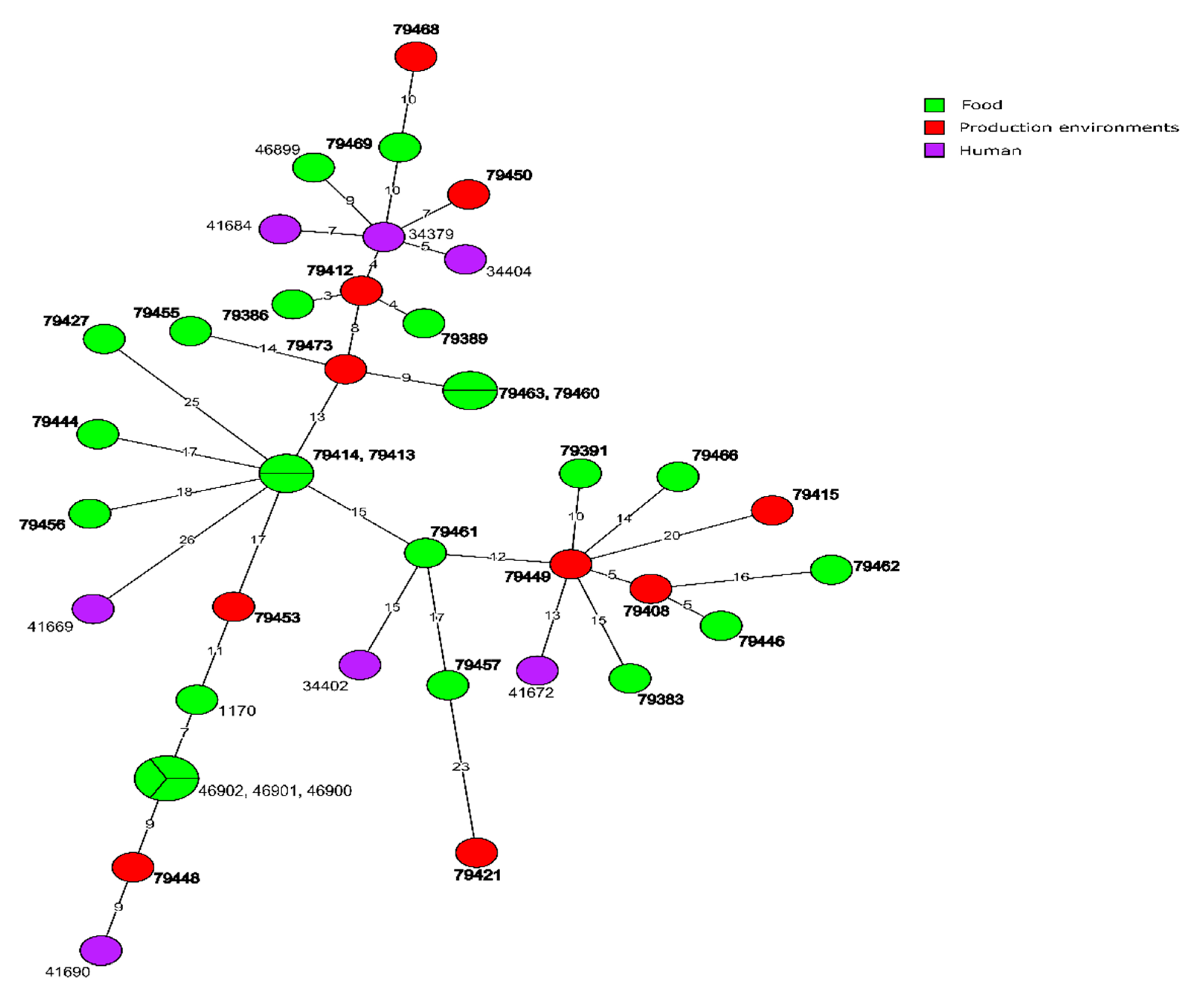
3.3.2. CC121 Isolates
3.3.3. CC8 Isolates
3.3.4. Isolates of the Remaining CCs
3.4. Molecular Relationships of the Current and Other Polish L. monocytogenes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burall, L.S.; Grim, C.J.; Mammel, M.K.; Datta, A.R. Whole genome sequence analysis using JSpecies tool establishes clonal relationships between Listeria monocytogenes strains from epidemiologically unrelated listeriosis outbreaks. PLoS ONE 2016, 11, e0150797. [Google Scholar] [CrossRef] [PubMed]
- Burall, L.S.; Grim, C.J.; Datta, A.R. A clade of Listeria monocytogenes serotype 4b variant strains linked to recent listeriosis outbreaks associated with produce from a defined geographic region in the US. PLoS ONE 2017, 12, e0176912. [Google Scholar] [CrossRef] [PubMed]
- Henri, C.; Leekitcharoenphon, P.; Carleton, H.A.; Radomski, N.; Kaas, R.S.; Mariet, J.F.; Felten, A.; Aarestrup, F.M.; Gerner Smidt, P.; Roussel, S.; et al. An assessment of different genomic approaches for inferring phylogeny of Listeria monocytogenes. Front. Microbiol. 2017, 8, 2351. [Google Scholar] [CrossRef] [PubMed]
- Hyden, P.; Pietzka, A.; Lennkh, A.; Murer, A.; Springer, B.; Blaschitz, M.; Indra, A.; Huhulescu, S.; Allerberger, F.; Ruppitsch, W.; et al. Whole genome sequence-based serogrouping of Listeria monocytogenes isolates. J. Biotechnol. 2016, 235, 181–186. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, 6971. [Google Scholar] [CrossRef]
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- Ranasinghe, R.A.S.S.; Satharasinghe, D.A.; Tang, J.Y.H.; Rukayadi, Y.; Radu, K.R.; New, C.Y.; Son, R.; Premarathne, J.M.K.J.K. Persistence of Listeria monocytogenes in food commodities: Foodborne pathogenesis, virulence factors, and implications for public health. Food Res. 2021, 5, 1–16. [Google Scholar] [CrossRef]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef]
- Matle, I.; Mbatha, K.R.; Madoroba, E. A review of Listeria monocytogenes from meat and meat products: Epidemiology, virulence factors, antimicrobial resistance and diagnosis. Onderstepoort J. Vet. Res. 2020, 87, a1869. [Google Scholar] [CrossRef]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tar, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, W.; Wang, J.; Xu, B.; Liu, H.; Dong, Q.; Zhang, X. 10-year molecular surveillance of Listeria monocytogenes using whole-genome sequencing in Shanghai, China, 2009–2019. Front. Microbiol. 2020, 11, 551020. [Google Scholar] [CrossRef]
- Alvarez-Molina, A.; Cobo-Díaz, J.F.; López, M.; Prieto, M.; de Toro, M.; Alvarez-Ordóñez, A. Unraveling the emergence and population diversity of Listeria monocytogenes in a newly built meat facility through whole genome sequencing. Int. J. Food Microbiol. 2021, 340, 109043. [Google Scholar] [CrossRef]
- Wang, Y.; Ji, Q.; Li, S.; Liu, M. Prevalence and genetic diversity of Listeria monocytogenes isolated from retail pork in Wuhan, China. Front. Microbiol. 2021, 12, 620482. [Google Scholar] [CrossRef]
- Hurley, D.; Luque-Sastre, L.; Parker, C.T.; Huynh, S.; Eshwar, A.K.; Nguyen, S.V.; Andrews, N.; Moura, A.; Fox, E.M.; Jordan, K.; et al. Whole-genome sequencing-based characterization of 100 Listeria monocytogenes isolates collected from food processing environments over a four-year period. mSphere 2019, 4, e00252-19. [Google Scholar] [CrossRef]
- Jagadeesan, B.; Baert, L.; Wiedmann, M.; Orsi, R.H. Comparative analysis of tools and approaches for source tracking Listeria monocytogenes in a food facility using whole-genome sequence data. Front. Microbiol. 2019, 10, 947. [Google Scholar] [CrossRef]
- Orsi, R.H.; Jagadeesan, B.; Baert, L.; Wiedmann, M. Identification of closely related Listeria monocytogenes isolates with no apparent evidence for a common source or location: A retrospective whole genome sequencing analysis. J. Food Prot. 2021, 84, 1104–1113. [Google Scholar] [CrossRef]
- Bergholz, T.M.; Shah, M.K.; Burall, L.S.; Rakic-Martinez, M.; Datta, A.R. Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis. Appl. Microbiol. Biotechnol. 2018, 102, 3475–3485. [Google Scholar] [CrossRef]
- Palma, F.; Brauge, T.; Radomski, N.; Mallet, L.; Felten, A.; Mistou, M.Y.; Brisabois, A.; Guillier, L.; Midelet-Bourdin, G. Dynamics of mobile genetic elements of Listeria monocytogenes persisting in ready-to-eat seafood processing plants in France. BMC Genom. 2020, 21, 130. [Google Scholar] [CrossRef]
- Sévellec, Y.; Torresi, M.; Félix, B.; Palma, F.; Centorotola, G.; Bilei, S.; Senese, M.; Terracciano, G.; Leblanc, J.C.; Pomilio, F.; et al. First report on the finding of Listeria mnocytogenes ST121 strain in a dolphin brain. Pathogens 2020, 9, 802. [Google Scholar] [CrossRef]
- Wagner, E.; Zaiser, A.; Leitner, R.; Quijada, N.M.; Pracser, N.; Pietzka, A.; Ruppitsch, W.; Schmitz-Esser, S.; Wagner, M.; Rychli, K. Virulence characterization and comparative genomics of Listeria monocytogenes sequence type 155 strains. BMC Genom. 2020, 21, 847. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Maiden, M.C.J. BIGSdb: Scalable analysis of bacterial 780 genome variation at the population level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Tourdjman, M.; Leclercq, A.; Hamelin, E.; Laurent, E.; Fredriksen, N.; Van Cauteren, D.; Bracq-Dieye, H.; Thouvenot, P.; Vales, G.; et al. Real-time whole-genome sequencing for surveillance of Listeria monocytogenes, France. Emerg. Infect. Dis. 2017, 23, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Schubert, W.D.; Urbanke, C.; Ziehm, T.; Beier, V.; Machner, M.P.; Domann, E.; Wehland, J.; Chakraborty, T.; Heinz, D.W. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 2002, 111, 825–836. [Google Scholar] [CrossRef]
- Franciosa, G.; Maugliani, A.; Scalfaro, C.; Floridi, F.; Aureli, P. Expression of internalin A and biofilm formation among Listeria monocytogenes clinical isolates. Int. J. Immunopathol. Pharmacol. 2009, 22, 183–193. [Google Scholar] [CrossRef]
- Meier, A.B.; Guldimann, C.; Markkula, A.; Pöntinen, A.; Korkeala, H.; Tasara, T. Comparative phenotypic and genotypic analysis of Swiss and Finnish Listeria monocytogenes isolates with respect to benzalkonium chloride resistance. Front. Microbiol. 2017, 8, 397. [Google Scholar] [CrossRef]
- Ortiz, S.; López-Alonso, V.; Rodríguez, P.; Martínez-Suárez, J.V. The connection between persistent, disinfectant-resistant Listeria monocytogenes strains from two geographically separate Iberian pork processing plants: Evidence from comparative genome analysis. Appl. Environ. Microbiol. 2016, 82, 308–317. [Google Scholar] [CrossRef]
- Kropac, A.C.; Eshwar, A.K.; Stephan, R.; Tasara, T. New insights on the role of the pLMST6 plasmid in Listeria monocytogenes biocide tolerance and virulence. Front. Microbiol. 2019, 10, 1538. [Google Scholar] [CrossRef]
- ISO 11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp.—Part 1: Detection Method. International Organization for Standardization: Geneva, Switzerland, 2017.
- Lachtara, B.; Osek, J.; Wieczorek, W. Molecular typing of Listeria monocytogenes IVb serogroup isolated from food and food production environments in Poland. Pathogens 2021, 10, 482. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: Aflexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.; Nikolenko, S.; Pham, S.; Prjibelski, A.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S.A. New perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef]
- Gilmour, M.W.; Graham, M.; Van Domselaar, G.; Tyler, S.; Kent, H.; Trout-Yakel, K.M.; Larios, O.; Allen, V.; Lee, B.; Nadon, C. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genom. 2010, 11, 120. [Google Scholar] [CrossRef]
- Harter, E.; Wagner, E.M.; Zaiser, A.; Halecker, S.; Wagner, M.; Rychli, K. Stress Survival Islet 2, predominantly present in Listeria monocytogenes strains of sequence type 121, is involved in the alkaline and oxidative stress responses. Appl. Environ. Microbiol. 2017, 83, e00827-17. [Google Scholar] [CrossRef]
- Ryan, S.; Begley, M.; Hill, C.; Gahan, C.G.M. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 2010, 109, 984–995. [Google Scholar] [CrossRef]
- Zhang, J.; Cao, G.; Xu, X.; Allard, M.; Li, P.; Brown, E.; Yang, X.; Pan, H.; Meng, J. Evolution and diversity of Listeria monocytogenes from clinical and food samples in Shanghai, China. Front. Microbiol. 2016, 7, 1138. [Google Scholar] [CrossRef]
- Kremer, P.H.; Lees, J.A.; Koopmans, M.M.; Ferwerda, B.; Arends, A.W.; Feller, M.M.; Schipper, K.; Valls Seron, M.; van der Ende, A.; Brouwer, M.C.; et al. Benzalkonium tolerance genes and outcome in Listeria monocytogenes meningitis. Clin. Microbial. Infect. 2017, 23, 265.e1–265.e7. [Google Scholar] [CrossRef]
- Møretrø, T.; Schirmer, B.; Heir, E.; Fagerlund, A.; Hjemli, P.; Langsrud, S. Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry. Int. J. Food Microbiol. 2017, 241, 215–224. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, 16–21. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards); Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Escámez, P.S.F.; Girones, R.; Herman, L.; Koutsoumanis, K.; et al. Scientific opinion on the Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018, 16, 5134. [Google Scholar] [CrossRef]
- Fischer, M.A.; Thürmer, A.; Flieger, A.; Halbedel, S. Complete genome sequences of three clinical Listeria monocytogenes sequence type 8 strains from recent German listeriosis outbreaks. Microbiol. Resour. Announc. 2021, 10, e00303-21. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Bai, L.; Fu, P.; Han, H.; Liu, J.; Guo, Y. The epidemiology of Listeria monocytogenes in China. Foodborne Pathog. Dis. 2018, 15, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Zhang, P.; Niu, Y.; Chen, Q.; Ma, X. Genomic characterization of clinical Listeria monocytogenes isolates in Beijing, China. Front. Microbiol. 2021, 12, 751003. [Google Scholar] [CrossRef]
- Kurpas, M.; Osek, J.; Moura, A.; Leclercq, A.; Lecuit, M.; Wieczorek, K. Genomic characterization of Listeria monocytogenes isolated from ready-to-eat meat and meat processing environments in Poland. Front. Microbiol. 2020, 11, 1412. [Google Scholar] [CrossRef]
- Shi, D.; Anwar, T.M.; Pan, H.; Chai, W.; Xu, S.; Yue, M. Genomic determinants of pathogenicity and antimicrobial resistance for 60 global Listeria monocytogenes isolates responsible for invasive infections. Front. Cell. Infect. Microbiol. 2021, 11, 718840. [Google Scholar] [CrossRef]
- Gouin, E.; Mengaud, J.; Cossart, P. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect. Immun. 1994, 62, 3550–3553. [Google Scholar] [CrossRef]
- Disson, O.; Moura, A.; Lecuit, M. Making sense of the biodiversity and virulence of Listeria monocytogenes. Trends Microbiol. 2021, 11, 811–822. [Google Scholar] [CrossRef]
- Parsons, C.; Lee, S.; Kathariou, S. Heavy metal resistance determinants of the foodborne pathogen Listeria monocytogenes. Genes 2019, 10, 11. [Google Scholar] [CrossRef]
- Chmielowska, C.; Korsak, D.; Szuplewska, M.; Grzelecka, M.; Maćkiw, E.; Stasiak, M.; Macion, A.; Skowron, K.; Bartosik, D. Benzalkonium chloride and heavy metal resistance profiles of Listeria monocytogenes strains isolated from fish, fish products and food-producing factories in Poland. Food Microbiol. 2021, 98, 103756. [Google Scholar] [CrossRef]
- Ratani, S.S.; Siletzky, R.M.; Dutta, V.; Yildirim, S.; Osborne, J.; Lin, W.; Hitchins, A.D.; Ward, T.J.; Kathariou, S. Heavy metal and disinfectant resistance of Listeria monocytogenes from foods and food processing plants. Appl. Environ. Microbiol. 2012, 78, 6938–6945. [Google Scholar] [CrossRef]
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to stress conditions encountered in food and food processing environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef]
- O’Byrne, C.P.; Karatzas, K.A. The role of sigma B (sB) in the stress adaptations of Listeria monocytogenes: Overlaps between stress adaptation and virulence. Adv. Appl. Microbiol. 2008, 65, 115–140. [Google Scholar] [CrossRef]
- Mohan, V.; Cruz, C.D.; van Vliet, A.H.M.; Pitman, A.R.; Visnovsky, S.B.; Rivas, L.; Gilpin, B.; Fletcher, G.C. Genomic diversity of Listeria monocytogenes isolates from seafood, horticulture and factory environments in New Zealand. Int. J. Food Microbiol. 2021, 347, 109166. [Google Scholar] [CrossRef]
- Mafuna, T.; Matle, I.; Magwedere, K.; Pierneef, R.E.; Reva, O.N. Whole genome-based characterization of Listeria monocytogenes isolates recovered from the food chain in South Africa. Front. Microbiol. 2021, 12, 669287. [Google Scholar] [CrossRef]
- Rychli, K.; Wagner, E.M.; Ciolacu, L.; Zaiser, A.; Tasara, T.; Wagner, M.; Schmitz-Esser, S. Comparative genomics of human and non-human Listeria monocytogenes sequence type 121 strains. PLoS ONE 2017, 12, e0176857. [Google Scholar] [CrossRef]
- Vu, H.T.K.; Benjakul, S.; Vongkamjan, K. Characterization of Listeria prophages in lysogenic isolates from foods and food processing environments. PLoS ONE 2019, 14, e0214641. [Google Scholar] [CrossRef]
- Casey, A.; Jordan, K.; Neve, H.; Coffey, A.; McAuliffe, O. A tail of two phages: Genomic and functional analysis of Listeria monocytogenes phages vB_LmoS_188 and vB_LmoS_293 reveal the receptor-binding proteins involved in host specificity. Front. Microbiol. 2015, 6, 1107. [Google Scholar] [CrossRef]
- Denes, T.; Vongkamjan, K.; Ackermann, H.W.; Moreno Switt, A.I.; Wiedmann, M.; den Bakker, H.C. Comparative genomic and morphological analyses of Listeria phages isolated from farm environments. Appl. Environ. Microbiol. 2014, 80, 4616–4625. [Google Scholar] [CrossRef] [PubMed]
- Matle, I.; Pierneef, R.; Mbatha, K.R.; Magwedere, K.; Madoroba, E. Genomic diversity of common sequence types of Listeria monocytogenes isolated from ready-to-eat products of animal origin in South Africa. Genes 2019, 10, 1007. [Google Scholar] [CrossRef] [PubMed]
- Kuenne, C.; Billion, A.; Mraheil, M.A.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genom. 2013, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Rabinovich, L.; Sigal, N.; Borovok, I.; Nir-Paz, R.; Herskovits, A.A. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 2012, 150, 792–802. [Google Scholar] [CrossRef]
- Duze, S.T.; Marimani, M.; Patel, M. Tolerance of Listeria monocytogenes to biocides used in food processing environments. Food Microbiol. 2021, 97, 103758. [Google Scholar] [CrossRef]
- Dutta, V.; Elhanafi, D.; Kathariou, S. Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl. Environ. Microbiol. 2013, 79, 6067–6074. [Google Scholar] [CrossRef]
- Müller, A.; Rychli, K.; Zaiser, A.; Wieser, C.; Wagner, M.; Schmitz-Esser, S. The Listeria monocytogenes transposon Tn6188 provides increased tolerance to various quaternary ammonium compounds and ethidium bromide. FEMS Microbiol. Lett. 2014, 361, 166–173. [Google Scholar] [CrossRef]
- Keeney, K.; Trmcic, A.; Zhu, Z.; Delaquis, P.; Wang, S. Stress survival islet 1 contributes to serotype-specific differences in biofilm formation in Listeria monocytogenes. Lett. Appl. Microbiol. 2018, 67, 530–536. [Google Scholar] [CrossRef]
- Matereke, L.T.; Okoh, A.I. Listeria monocytogenes virulence, antimicrobial resistance and environmental persistence: A review. Pathogens 2020, 9, 528. [Google Scholar] [CrossRef]
- Palaiodimou, L.; Fanning, S.; Fox, E.M. Genomic insights into persistence of Listeria species in the food processing environment. J. Appl. Microbiol. 2021, 131, 2082–2094. [Google Scholar] [CrossRef]
- Gilmartin, N.; Gião, M.S.; Keevil, C.W.; O’Kennedy, R. Differential internalin A levels in biofilms of Listeria monocytogenes grown on different surfaces and nutrient conditions. Int. J. Food Microbiol. 2016, 219, 50–55. [Google Scholar] [CrossRef]
- Nightingale, K.K.; Windham, K.; Martin, K.E.; Yeung, M.; Wiedmann, M. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 2005, 71, 8764–8772. [Google Scholar] [CrossRef]
- Naditz, A.L.; Dzieciol, M.; Wagner, M.; Schmitz-Esser, S. Plasmids contribute to food processing environment-associated stress survival in three Listeria monocytogenes ST121, ST8, and ST5 strains. Int. J. Food Microbiol. 2019, 299, 39–46. [Google Scholar] [CrossRef]
- Schmitz-Esser, S.; Müller, A.; Stessl, B.; Wagner, M. Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front. Microbiol. 2015, 6, 380. [Google Scholar] [CrossRef]
- Horlbog, J.A.; Kent, D.; Stephan, R.; Guldimann, C. Surviving host and food relevant stresses: Phenotype of L. monocytogenes strains isolated from food and clinical sources. Sci. Rep. 2018, 8, 12931. [Google Scholar] [CrossRef]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 2488. [Google Scholar] [CrossRef]
- Kovacevic, J.; Ziegler, J.; Wałecka-Zacharska, E.; Reimer, A.; Kitts, D.D.; Gilmour, M.W. Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 2016, 82, 939–953. [Google Scholar] [CrossRef]
- Fagerlund, A.; Langsrud, S.; Schirmer, B.C.T.; Møretrø, T.; Heir, E. Genome analysis of Listeria monocytogenes sequence type 8 strains persisting in salmon and poultry processing environments and comparison with related strains. PLoS ONE 2016, 11, e0151117. [Google Scholar] [CrossRef]
- Wieczorek, K.; Bomba, A.; Osek, J. Whole-genome sequencing-based characterization of Listeria monocytogenes from fish and fish production environments in Poland. Int. J. Mol. Sci. 2020, 21, 9419. [Google Scholar] [CrossRef]
- Bergholz, T.M.; den Bakker, H.C.; Katz, L.S.; Silk, B.J.; Jackson, K.A.; Kucerova, Z.; Joseph, L.A.; Turnsek, M.; Gladney, L.M.; Halpin, J.L.; et al. Determination of evolutionary relationships of outbreak-associated Listeria monocytogenes strains of serotypes 1/2a and 1/2b by whole-genome sequencing. Appl. Environ. Microbiol. 2016, 82, 928–938. [Google Scholar] [CrossRef][Green Version]
- European Centre for Disease Prevention and Control and European Food Safety Authority. Multi-Country Outbreak of Listeria monocytogenes Clonal Complex 8 Infections Linked to Consumption of Cold-Smoked Fish Products—4 June 2019; ECDC: Stockholm, Sweden; EFSA: Parma, Italy, 2019. [CrossRef]
- European Centre for Disease Prevention and Control and European Food Safety Authority. Multi-Country Outbreak of Listeria monocytogenes Sequence Type 8 Infections Linked to Consumption of Salmon Products—25 October 2018; ECDC: Stockholm, Sweden; EFSA: Parma, Italy, 2018. [CrossRef]
- Borucki, M.K.; Peppin, J.D.; White, D.; Loge, F.; Call, D.R. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7336–7342. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Miya, S.; Igarashi, K.; Suda, T.; Kuramoto, S.; Kimura, B. Biofilm formation ability of Listeria monocytogenes isolates from raw ready-to-eat seafood. J. Food Prot. 2009, 72, 1476–1480. [Google Scholar] [CrossRef] [PubMed]
- Raschle, S.; Stephan, R.; Stevens, M.; Cernela, N.; Zurfluh, K.; Muchaamba, F.; Nüesch-Inderbinen, M. Environmental dissemination of pathogenic Listeria monocytogenes in flowing surface waters in Switzerland. Sci. Rep. 2021, 11, 9066. [Google Scholar] [CrossRef] [PubMed]
- Šteingolde, Ž.; Meistere, I.; Avsejenko, J.; Ķibilds, J.; Bergšpica, I.; Streikiša, M.; Gradovska, S.; Alksne, L.; Roussel, S.; Terentjeva, M.; et al. Characterization and genetic diversity of Listeria monocytogenes isolated from cattle abortions in Latvia, 2013–2018. Vet. Sci. 2021, 8, 195. [Google Scholar] [CrossRef]



| Source of Isolates | No. of Total Types | Most Common Types (No. of Isolates) | No. of Types Unique for the Source | Common to All Sources | ||||
|---|---|---|---|---|---|---|---|---|
| ST | CT | ST | CT | ST | CT | ST | CT | |
| Raw meat (n = 19) | 3 | 10 | ST155 (14) | CT1170 (6) | 0 | 6 | ST8; ST121; ST155 | CT750; CT1170 |
| RTE food (n = 48) a | 8 | 36 | ST121 (18) | CT1170 (6) | 3 | 31 | ||
| FPEs (n = 33) b | 5 | 22 | ST155 (13) | CT9831 (6) | 0 | 17 | ||
| Trait | Gene | No. of Isolates | |||
|---|---|---|---|---|---|
| CC155 (n = 33) | CC121 (n = 32) | CC8 (n = 28) | |||
| ST155 (n = 33) | ST121 (n = 30) | ST1398 (n = 2) | ST8 (n = 28) | ||
| Metal & disinfectants resistance | bcrA | 12 | 0 | 0 | 0 |
| bcrB | 12 | 0 | 0 | 0 | |
| bcrC | 12 | 0 | 0 | 0 | |
| ermC (Tn6118_qac) | 0 | 13 | 0 | 2 | |
| Stress Islands | SSI1_lmo0444 | 33 | 0 | 0 | 27 |
| SSI1_lmo0445 | 33 | 0 | 0 | 28 | |
| SSI1_lmo0446 | 33 | 0 | 0 | 28 | |
| SSI1_lmo0447 | 33 | 0 | 0 | 28 | |
| SSI1_lmo0448 | 32 | 0 | 0 | 28 | |
| SSI2_lin0464 | 0 | 30 | 2 | 0 | |
| SSI2_lin0465 | 0 | 30 | 2 | 0 | |
| Internalins | inlA | 33 | 29 | 2 | 27 |
| inlB | 33 | 30 | 2 | 28 | |
| inlC | 33 | 30 | 2 | 28 | |
| inlE | 33 | 30 | 2 | 28 | |
| inlF | 33 | 0 | 0 | 28 | |
| inlG | 33 | 0 | 0 | 28 | |
| inlH | 33 | 30 | 2 | 28 | |
| inlJ | 33 | 30 | 2 | 27 | |
| inlK | 33 | 30 | 2 | 27 | |
| LIPI-1 | prfA | 33 | 30 | 2 | 28 |
| plcA | 33 | 27 | 2 | 28 | |
| hly | 33 | 30 | 2 | 28 | |
| mpl | 33 | 29 | 2 | 27 | |
| actA | 33 | 30 | 2 | 28 | |
| plcB | 33 | 30 | 2 | 26 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lachtara, B.; Wieczorek, K.; Osek, J. Genetic Diversity and Relationships of Listeria monocytogenes Serogroup IIa Isolated in Poland. Microorganisms 2022, 10, 532. https://doi.org/10.3390/microorganisms10030532
Lachtara B, Wieczorek K, Osek J. Genetic Diversity and Relationships of Listeria monocytogenes Serogroup IIa Isolated in Poland. Microorganisms. 2022; 10(3):532. https://doi.org/10.3390/microorganisms10030532
Chicago/Turabian StyleLachtara, Beata, Kinga Wieczorek, and Jacek Osek. 2022. "Genetic Diversity and Relationships of Listeria monocytogenes Serogroup IIa Isolated in Poland" Microorganisms 10, no. 3: 532. https://doi.org/10.3390/microorganisms10030532
APA StyleLachtara, B., Wieczorek, K., & Osek, J. (2022). Genetic Diversity and Relationships of Listeria monocytogenes Serogroup IIa Isolated in Poland. Microorganisms, 10(3), 532. https://doi.org/10.3390/microorganisms10030532






