A Novel lncRNA SAAL Suppresses IAV Replication by Promoting Innate Responses
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Virus
2.2. Virus Infection
2.3. qRT-PCR Analysis
2.4. Hemagglutination Assay
2.5. Plaque Assay
2.6. Bioinformatics Analysis of Noncoding Potential
2.7. Statistical Analysis
3. Results
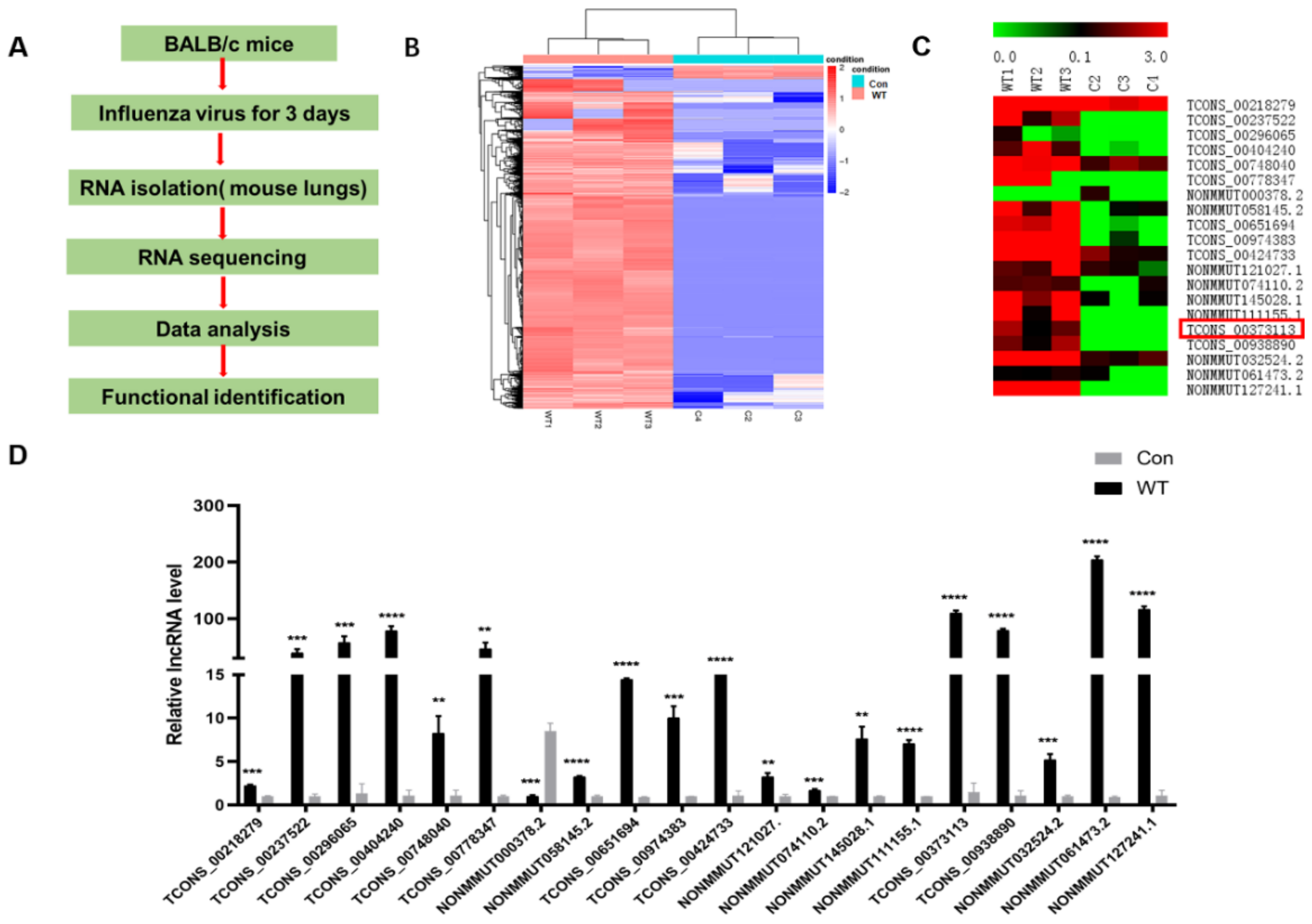
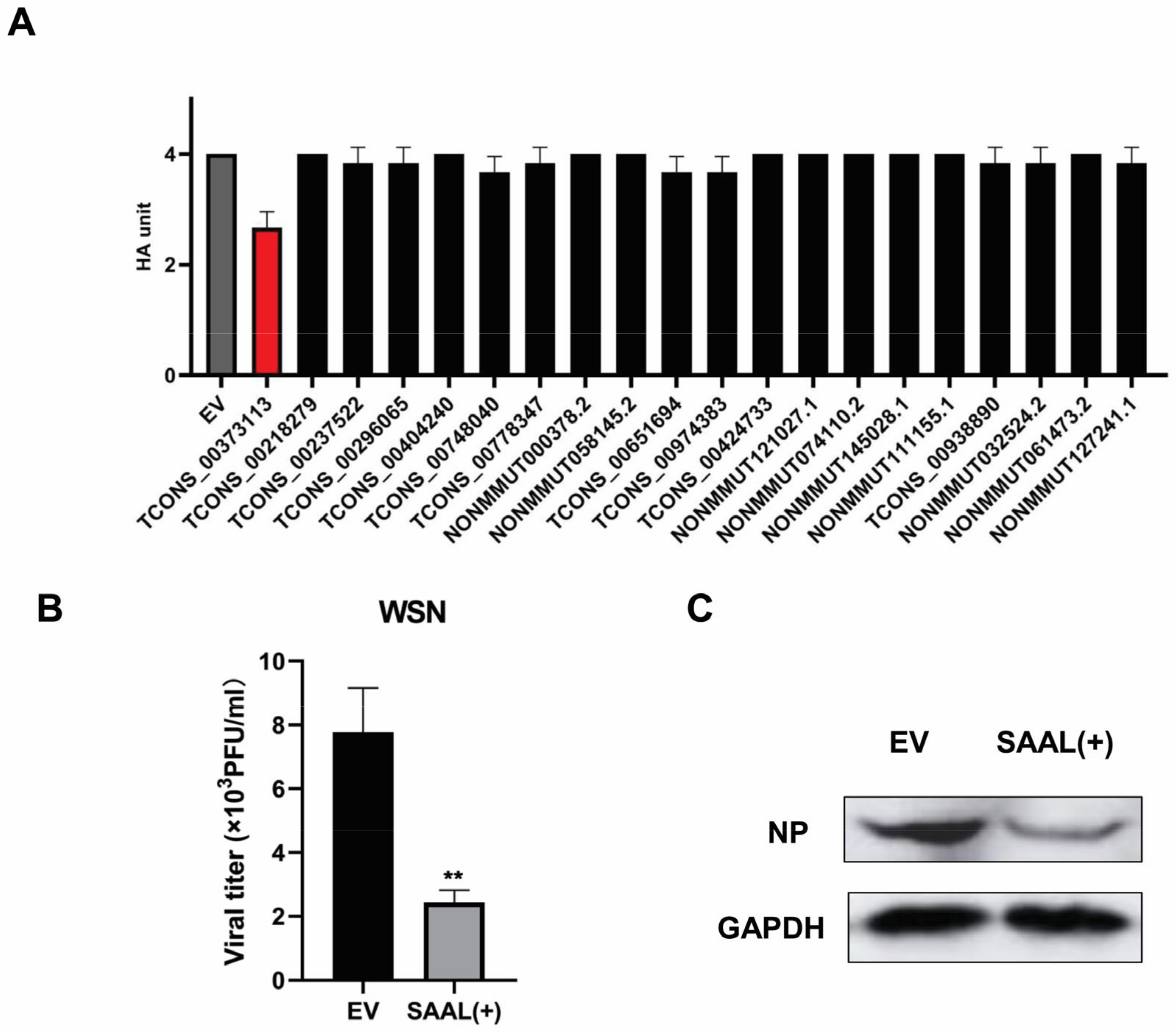
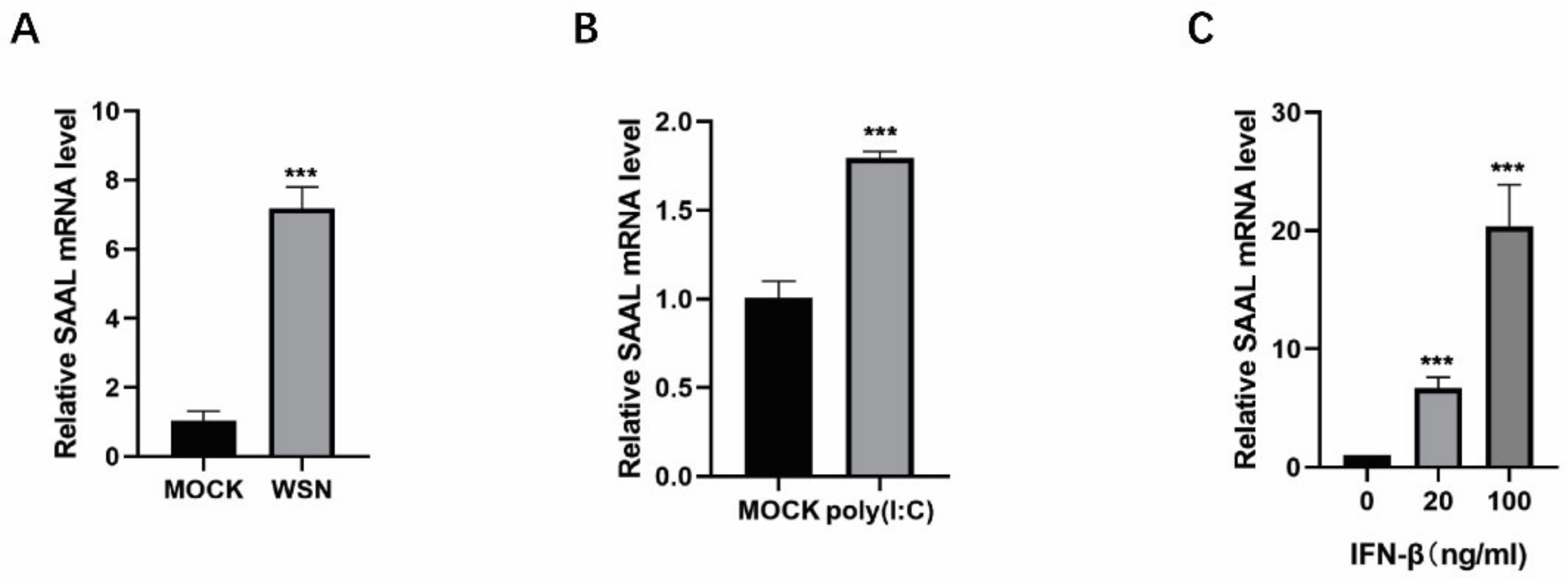
3.1. IAV Infection Induces lncRNA SAAL Expression in Mice
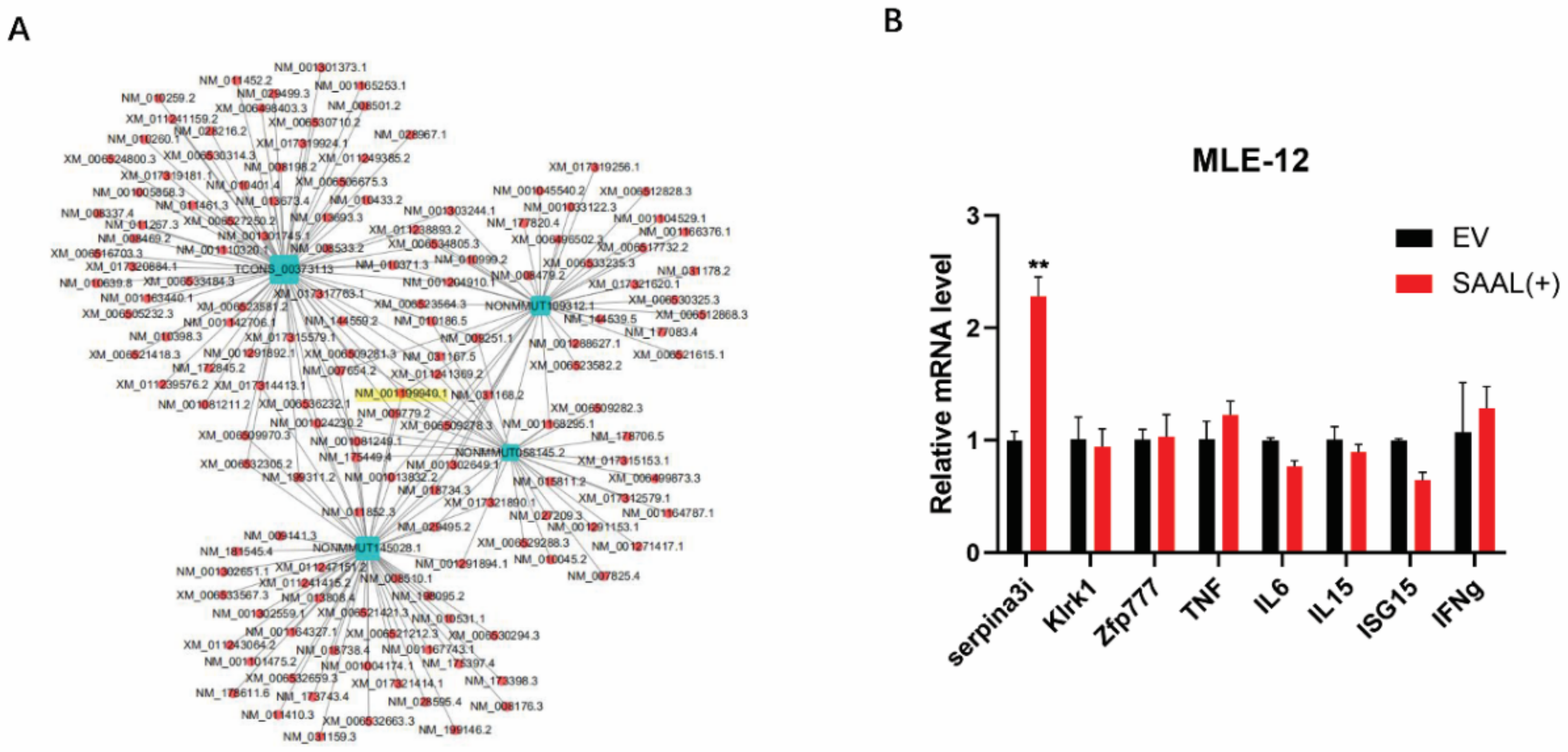
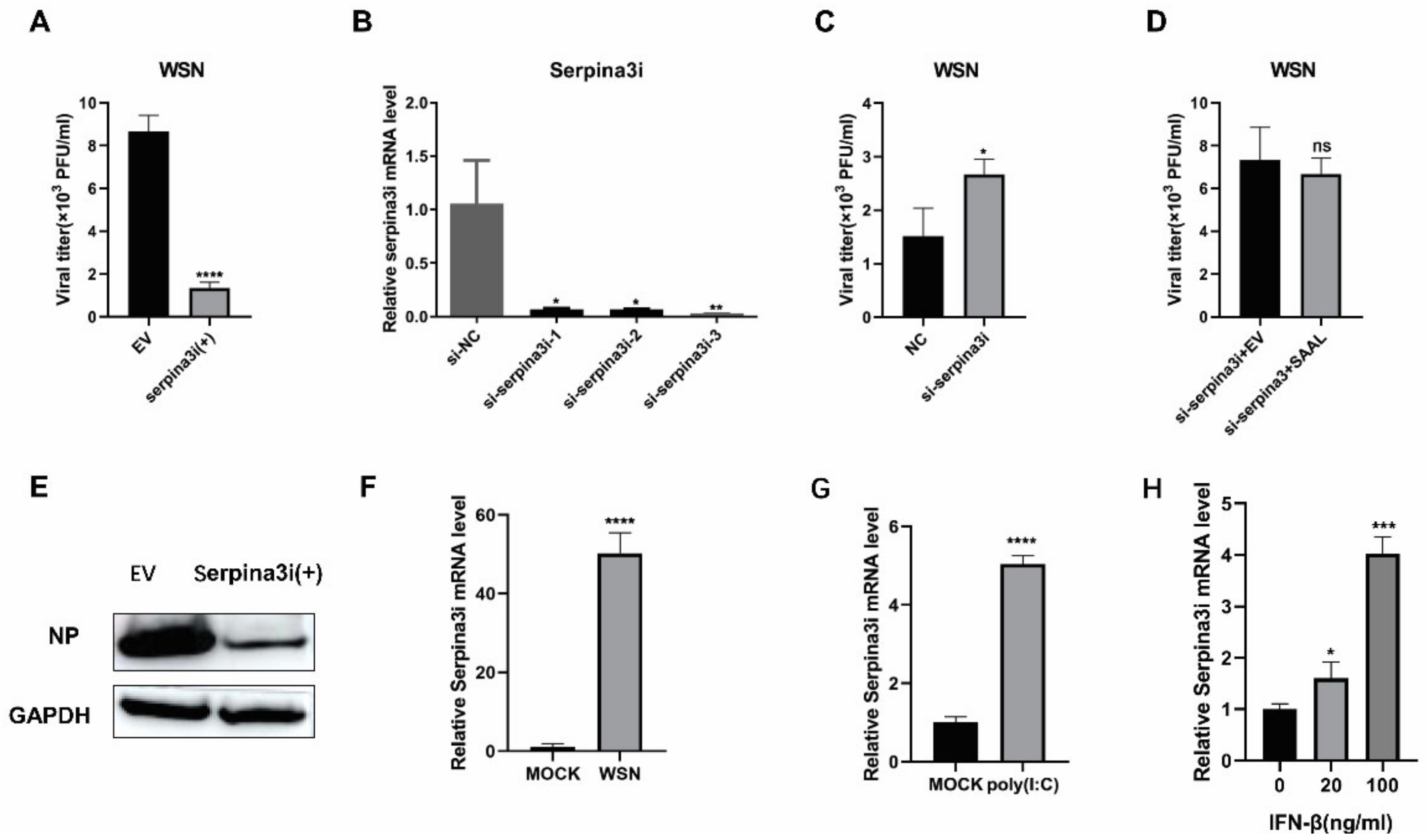
3.2. Serpina3i Is Coexpressed with SAAL
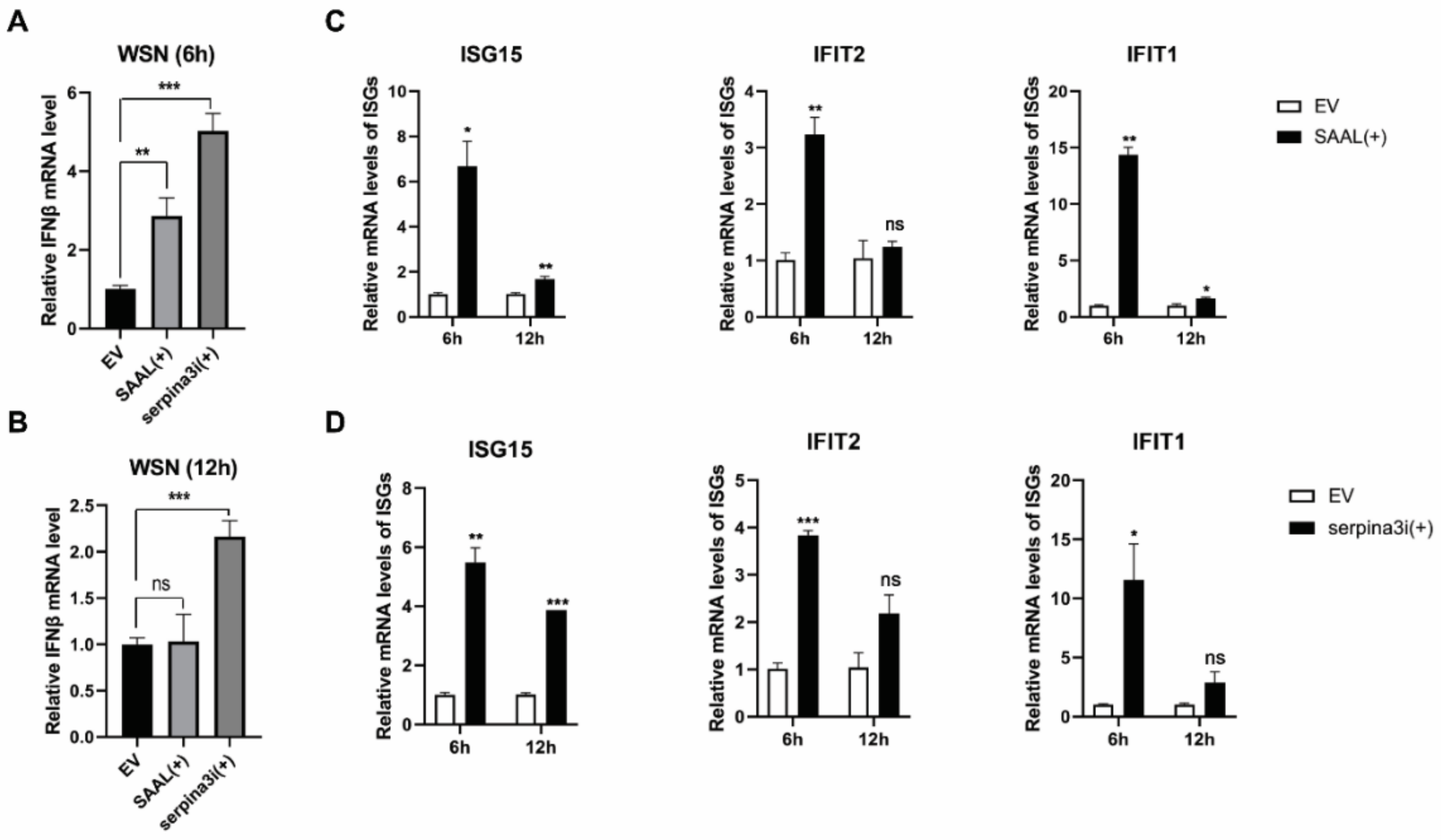
3.3. SAAL- and Serpina3i-Induced mRNA Level of IFN-β and ISGs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Long, J.S.; Mistry, B.; Haslam, S.M.; Barclay, W.S. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019, 17, 67–81. [Google Scholar] [CrossRef]
- Wang, G.; Jiang, L.; Wang, J.; Zhang, J.; Kong, F.; Li, Q.; Yan, Y.; Huang, S.; Zhao, Y.; Liang, L.; et al. The G Protein-Coupled Receptor FFAR2 Promotes Internalization during Influenza A Virus Entry. J. Virol. 2020, 94, e01707-19. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, H. Enhancement of influenza virus transmission by gene reassortment. Curr. Top Microbiol. Immunol. 2014, 385, 185–204. [Google Scholar] [PubMed]
- Wang, G.; Zhao, Y.; Zhou, Y.; Jiang, L.; Liang, L.; Kong, F.; Yan, Y.; Wang, X.; Wang, Y.; Wen, X.; et al. PIAS1-mediated SUMOylation of influenza A virus PB2 restricts viral replication and virulence. PLoS Pathog. 2022, 18, e1010446. [Google Scholar] [CrossRef] [PubMed]
- Pinsent, A.; Fraser, C.; Ferguson, N.M.; Riley, S. A systematic review of reported reassortant viral lineages of influenza A. BMC Infect. Dis. 2016, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Reid, A.H.; Krafft, A.E.; Bijwaard, K.E.; Fanning, T.G. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science 1997, 275, 1793–1796. [Google Scholar] [CrossRef] [PubMed]
- Tanner, W.D.; Toth, D.J.A.; Gundlapalli, A.V. The pandemic potential of avian influenza A(H7N9) virus: A review. Epidemiol. Infect. 2015, 143, 3359–3374. [Google Scholar] [CrossRef]
- Li, C.; Chen, H. H7N9 Influenza Virus in China. Cold Spring Harb. Perspect. Med. 2021, 11, a038349. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Ouyang, J.; Wei, J.; Maarouf, M.; Chen, J.-L. Involvement of Host Non-Coding RNAs in the Pathogenesis of the Influenza Virus. Int. J. Mol. Sci. 2016, 18, 39. [Google Scholar] [CrossRef]
- Qu, Z.; Meng, F.; Shi, J.; Deng, G.; Zeng, X.; Ge, J.; Li, Y.; Liu, L.; Chen, P.; Jiang, Y.; et al. A Novel Intronic Circular RNA Antagonizes Influenza Virus by Absorbing a microRNA That Degrades CREBBP and Accelerating IFN-β Production. mBio 2021, 12, e0101721. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Han, L.; Wei, Y.; Zhang, B.; Wang, Q.; Liu, J.; Liu, M.; Chen, Z.; Wang, Z.; Chen, H.; et al. MicroRNA-200c-targeted contactin 1 facilitates the replication of influenza A virus by accelerating the degradation of MAVS. PLoS Pathog. 2022, 18, e1010299. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Liu, X.; Su, Z.Z.; Hsu, A.C.-Y.; Foster, P.S.; Yang, M. Potential Role of MicroRNAs in the Regulation of Antiviral Responses to Influenza Infection. Front. Immunol. 2018, 9, 1541. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Lee, Y.-S.; Sung, S. Epigenetic regulation by long noncoding RNAs in plants. Chromosome Res. 2013, 21, 685–693. [Google Scholar] [CrossRef]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Heward, J.A.; Lindsay, M.A. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014, 35, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Rapicavoli, N.A.; Qu, K.; Zhang, J.; Mikhail, M.; Laberge, R.M.; Chang, H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2013, 2, e00762. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.A.; Wapinski, O.L.; Yang, Y.W.; Bureau, J.-F.; Gopinath, S.; Monack, D.M.; Chang, H.Y.; Brahic, M.; Kirkegaard, K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 2013, 152, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Zhu, X.; Chen, Y.; Wei, H.; Chen, Q.; Chi, X.; Qi, B.; Zhang, L.; Zhao, Y.; Gao, G.F.; et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 2014, 16, 616–626. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Zhou, R.; Zhao, J.; Zhang, Y.; Yi, D.; Li, Q.; Zhou, J.; Guo, F.; Liang, C.; et al. Host Long Noncoding RNA lncRNA-PAAN Regulates the Replication of Influenza A Virus. Viruses 2018, 10, 330. [Google Scholar] [CrossRef] [PubMed]
- More, S.; Zhu, Z.; Lin, K.; Huang, C.; Pushparaj, S.; Liang, Y.; Sathiaseelan, R.; Yang, X.; Liu, L. Long non-coding RNA PSMB8-AS1 regulates influenza virus replication. RNA Biol. 2019, 16, 340–353. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, S.; Yang, Z.; Lin, H.; Zhu, J.; Liu, L.; Wang, W.; Liu, S.; Liu, W.; Ma, Y.; et al. Self-Recognition of an Inducible Host lncRNA by RIG-I Feedback Restricts Innate Immune Response. Cell 2018, 173, 906–919.e13. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xu, J.; Wang, Y.; Cao, X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 2017, 358, 1051–1055. [Google Scholar] [CrossRef] [PubMed]
- Chai, W.; Li, J.; Shangguan, Q.; Liu, Q.; Li, X.; Qi, D.; Tong, X.; Liu, W.; Ye, X. Lnc-ISG20 Inhibits Influenza A Virus Replication by Enhancing ISG20 Expression. J. Virol. 2018, 92, e00539-18. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Neumann, G.; Kawaoka, Y.; Hobom, G.; Webster, R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 2000, 97, 6108–6113. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, H.; Chen, Y.; Wei, H.; Gao, G.F.; Liu, H.; Huang, S.; Chen, J.L. Transport of influenza virus neuraminidase (NA) to host cell surface is regulated by ARHGAP21 and Cdc42 proteins. J. Biol. Chem. 2012, 287, 9804–9816. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, N.; Tian, M.; Fan, M.; Liu, Q.; Liu, Z.; Sun, T.; Huang, J.; Xia, H.; Zhao, Y.; et al. Integrated Analysis of microRNA-mRNA Expression in Mouse Lungs Infected with H7N9 Influenza Virus: A Direct Comparison of Host-Adapting PB2 Mutants. Front. Microbiol. 2020, 11, 1762. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Li, Q.; Zhao, J.; Yi, D.; Ding, J.; Zhao, F.; Hu, S.; Zhou, J.; Deng, T.; et al. Influenza Virus Exploits an Interferon-Independent lncRNA to Preserve Viral RNA Synthesis through Stabilizing Viral RNA Polymerase PB1. Cell Rep. 2019, 27, 3295–3304.e4. [Google Scholar] [CrossRef]
- Sui, B.; Chen, D.; Liu, W.; Wu, Q.; Tian, B.; Li, Y.; Hou, J.; Liu, S.; Xie, J.; Jiang, H.; et al. A novel antiviral lncRNA, EDAL, shields a T309 O-GlcNAcylation site to promote EZH2 lysosomal degradation. Genome Biol. 2020, 21, 228. [Google Scholar] [CrossRef]
- Guo, X.-Z.J.; Thomas, P.G. New fronts emerge in the influenza cytokine storm. Semin. Immunopathol. 2017, 39, 541–550. [Google Scholar] [CrossRef] [PubMed]
- García-Sastre, A.; Biron, C.A. Type 1 interferons and the virus-host relationship: A lesson in détente. Science 2006, 312, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Zhao, Z.; Liao, Y.; Chiu, S.-H.; Wang, S.; Chen, B.; Chen, N.; Chen, Y.; Chen, J.-L. Identification of an Interferon-Stimulated Long Noncoding RNA (LncRNA ISR) Involved in Regulation of Influenza A Virus Replication. Int. J. Mol. Sci. 2019, 20, 5118. [Google Scholar] [CrossRef] [PubMed]
| lncRNA ID | RNA Size | ORF Size | Ficket Score | Hexamer Score | Coding Probability | Coding Label |
|---|---|---|---|---|---|---|
| TCONS_00373113 | 544 | 186 | 0.904 | −0.239 | 0.041 | no |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Yang, H.; Zhao, L.; Huang, N.; Ping, J. A Novel lncRNA SAAL Suppresses IAV Replication by Promoting Innate Responses. Microorganisms 2022, 10, 2336. https://doi.org/10.3390/microorganisms10122336
Liu Q, Yang H, Zhao L, Huang N, Ping J. A Novel lncRNA SAAL Suppresses IAV Replication by Promoting Innate Responses. Microorganisms. 2022; 10(12):2336. https://doi.org/10.3390/microorganisms10122336
Chicago/Turabian StyleLiu, Qingzheng, Hongjun Yang, Lingcai Zhao, Nan Huang, and Jihui Ping. 2022. "A Novel lncRNA SAAL Suppresses IAV Replication by Promoting Innate Responses" Microorganisms 10, no. 12: 2336. https://doi.org/10.3390/microorganisms10122336
APA StyleLiu, Q., Yang, H., Zhao, L., Huang, N., & Ping, J. (2022). A Novel lncRNA SAAL Suppresses IAV Replication by Promoting Innate Responses. Microorganisms, 10(12), 2336. https://doi.org/10.3390/microorganisms10122336






