Flavacol and Its Novel Derivative 3-β-Hydroxy Flavacol from Streptomyces sp. Pv 4-95 after the Expression of Heterologous AdpA
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Growth Conditions
2.2. DNA Extraction, Amplification and Sequencing
2.3. Phylogenetic Analysis of the Pv 4-95 Strain
2.4. Construction of the Pv 4-95 Strain with AdpA Expression
2.5. Secondary Metabolite Extraction and Analysis
2.6. Secondary Metabolite Purification
2.7. Nuclear Magnetic Resonance Spectroscopy (NMR)
3. Results and Discussion
3.1. Activation of Secondary Metabolite Production in the Pv 4-95 Strain
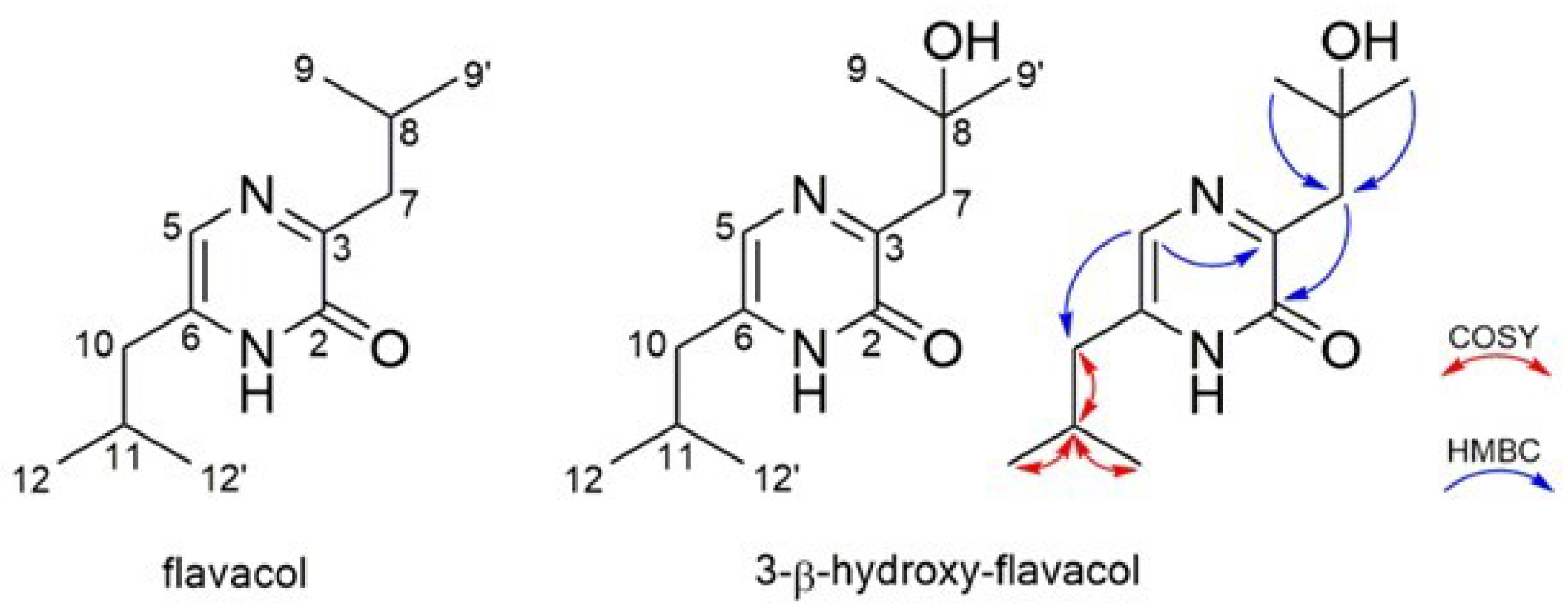
3.2. Purification and Structure Elucidation of the Activated Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Selim, M.S.M.; Abdelhamid, S.A.; Mohamed, S.S. Secondary metabolites and biodiversity of actinomycetes. J. Genet. Eng. Biotechnol. 2021, 19, 72. [Google Scholar] [CrossRef] [PubMed]
- Watve, M.G.; Tickoo, R.; Jog, M.M.; Bhole, B.D. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 2001, 176, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 2008, 8, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Sekurova, O.N.; Schneider, O.; Zotchev, S.B. Novel bioactive natural products from bacteria via bioprospecting, genome mining and metabolic engineering. Microb. Biotechnol. 2019, 12, 828–844. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; International Natural Product Sciences Taskforce; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Higo, A.; Hara, H.; Horinouchi, S.; Ohnishi, Y. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 2012, 19, 259–273. [Google Scholar] [CrossRef]
- Makitrynskyy, R.; Ostash, B.; Tsypik, O.; Rebets, Y.; Doud, E.; Meredith, T.; Luzhetskyy, A.; Bechthold, A.; Walker, S.; Fedorenko, V. Pleiotropic regulatory genes bldA, adpA and absB are implicated in production of phosphoglycolipid antibiotic moenomycin. Open Biol. 2013, 3, 130121. [Google Scholar] [CrossRef]
- Nguyen, K.T.; Tenor, J.; Stettler, H.; Nguyen, L.T.; Nguyen, L.D.; Thompson, C.J. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J. Bacteriol. 2003, 185, 7291–7296. [Google Scholar] [CrossRef][Green Version]
- Akanuma, G.; Hara, H.; Ohnishi, Y.; Horinouchi, S. Dynamic changes in the extracellular proteome caused by absence of a pleiotropic regulator AdpA in Streptomyces griseus. Mol. Microbiol. 2009, 73, 898–912. [Google Scholar] [CrossRef]
- Ostash, B.; Yushchuk, O.; Tistechok, S.; Mutenko, H.; Horbal, L.; Muryn, A.; Dacyuk, Y.; Kalinowski, J.; Luzhetskyy, A.; Fedorenko, V. The adpA-like regulatory gene from Actinoplanes teichomyceticus: In silico analysis and heterologous expression. World J. Microbiol. Biotechnol. 2015, 31, 1297–1301. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, J.; Zhuo, J.; Li, Y.; Tian, Y.; Tan, H. Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J. Biol. Chem. 2017, 292, 19708–19720. [Google Scholar] [CrossRef] [PubMed]
- Yushchuk, O.; Ostash, I.; Mösker, E.; Vlasiuk, I.; Deneka, M.; Rückert, C.; Busche, T.; Fedorenko, V.; Kalinowski, J.; Süssmuth, R.D.; et al. Eliciting the silent lucensomycin biosynthetic pathway in Streptomyces cyanogenus S136 via manipulation of the global regulatory gene adpA. Sci. Rep. 2021, 11, 3507. [Google Scholar] [CrossRef] [PubMed]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics. A Laboratory Manual; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Blodgett, J.A.; Thomas, P.M.; Li, G.; Velasquez, J.E.; van der Donk, W.A.; Kelleher, N.L.; Metcalf, W.W. Unusual transformations in the biosynthesis of the antibiotic phosphinothricin tripeptide. Nat. Chem. Biol. 2007, 3, 480–485. [Google Scholar] [CrossRef][Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1980, 39, 783–791. [Google Scholar] [CrossRef]
- Ostash, B.; Gren, T.; Hrubskyy, Y.; Tistechok, S.; Beshley, S.; Baranov, V.; Fedorenko, V. Cultivable actinomycetes from rhizosphere of birch (Betula pendula) growing on a coal mine dump in Silets, Ukraine. J. Basic. Microbiol. 2013, 54, 851–857. [Google Scholar] [CrossRef]
- Raju, R.; Gromyko, O.; Fedorenko, V.; Luzketskyy, A.; Müller, R. Albaflavenol B, a new sesquiterpene isolated from the terrestrial actinomycete, Streptomyces sp. J. Antibiot. 2015, 68, 286–288. [Google Scholar] [CrossRef] [PubMed]
- Raju, R.; Gromyko, O.; Fedorenko, V.; Herrmann, J.; Luzhetskyy, A.; Muller, R. Rubimycinone A, a new anthraquinone from a terrestrial Streptomyces sp. Tetrahedron Lett. 2013, 54, 900–902. [Google Scholar] [CrossRef]
- Paulus, C.; Gromyko, O.; Luzhetskyy, A. New kendomycin derivative isolated from Streptomyces sp. Cl 58-27. Molecules 2021, 26, 6834. [Google Scholar] [CrossRef] [PubMed]
- Ochi, K. Insights into microbial cryptic gene activation and strain improvement: Principle, application and technical aspects. J. Antibiot. 2017, 70, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, J. Dictionary of Natural Products on CD-ROM. Version 13:2; Chapman and Hall: London, UK, 2005. [Google Scholar]
- Dunn, G.; Newbold, G.T.; Spring, F.S. Synthesis of flavacol, a metabolic product of Aspergillus flavus. J. Chem. Soc. 1949, 545, 2586–2587. [Google Scholar] [CrossRef]
- Micetich, R.G.; Macdonald, J.C. Biosynthesis of neoaspergillic and neohydroxyaspergillic acids. J. Biol. Chem. 1965, 240, 1692–1695. [Google Scholar] [CrossRef]
- Xu, X.; He, F.; Zhang, X.; Bao, J.; Qi, S. New mycotoxins from marine-derived fungus Aspergillus sp. SCSGAF0093. Food Chem. Toxicol. 2013, 53, 46–51. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T. Identification and bioactivity of compounds from the fungus Penicillium sp. CYE-87 isolated from a marine tunicate. Mar. Drugs. 2015, 13, 1698–1709. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T.A.; Badr, J.M.; Harakeh, S.M. Bioactive 2(1H)-pyrazinones and diketopiperazine alkaloids from a tunicate-derived actinomycete Streptomyces sp. Molecules 2016, 21, 1116. [Google Scholar] [CrossRef]
- López-Gresa, M.P.; González, M.C.; Primo, J.; Moya, P.; Romero, V.; Estornell, E. Circumdatin H, a new inhibitor of mitochondrial NADH oxidase, from Aspergillus ochraceus. J. Antibiot. 2005, 58, 416–419. [Google Scholar] [CrossRef]
- Rajini, K.S.; Aparna, P.; Sasikala, C.H.; Ramana, C. Microbial metabolism of pyrazines. Crit. Rev. Microbiol. 2011, 37, 99–112. [Google Scholar] [CrossRef]
- Mortzfeld, F.B.; Hashem, C.; Vranková, K.; Winkler, M.; Rudroff, F. Pyrazines: Synthesis and industrial application of these valuable flavor and fragrance compounds. Biotechnol. J. 2020, 15, 2000064. [Google Scholar] [CrossRef]
- Rizzi, G.P. The biogenesis of food-related Pyrazines. Food Rev. Int. 1988, 4, 375–400. [Google Scholar] [CrossRef]


| 3-β-Hydroxy-Flavacol (2) | Flavacol (1) | ||||
|---|---|---|---|---|---|
| Atom # | δC, mult. | ΔH, mult. (J in Hz) | Atom # | δC, mult. | ΔH, mult. (J in Hz) |
| 1 | NH | - | 1 | NH | - |
| 2 | 159.8, C | - | 2 | 159.9, C | - |
| 3 | 155.8, C | - | 3 | 158.0, C | - |
| 4 | N | - | 4 | N | - |
| 5 | 124.0, CH | 7.2, s | 5 | 123.5, CH | 7.1, s |
| 6 | 141.0, C | - | 6 | 140.4, C | - |
| 7 | 46.3, CH2 | 3.0, s | 7 | 42.6, CH2 | 2.6, d (7.25) |
| 8 | 72.6, C | - | 8 | 28.2, CH | 2.2, m |
| 9/9′ | 29.8, CH3 | 1.2, s | 9/9′ | 23.1, CH3 | 0.9, d (6.62) |
| 10 | 40.2, CH2 | 2.4, d (7.35) | 10 | 40.2, CH2 | 2.4, d (7.25) |
| 11 | 29.7, CH | 2.0, m | 11 | 29.6-, CH | 2.0, m |
| 12/12′ | 22.5, CH3 | 1.0, d (6.65) | 12/12′ | 22.5, CH3 | 1.0, d (6.62) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tistechok, S.; Stierhof, M.; Kachor, A.; Myronovskyi, M.; Gromyko, O.; Luzhetskyy, A. Flavacol and Its Novel Derivative 3-β-Hydroxy Flavacol from Streptomyces sp. Pv 4-95 after the Expression of Heterologous AdpA. Microorganisms 2022, 10, 2335. https://doi.org/10.3390/microorganisms10122335
Tistechok S, Stierhof M, Kachor A, Myronovskyi M, Gromyko O, Luzhetskyy A. Flavacol and Its Novel Derivative 3-β-Hydroxy Flavacol from Streptomyces sp. Pv 4-95 after the Expression of Heterologous AdpA. Microorganisms. 2022; 10(12):2335. https://doi.org/10.3390/microorganisms10122335
Chicago/Turabian StyleTistechok, Stepan, Marc Stierhof, Anna Kachor, Maksym Myronovskyi, Oleksandr Gromyko, and Andriy Luzhetskyy. 2022. "Flavacol and Its Novel Derivative 3-β-Hydroxy Flavacol from Streptomyces sp. Pv 4-95 after the Expression of Heterologous AdpA" Microorganisms 10, no. 12: 2335. https://doi.org/10.3390/microorganisms10122335
APA StyleTistechok, S., Stierhof, M., Kachor, A., Myronovskyi, M., Gromyko, O., & Luzhetskyy, A. (2022). Flavacol and Its Novel Derivative 3-β-Hydroxy Flavacol from Streptomyces sp. Pv 4-95 after the Expression of Heterologous AdpA. Microorganisms, 10(12), 2335. https://doi.org/10.3390/microorganisms10122335







